Abstract
Background
The family of RecQ DNA helicases plays an important role in the maintenance of genomic integrity. Mutations in three of the five known RecQ family members in humans, BLM, WRN and RecQ4, lead to disorders that are characterized by predisposition to cancer and premature aging.
Methodology/Principal Findings
To address the in vivo functions of Drosophila RecQ4 (dRecQ4), we generated mutant alleles of dRecQ4 using the targeted gene knock-out technique. Our data show that dRecQ4 mutants are homozygous lethal with defects in DNA replication, cell cycle progression and cell proliferation. Two sets of experiments suggest that dRecQ4 also plays a role in DNA double strand break repair. First, mutant animals exhibit sensitivity to gamma irradiation. Second, the efficiency of DsRed reconstitution via single strand annealing repair is significantly reduced in the dRecQ4 mutant animals. Rescue experiments further show that both the N-terminal domain and the helicase domain are essential to dRecQ4 function in vivo. The N-terminal domain is sufficient for the DNA repair function of dRecQ4.
Conclusions/Significance
Together, our results show that dRecQ4 is an essential gene that plays an important role in not only DNA replication but also DNA repair and cell cycle progression in vivo.
Introduction
RecQ4 encodes a DNA helicase that belongs to the RecQ family; in humans, this family consists of five members [1]–[5]. Unlike other RecQ family members such as BLM and WRN [6]–[12], the biological functions of RecQ4 remain relatively less clear and more controversial [13]–[25]. For example, various studies have led to contradictory conclusions on where RecQ4 is localized [10], [25], [26]. Furthermore, the sensitivity of RecQ4 deficient cells or organisms to treatments that block DNA replication or cause DNA damage, e.g., ionizing radiation, remains poorly resolved [27]–[29].
Cancer predisposition of either human patients or mice models with RecQ4 mutations represent another unresolved issue (for review, see [2]). Mutations in the human RecQ4 gene have been found to contribute to three rare syndromes: Rothmund-Thomson syndrome [5], [30]–[32], RAPADILINO syndrome [22], [25] and Baller-Gerold syndrome [2], [23]. Currently there is no common conclusion on whether these three syndromes are independent disorders or represent one syndrome with different symptoms. Several labs have developed mice models with different RecQ4 mutations, but these mice show different phenotypes that range from embryonic lethality to defects restricted to adult mice, some of which resemble the symptoms of human patients [19], [28], [33].
Several recent studies have revealed new insights concerning the role of RecQ4 in DNA replication initiation [18], [20], [21], [24]. Cut5, the metazoan homolog of Saccharomyces cerevisiae Dpb11, which is required for loading DNA polymerases onto chromatin, was shown to interact with the Xenopus RecQ4 (xRecQ4) both in vitro and in vivo [20], [21]. Purified N-terminal fragments of xRTS/xRecQ4 were able to rescue the DNA replication defects of xRecQ4-depleted Xenopus egg extracts [20]. In mammalian cells, RecQ4 has been shown to interact with RAD51 and PARP1, suggesting that it may also participate in DNA repair [2], [10], [34], [35]. However, the role of RecQ4 in DNA repair has not been fully characterized, particularly in the context of an in vivo system.
Unlike in mammals, the fruit fly genome encodes three complete RecQ helicases, namely dBLM, dRecQ4 and dRecQ5 [24], [36]–[43]. In addition, DmWRNexo was recently identified as the Drosophila homologue of human WRN exonuclease domain [44], [45]. In order to develop a model system more amenable to genetic analysis of RecQ4 function in vivo which would also help to clarify, at least, some of the controversies about RecQ4, we set out to characterize RecQ4 in Drosophila.
In this report, we describe the generation and phenotypic analyses of dRecQ4 mutants in Drosophila. Our results show that the dRecQ4 mutants exhibit defects in DNA replication. They are also selectively sensitive to paraquat and gamma irradiation. Mutant animals exhibit lower efficiency of double strand break (DSB) repair as assayed by reconstitution of the DsRed transgene in vivo [46]. Rescue experiments with various truncated dRecQ4 proteins suggest that the N-terminal domain of dRecQ4 is essential for DSB repair, whereas both the N-terminal domain and the helicase domain are indispensable for DNA replication and animal viability.
Results
dRecQ4 is essential for development
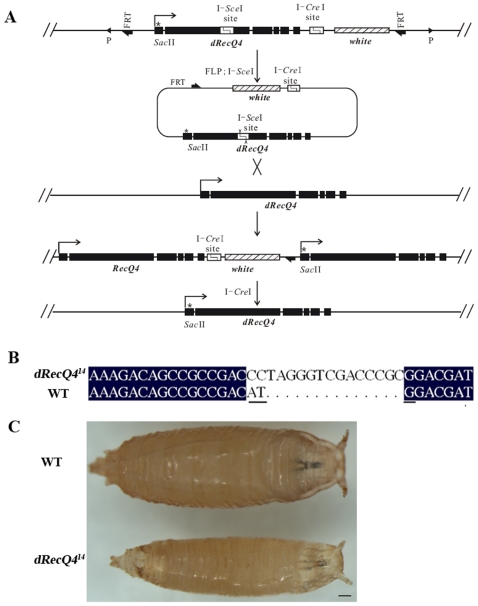
Prior to the report of Wu et al., there were no transposable elements inserted within or nearby the dRecQ4 locus [24]. We took advantage of the targeted knockout technique to generate dRecQ4 mutants through the replacement of the endogenous locus with an engineered mutant form via homologous recombination. Specifically, an 8 kb genomic fragment was modified by replacing the start codon ATG with CCTAGGGTCGACCCGCG and inserting an I-SceI recognition site into the second exon of dRecQ4 (Figure 1A; see Materials and methods for details). Targeting of the dRecQ4 locus was achieved by a modified procedure described by Rong and Golic [47] and Egli and colleagues [48], [49]. Four mutant candidates were obtained and confirmed by restriction enzyme digestions; all four alleles showed similar phenotypes in the viability test (see below). One of these alleles, dRecQ414, was further confirmed by DNA sequencing (Figure 1B) revealing that the start codon mutation and open reading frame shift are as designed. This allele, which can be fully rescued by a genomic rescue transgene as judged by adult flies' viability (see below), was used for detailed phenotypic analysis throughout this study.
Figure 1. Generation of dRecQ414 mutants: strategy and identification.
(A) Schematic view of the dRecQ4 locus and targeting strategy. A transgene containing a mutant dRecQ4 and the marker gene w+ is circularized from the genome by FLP recombinase and linearized by the yeast restriction endonuclease I-SceI. Alignment of the targeting DNA and the resident dRecQ4 locus by ‘ends-in’ recombination results in a duplication of dRecQ4. Then the genomic DNA is cut by another rare-cutter I-CreI, repaired by homologous recombination, leading to a single copy of dRecQ4. * indicates mutation of the start codon. (B) Sequence comparison of the mutant (dRecQ414) and the wild type (WT) indicates that dRecQ414 harbors the expected changes as was designed. The translation start codon ATG (underlined in wild type sequence) is disrupted and the open reading frame is also shifted for dRecQ414 mutant. (C) dRecQ414 mutants are homozygous lethal and die at early pupal stage when raised at 25°C. A y w pupa, serving as a wild type control, and a dRecQ414 mutant are shown. Scale bar = 150 µm
dRecQ414 mutants are homozygous lethal, indicating that dRecQ4 is an essential gene. Using a GFP marked balancer chromosome, we separated the homozygous from the heterozygous dRecQ414 animals. Nearly all the homozygous mutants survive for up to 8 days under normal culture conditions. However, they exhibit developmental delays when compared with heterozygous siblings or y w wild type flies and eventually die at early pupal stage (Figure 1C). The lethal phenotype of the dRecQ414 mutants can be fully rescued by either a genomic fragment of dRecQ4 or a UAS mediated dRecQ4 expression (data not shown), further indicating that the lethality phenotype was a direct consequence of the dRecQ4 mutation.
To determine the role of maternal contributions to development, we set out to generate dRecQ4 germline clones in females that carry FRT combined dRecQ4 mutation and ovoD chromosomes (Table 1). However, when these females were crossed to y w; dRecQ414/TM3, Kr-GFP males, no eggs were obtained (Table 1). The oocytes from dRecQ414 FRT2A/FRT2A ovoD females failed to go beyond stage 6 of oogenesis (data not shown). These results suggest that dRecQ4 is essential for oogenesis, further supporting the conclusion that dRecQ4 is an essential gene critical to cell viability.
Table 1. Statistics of germline clone analysis.
| Maternal genotypes | Number of mothers tested | Number of mothers with eggs |
| FRT2A/FRT2A ovoD | 59 | 12 |
| dRecQ414 FRT2A/FRT2A ovoD | 85 | 0 |
dRecQ4 loss-of-function affects endogenous DNA replication both in the salivary glands and the late larval brain
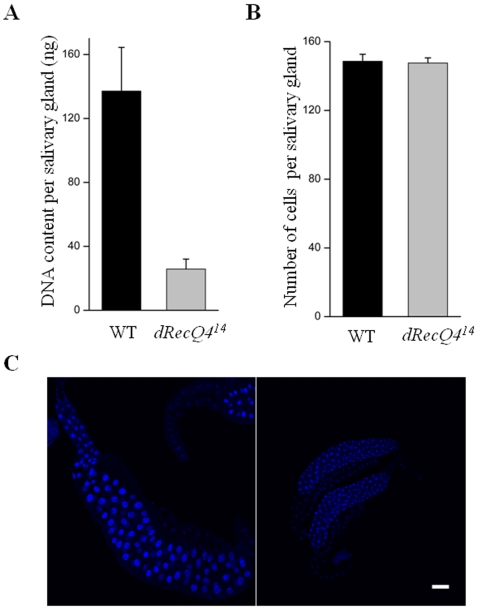
It has been shown in Xenopus egg extracts that DNA replication is blocked when RecQ4 is depleted [20], [21]. To specifically determine whether dRecQ4 is essential for DNA replication in vivo, we measured both DNA content and cell numbers of salivary glands from wild type and dRecQ4 mutant animals. Figure 2B shows that salivary glands from wild type and mutants have similar cell numbers at third instar larval stage (5 days AED). However, the total amount of DNA from each salivary gland at this stage differs significantly between wild type and mutants (Figure 2A). The amount of DNA normalized by cell number is much lower in mutant cells than in wild type cells (∼0.18 ng/cell and ∼0.92 ng/cell, respectively), indicating a defect in DNA accumulation, presumably reflecting an under replication of DNA. Salivary gland increases its DNA content through cell cycle independent endoreplication. Inefficient DNA endoreplication of salivary gland cells is also consistent with the finding of small cells and nuclei in this tissue of mutant animals (Figure 2C).
Figure 2. dRecQ4 mutant cells contain less genomic DNA than wild type cells.
(A) DNA content of the salivary glands of dRecQ414 mutants is lower than that of wild type. P<0.05. (B) The cell number of salivary glands from dRecQ414 mutants and wild type remains unchanged. P<0.001. Error bars represent the standard deviation of the mean value of three independent experiments. (C) 5 days old salivary glands from wild type and the mutant larvae were stained with DAPI. Note that the mutant nuclei are smaller. Scale bar = 100 µm
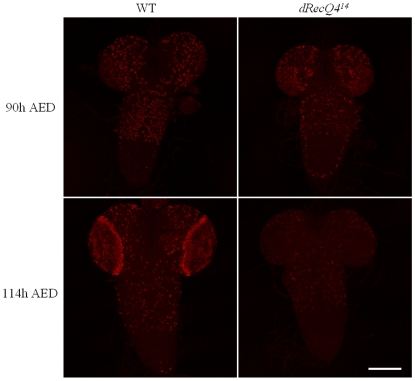
To directly investigate whether dRecQ4 is required for DNA replication, particularly in the non-endoreplicating cells, we performed BrdU labeling followed by anti-BrdU immuno-staining on the larval brain. Figure 3 shows that at the stage of four days after egg deposition (AED), wild type and dRecQ4 mutants have comparable BrdU incorporation likely reflecting maternal contributions of dRecQ4 protein in the mutants. However, at five days AED the mutant brain incorporates much less BrdU than the wild type, which is consistent with the observed reduction of DNA accumulation in salivary glands (see above). Our data are consistent with those of Wu (Wu et al., 2008) and together they demonstrate that dRecQ4 is involved in DNA replication in Drosophila.
Figure 3. Incorporation of BrdU is significantly reduced in dRecQ4 mutants as compared to wild type.
The incorporation of BrdU was visualized by staining with anti-BrdU. As shown in the upper panel, at 90 hours AED, the BrdU incorporation in the brain is only slightly different while at 114 hours AED, mutant larvae incorporate significantly less BrdU as shown in the lower panel. Scale bar = 100 µm
dRecQ4 is involved in double strand breaks repair
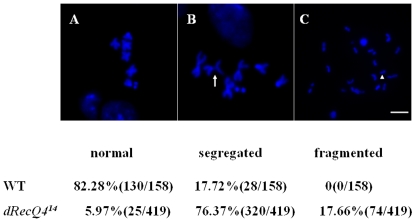
Inactivation of RecQ4 in mouse results in defective sister chromatid cohesion and aneuploidy [50]. To determine whether dRecQ4 deficiency causes a similar effect on chromosomal behavior in Drosophila cells, we analyzed metaphase spreads from wild type and mutant brain cells. In dRecQ4 mutants, the spreads' patterns fall into three major categories (Figure 4A–C): normal pattern (Figure 4A), segregated (Figure 4B) and fragmented (Figure 4C) aberrant patterns. The segregated patterns are indicative of a failure of sister chromatids association (arrows in Figure 4B). The fragmented patterns have broken chromosomes, often with the broken ends fused together (arrow heads in Figure 4C). Statistical analysis shows that the frequency of aberrant patterns in dRecQ414 mutants is much higher than in wild type control. Specifically, mutant cells have less than 10% of normal patterns, with the aberrant segregated and fragmented patterns representing the majority of the mitotic cell population. In wild type cells, over 80% of the cells have normal patterns, with only less than 20% being the mildly abnormal segregated patterns. These results suggest that dRecQ4 is important for maintaining genome integrity.
Figure 4. Chromosomal aberrations in dRecQ414 mutant cells.
Mitotic chromosome patterns of wild type and dRecQ414mutant larval brains are shown in (A), (B) and (C). (A) Normal pattern of metaphase chromosomes for wild type cells. (B) and (C) show typical metaphase chromosomes of dRecQ414 mutant cells. In (B), sister chromatids are precociously separated, while in (C) chromosomes are mostly broken into smaller fragments. The percentage of cells for the three categories in wild type and dRecQ414 mutants is shown below. Total cell numbers analyzed in each case are indicated in parenthesis. Scale bar = 5 µm
To investigate DNA repair pathways in which dRecQ4 participates, we treated mutants with various DNA-damaging mutagens including hydroxyurea (HU), methyl methane sulfonate (MMS), paraquat and gamma irradiation (Table 2). These mutagens exert their effects through distinct mechanisms and, thus, can provide insights into DNA repair defects in dRecQ4 mutants. Our results show sensitivity of dRecQ414 mutants to paraquat and gamma irradiation (Table 2). Paraquat mainly causes single-base damage which is corrected through base excision repair pathway. Using T7 phage display screen, human RecQ4 was found to interact with poly(ADP-ribose) polymerase-1 (PARP-1), an enzyme that maintains genome stability through its involvement in base excision repair pathway ([34]). Together with our in vivo data, they demonstrate that RecQ4 is involved in the base excision repair pathway. DNA double strand break (DSB) is the major type of DNA damage after gamma irradiation, the sensitivity of dRecQ414 mutants to gamma irradiation suggests that dRecQ4 is also involved in the DSB repair pathways.
Table 2. Sensitivity of dRecQ414 mutant flies to mutagens.
| Mutagen | N. hetero./N. homo. | Relative survival | dRecQ414 mutant response |
| Nothing | 2.12(403/190) | 94.3% | N.A. |
| HU (6.4 mM) | 2.11(327/155) | 94.8% | not sensitive |
| MMS (0.1%) | 2.24(470/209) | 88.9% | not sensitive |
| Paraquat (10 mM) | 3.77(490/130) | 53.1% | sensitive |
| Gamma irradiation (9 Gy) | 2.91(918/315) | 68.6% | sensitive |
See Materials and methods for details of the experiment.
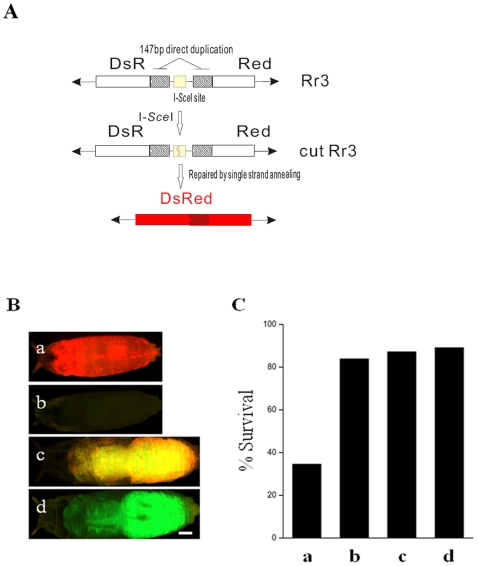
To demonstrate more directly that dRecQ4 participates in DSB repair, we employed the in vivo inducible DSB break-repair system in Drosophila (Fig. 5A; [46]). In this system, the reporter construct, Rr3, consists of a DsRed gene interrupted by the recognition sequence for the rare-cutting endonuclease, I-SceI. UIE is a transgene that expresses the enzyme I-SceI under the control of the ubiquitin gene promoter. The intact Rr3 element does not express a functional DsRed gene product owing to the presence of the cutting site. However, when a DSB is formed at the I-SceI cutting site, repair via the single-strand annealing pathway results in a functional DsRed gene (Figure 5B a and c). We tested the repair efficiency of the induced DSBs both in the presence and in the absence of dRecQ414 mutation. When Rr3 is not cut, the heterozygous control and homozygous mutant animals have similar survival ratios (Figure 5C. category b and d), which serves as a system control. However, when Rr3 is cut, the survival ratio is significantly reduced in dRecQ4 homozygous mutant background compared with heterozygous animals (Figure 5C. category a and c), indicating a reduced efficiency of DSB repair in the absence of dRecQ4 gene function. These results provide additional support to our conclusion that dRecQ4 is involved in DSB repair in vivo.
Figure 5. Repair efficiency of DSBs in dRecQ414 mutants is lowered.
(A) The repair reporter construct, Rr3, consists of a DsRed gene with an I-SceI recognition site in the middle. Flanking the cut site is a 147 bp direct duplication of a part of the DsRed gene sequence. The modified DsRed gene is put into a P element. The entire Rr3 does not express a functional DsRed protein, however, when a DSB is generated by the I-SceI enzyme, repair through the single strand annealing (SSA) pathway results in a functional DsRed gene. A GFP marked balancer chromosome was used to separate the homozygous (panel B, a and b) from the heterozygous (panel B c and d) dRecQ414 animals. The heterozygous mutants served as control. (C) When Rr3 is not cut, control (d, GFP+ and DsRed−) and homozygous dRecQ414 mutant (b, GFP− and DsRed−) animals exhibit similar survival ratios; when Rr3 is cut, the survival ratio is significantly reduced in dRecQ4 mutants (a, GFP− and DsRed+) compared with the control (c, GFP+ and DsRed+). The relative survival ratio (number of pupae/number of first instar larvae) of each category is shown in (C). More than 120 animals were counted for each category. Scale bar = 250 µm
dRecQ4 loss-of-function leads to mitotic (M) phase arrest and reduction of cell proliferation in late wing and eye imaginal discs
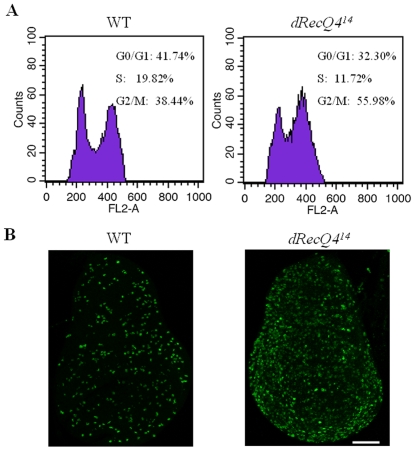
Cell cycle progression is strictly controlled by a strong checkpoint system that arrests progression of the cell cycle until either DNA replication is completed or DNA damage is repaired [51]. To test whether DNA replication and/or repair defects in dRecQ414 mutants lead to cell cycle arrest in vivo, wing imaginal discs from wild type and mutant larvae were dissected, trypsinized and stained with propidium iodide (PI), followed by fluorescence activated cell sorting (FACS). Compared with wild type, more cells from dRecQ414 mutants are accumulated at G2/M phase at the expense of G1/G0 and S phase cells (Figure 6A). It is notable that the second peak is shifted with its position closer to the G1 peak in the dRecQ4 mutant cells comparing with that in the WT cells (Figure 6A). A possible explanation could be that in the absence of dRecQ4, the S-M checkpoint becomes defective, which allows the mutant cells to enter mitosis with incompletely replicated DNA. Immunostaining using antibody against phospho-histone H3, which serves as M phase marker, shows a significant increase of M phase cells in the wing discs of dRecQ414 larvae compared with that of wild type (Figure 6B).
Figure 6. The wing imaginal discs of the dRecQ4 mutants have more M phase cells than wild type.
(A) FACS analysis showing cell cycle profiles of the wing discs of WT and dRecQ414 animals. dRecQ414 mutant discs have more G2/M phase cells at the expense of G1 and S phase cells. (B) WT and dRecQ414 third-instar larvae stained with anti-phospho histone H3 (Ser 10) antibody (mitotic marker). Note that mutant wing discs exhibits higher levels of M phase cells compared with wild-type discs. Scale bar = 100 µm
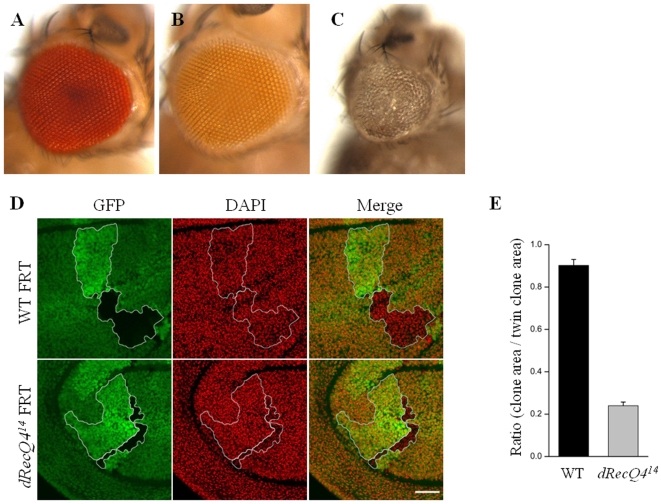
To test whether the cell cycle aberration in the absence of dRecQ4 function leads to cell proliferation defect, two strategies were employed. First, we conducted the tissue specific knock-out of dRecQ4 function using tissue specific flipase that acts on a FRT-flanking genomic rescue transgene. We generated a transgenic line, pTARG-dRecQ4, which harbors a dRecQ4 genomic segment flanked by two FRTs (see Materials and methods for details). This transgene is able to completely rescue the dRecQ4 mutant animals to adulthood without any observable defects (Figure 7B). Taking advantage of such rescued flies, we used ey-FLP to specifically delete the rescuing genomic fragment in the eye-antenna primordia. Figure 7C shows that tissue-specific knockout of dRecQ4 in the eye leads to small rough eyes with disorganized and fewer ommatidia, phenotypes that are indicative of reduced cell proliferation.
Figure 7. dRecQ4 mutation affects eye development and cell proliferation in the wing discs.
dRecQ414 mutant flies that are rescued by the pTARG-dRecQ4[rescue] transgene have as normal eyes (B) as wild type (A). When crossed to flies that carry ey-FLP, the rescuing transgene of dRecQ4 is removed specifically in the eye, leading to smaller and rough eyes (C). (D) Upper panel, wild type sister clones marked by either absence of GFP (GFP-, dark region) or two copies of the GFP (2XGFP, bright region). Lower panel, sister clones of a homozygous dRecQ414 mutant clone marked by GFP- (dark region) and a homozygous wild type twin clone marked by 2XGFP. The dRecQ414 mutant clone has fewer cells than its wild-type twin clone. (E) Statistical analysis indicates that dRecQ4 mutant clones occupy less area than their wild type twin clones. Error bars represent S.E.M. n = 29. P<0.001. Scale bar = 30 µm
In a second strategy to determine whether cell proliferation is affected in the absence of dRecQ4, normal somatic clone analysis was employed (see Materials and methods for details). As shown in Figure 7D, dRecQ4 mutant clones can be detected adjacent to their twin clones (lower row) 72 hours after the induction of FLP expression by heat shock. Similar to the wild type (GFP−) clones (upper row), the total number of dRecQ4 clones (GFP−, lower row) is similar to their twin clones (2XGFP). However, the dRecQ4 mutant clones (GFP−, lower row) are smaller than the 2XGFP twin clones (lower row), while the wild type (GFP−, upper row) clones are similar in size to their 2XGFP twin clones (upper row). The dRecQ4 mutant clones on average occupy about 20% of the territory that their twin clones occupy (2XGFP); in wild type clones, the size of GFP− clones is similar to that of their twin clones (Figure 7E). Taken together, these data suggest that dRecQ4 mutant cells are aberrant in cell cycle progression, which may have resulted in cell proliferation defects.
Both the N-terminal domain and the helicase domain of dRecQ4 are essential to its in vivo function
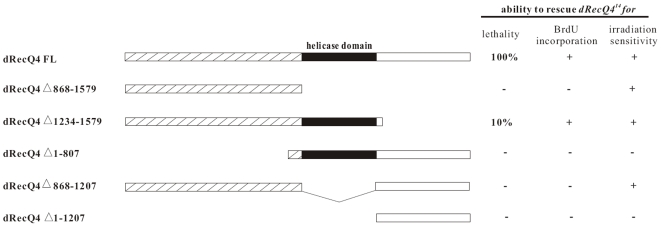
Our studies described thus far suggest that dRecQ4 plays a role in multiple processes including DNA replication, DNA repair and cell cycle progression. To clarify which function is primarily responsible for its essentiality and the protein domain for such function, we performed dRecQ4 functional domain dissection experiments. The dRecQ4 protein consists of 1579 amino acids including a helicase domain extending from aa 867 to 1208. A series of deletion mutants of the dRecQ4 coding sequence were generated, and the resulting truncated proteins are as shown in Figure 8. The corresponding transgenic flies were generated and analyzed for their ability to rescue the mutants' phenotypes including lethality, BrdU incorporation and sensitivity to DNA damaging reagents. The full length dRecQ4 is able to fully rescue dRecQ414 mutant to adulthood without any obvious defects. The C-terminal deletion form, dRecQ4[Δ1234–1579], can also rescue dRecQ414 animal to adulthood, but only at an efficiency of about 10%. Nevertheless, it can fully rescue the BrdU incorporation defects and the sensitivity to gamma irradiation of the mutant. Neither of the other truncations, lacking either the N terminal or the helicase domain or both, exhibited any ability to rescue the mutant animals to eclosed flies (Figure 8). However, gamma irradiation sensitivity can be rescued fully by truncated forms that contain the N-teminal domain, namely dRecQ4Δ868–1579, dRecQ4Δ1234–1579 and dRecQ4Δ868–1207, in addition to the full-length protein. These results indicate that both the N-terminal and helicase domain of dRecQ4 are indispensable for animal viability, although the N-terminal domain alone is sufficient to rescue the mutants' sensitivity to gamma irradiation.
Figure 8. Functional dissection of dRecQ4 protein in vivo.
Constructs that express full-length and different truncated forms of the dRecQ4 protein are shown. Full length dRecQ4 protein that consists of 1579 amino acids including a helicase domain extending from aa 867 to 1208 can rescue 100% of dRecQ414 mutants to adulthood. The C terminal deletion form (dRecQ4Δ1234–1579) of dRecQ4 can rescue dRecQ414 animals to adulthood with an efficiency of only 10%, but fully rescue the BrdU incorporation defects and gamma-irradiation sensitivity of the mutants. None of the other deletion forms can rescue any dRecQ4 mutant animals to adulthood. However, gamma-irradiation sensitivity can be rescued fully by truncated forms that contain the N-teminal domain, namely dRecQ4Δ868–1579, dRecQ4Δ1234–1579 and dRecQ4Δ868–1207, in addition to the full-length protein.
Discussion
The studies described in this report demonstrate that dRecQ4 is essential for Drosophila development. Loss-of-function of dRecQ4 leads to a failure of proper DNA replication and inappropriate cell cycle progression. The dRecQ4 mutant animals show a preferential sensitivity to gamma irradiation. Because gamma irradiation mainly causes DNA double strand breaks, this suggests an in vivo role of dRecQ4 in DSB repair, which is further supported by DsRed reconstitution experiment in vivo. While the C-terminal domain of dRecQ4 is not essential to its function, the N-terminal domain and the helicase domain are indispensable in vivo. Considering our combined results and what is known up-to-date about RecQ4, the following issues are worth further discussion.
(1) dRecQ4 is the only essential RecQ helicase in flies. The three Drosophila helicase genes, dBLM, dRecQ4 and dRecQ5, as well as the Drosophila homologue of human WRN exonuclease gene, dWRNexo, have now all been studied genetically [37], [38], [40], [45] (Chen and Jiao, unpublished). Interestingly, among all the mutants of these dRecQ genes, only dRecQ4 mutants exhibit a homozygous lethal phenotype; mutants of the other dRecQ family members only show either fertility defects or various defects in DNA repair, including recombinational repair. There are several possibilities why dRecQ4 mutants show a more severe phenotype (i.e., lethality) than the other dRecQ mutants. First, dRecQ4 mutants clearly have defects in DNA replication. The defects occur not only in multiploid cells during endoreplication, but also in normal diploid cells. Interestingly, endoreplication defects do not result in total cell number changes although it does affect cell growth, while replication defects in normal diploid cells lead to cell proliferation abnormality. The simplest explanation for this observation is that, at late embryonic stages/early larval stage, the maternal contribution of dRecQ4 helps the mutant animal acquire proper cell number of the salivary glands before loss of zygotic dRecQ4 takes effect. However, by the time of second and third instar larval stages when the cells in the wing imaginal discs proliferate extensively, the maternal dRecQ4 has been diluted and/or degraded to a level that is insufficient to promote cell cycle progression (Figure 6A). The failure of proper cell cycle progression and proliferation is likely to be the cause of lethality.
If the DNA replication defects were the primary cause of animal lethality, one would expect that a dRecQ4 domain that is responsible for DNA replication should rescue the lethality. However, our results show that the N-terminal domain of dRecQ4, dRecQ4△868–1579 (Figure 8), which is homologous to the xRecQ4 N-terminal domain, necessary and sufficient for DNA replication in Xenopus [20], [21], does not rescue the animal's lethality. As shown in this study, dRecQ4 also plays an important role in DSB repair (Figure 5). If the DNA repair defects were the primary cause for lethality of dRecQ4 mutants, one might expect that the other fly RecQ mutants that have DSB repair defects, such as dBLM, would also be lethal, which in fact is not the case [38]. Together, these considerations suggest that the lethality of the dRecQ4 mutants is likely a consequence of loss of both of its primary functions, namely DNA replication and DNA repair.
(2) RecQ4 deficient cells and animals show differential sensitivity to genotoxic agents. It has been controversial in the literature regarding the mutagen sensitivity for RecQ4 deficient cells and/or model organisms [4], [15], [26]–[28], [34], [52]–[54]. For example, it has been shown by two independent groups that RecQ4 is involved in UV-induced damage repair in human cells [15], [52]. However, using fibroblasts derived from different RTS patients, Cabral et al. found that these cells are not sensitive to a wide variety of genotoxic agents including ionizing or UV irradiation, H2O2 and HU [53]. Jin et al. showed very recently that RecQ4 deficient human cells have increased sensitivity to HU, camptothecin (CPT) and doxorubicin (DOX), modest sensitivity to UV or ionizing irradiation[27]. Werner et al. showed that RecQ4-deficient human cells are hypersensitive to oxidative stress such as H2O2 [26], while Woo et al. found changes in the subcellular localization of RecQ4 after exposure to oxidative stress and identify an interaction of RecQ4 with PARP-1 [34]. At the animal level, the mouse model generated by Hoki et al. which bears in frame deletion of exon 13 RecQ4 shows normal sensitivity to IR and UV irradiation [28]. However, our Drosophila RecQ4 mutants are strongly sensitive to ionizing radiation, which suggests a role for dRecQ4 in DSB repair that is in agreement with what has been found in Xenopus [54]. The possible explanations for the conflicting sensitivity results of RecQ4 mutants reported thus far could be as follows: (i) it is possible that RecQ4 plays a more important role in DSB repair pathways in flies than in humans. There are five RecQ helicases in humans while there are only three in flies. The functions for RecQ members may be more specialized in humans than in flies. For example, in humans BLM and WRN are primarily involved in different DSB repair pathways with BLM more in homologous recombinational repair and WRN more in non-homologous end joining repair [4], [6]–[8]. Since there is only dWRNexo in flies, the homologous function of human WRN in flies may have been incorporated in dRecQ4 protein; (ii) more likely, cells derived from different human patients with different mutations or the same mutations in different genetic backgrounds could be also the causes of differential sensitivities. Xu and Liu recently found that the N terminal region and helicase domain of human RecQ4 both possess helicase activity [55], which argues for the possibility that human patients or mutant mouse with intact N terminal region have less severe sensitivity to mutagens. This is very well evidenced by our domain dissection study in Drosophila. The N terminal domain is sufficient to rescue dRecQ4 mutant's sensitivity to gamma irradiation.
(3) Although we expected dRecQ414 allele to be null based on the designed mutations of the translation start codon ATG and the frame shift of the coding sequence, the phenotypes are generally weaker than Wu's null mutant; for example, our mutants die at early pupal stage, but theirs at early larval stage. It is possible that our mutant allele is not completely null that may express a truncated form of dRecQ4; we note that the 1017th codon (for methionine) is followed by a 562 amino acids in-frame reading frame of dRecQ4 (there is currently no appropriate antibodies available to detect this possible truncated protein of dRecQ4). Unlike the null allele of Wu that could not generate mutant somatic clones in the wing discs, our mutant is capable to support limited cell proliferation (Figure 7D). Our mutant may thus represent a potentially useful tool in further mechanistic studies of DNA repair in vivo.
(4) Functional domain dissection combined with rescue experiments suggests that the essential functions of the dRecQ4 protein reside in the N-terminal and the helicase domain. A very recent report by Xu and Liu [56] has shown for the first time that human RecQ4 exhibits dual DNA helicase activity. Two distinct regions of the protein, the conserved helicase motifs and the Sld2-like N-terminal domain, display independent ATP-dependent DNA unwinding activity. Although the N-terminal domain of RecQ4 is sufficient for DNA replication initiation in Xenopus ([20]), our in vivo data clearly suggests the helicase domain is required for proper DNA replication in Drosophila. The C-terminal domain of dRecQ4 is dispensable for its essentiality, but the rescue efficiency of the truncated protein that lacks the C-terminal is only about 10% compared with the full length dRecQ4 protein. It is possible that the C-terminal domain modulates the protein activity of dRecQ4, possibly via amino acids modifications and/or interactions with other proteins.
Materials and Methods
Ethics statement
N/A.
DNA constructs
5 kb dRecQ4 genomic fragment (coding region of the gene) and 3 kb 5′ regulatory sequences with intended modifications were cloned in the pTARG vector [49] to make the gene targeting construct,pTARG-dRecQ4. Changes were introduced by PCR with the following oligos (altered bases for either restriction sites and/or mutations are highlighted by underlining). The primers used to amplify the 5 kb dRecQ4 genomic sequence were 5′-TCCCCGCGGACGATTCGGTGTTCAAGCTAAAAT-3′ and 5′-GGACTAGTGCAGGATGCGATTGAAATCCACTT-3′. The primers for amplifying the upstream 3 kb fragment were 5′-ATAAGAATGCGGCCGCGCTCTCCATCGTGATGGGCCT-3′ and 5′-GGCC TAGGGTCGGCGGCTGTCTTTAATTGTCAATA -3′. Mutation of ATGG to CCTAGGGTCGACCCGCGG generates a new restriction site (SacII) for identification of mutant DNA. Oligos used to introduce the I-SceI cleavage sequence at the MfeI cutting site were 5′-AATTTAGGGATAACAGGGTAAT-3′and 5′-AATTATTACCCTGTTATCCCTA-3′.
For constructing UAS-dRecQ4, primers 5′-ATAAGAATGCGGCCGCACATGGACGATTCGGTGTTC-3′ and 5′-GGGGT ACCTCACGTACGCCTCTTGATAA3′ were used to PCR the genomic DNA that spans from the start to the stop codons of dRecQ4 gene's coding region for putting into pUAST vector at Not I and Kpn I sites (start and stop codons are underlined).
pTARG-dRecQ4[rescue] construct contains 2.1 kb upstream of the ATG and 1.5 kb downstream after stop codon sequence in addition to the entire coding region. It was constructed by putting two PCR products into the pTARG vector at Not I and Avr II sites. The two pairs of primers used for PCR were as follows: 5′-AATGAATTGCGGCCGCGTCGGGAACTACAGTCCAACCT-3′/5′-CGAAACCGGTTGGCTTAGGGAAGCTTCG-3′ and 5′-GCCAACCGGTTTCGCAAGAGAAAGCAGC-3′/5′-TAGACCTAGGATGAAGGAGCACGGCCAAATGCCAG-3′. Restriction sites for cloning are highlighted in italics.
For primers that are used for generating domain dissection constructs, please see Table 3. Detailed cloning strategies are available upon request.
Table 3. Forward and reverse primers used to generate pUAST-dRecQ4△ constructs that produce truncated proteins used in Fig. 8 #.
| Construct | Primers |
| pUAST-dRecQ4 | GGGctcgagATGGACTACAAAGACCATGA |
| GGggtaccTTACTTGTCATCGTCATCCTTGT | |
| pUAST-dRecQ4△868-1579 | AAGGAAAAgcggccgcATGGACGATTCGGTGTTCAAGCT |
| GGGctcgagCCCGAACATGTGGAGTGCCTCTA | |
| pUAST-dRecQ4△1234-1579 | AAGGAAAAgcggccgcATGGACGATTCGGTGTTCAAGCT |
| GGGctcgagAGAATACACATGGCGACGCAGCT | |
| pUAST-dRecQ4△1-807 | AAGGAAAAgcggccgcATGACATACGTCGGCCACAAGATTCC |
| GGGctcgagCGTACGCCTCTTGATAATAGCCA | |
| pUAST-dRecQ4△868-1207 * | ACGCgtcgacATGTTGCCTTCCCACTGTCACCTCTT |
| GGGggtaccTTACTTGTCATCGTCATCCTTGT | |
| pUAST-dRecQ4△1-1207 | AAGGAAAAgcggccgcATGTTGCCTTCCCACTGTCACCTCTT |
| GGGctcgagCGTACGCCTCTTGATAATAGCCA |
All proteins resulted from above constructs are Flag-tagged.
For constructing pUAST-dRecQ4△868–1207, first PCR using pUAST-dRecQ4△1–1207 as template, with primers listed as in the table, digested with Sal I and Kpn I, then ligated into pUAST-dRecQ4△868–1579 vector which had been digested with Xho I and Kpn I.
Fly stocks and genetics
Flies were cultured at 25°C for all experiments. For generation of germline clones (GLCs), we used the FLP-DFS system as described [57]. Briefly, the dRecQ414 mutation was recombined with the third chromosomal FRT insertion 2A, balanced with TM6B, Tb balancer and crossed with ovoD, FRT2A males. Offspring of this cross were given a heat shock (37°C, 2 hrs) at late third instar larval stage and virgin females with correct genotypes (Table 1) were crossed with heterozygous mutant males.
Listed below are fly stocks used in this study:
y w
Canton S
w; actin-Gal4
y w; ey-FLP; MKRS/TM2, y+
y w; hs-I SceI, hs-FLP, Sco/CyO
w1118; hs-I-CreI, Sb/TM6
FRT2A (kindly provided by Dr. Xinhua Lin)
y w, hs-FLP; FRT2A ovoD/TM3, Sb (kindly provided by Dr. Xinhua Lin)
y w; actin-Gal4/TM3, Ser
y w; pTARG-dRecQ4[rescue]
w1118; P{XP}d02769 P{neoFRT}80B (Bloomington Drosophila Stock Center)
y w, hs-FLP; If/CyO; Ubi-GFP FRT80B/TM6B, Tb (kindly provided by Dr. Zhaohui Wang)
Sp P[Rr3] 48C L/CyO (Kindly provided by Dr. William R. Engels)
Sco/CyO P[UIE] 53D (Kindly provided by Dr. William R. Engels)
Generation of dRecQ414 mutant
For generation of the dRecQ4 mutant, we used the ends-in gene targeting method [47], [48]. Donor transgenic flies that bear the targeting construct on the second chromosome were crossed to flies that contain hs-I SceI and hs-FLP transgenes. Three heat shocks (38°C, 1 hr each) were applied on days 2, 3 and 4 after egg laying. Heat-shocked virgins were singly crossed to y w; ey-FLP; MKRS/TM2, y+ males, and females were screened for targeted integration of targeting construct indicated by the w+ marker. Reduction of two dRecQ4 copies (one wild type and one mutant copy) by I-CreI was performed by crossing the targeted alleles to w1118; hs-I-CreI, Sb/TM6. The offspring were given a single heat shock (36°C, 1 hr) at the third instar larval stage. w− males were crossed individually to y w; actin-Gal4/TM3, Ser to make stocks. The allele, we designated dRecQ414, was further characterized by DNA sequencing for the intended mutations, the primers for PCR were 5′-TCCCAGCATGTGATAGTCTG-3′ and 5′-TCCTCAAGATTACCAG AGCTC-3′. The resulting data has been deposited in GenBank (accession number GQ128383).
Generation of somatic clones
Loss-of-function somatic clones were induced using FLP/FRT mediated mitotic recombination [58]. To induce the clones, first instar larvae with correct genotypes were heat shocked for 1 hour at 38°C and then dissected at third instar larval stage. Mutant clones and twin spot areas were measured with confocal images using the histogram function of Adobe Photoshop.
Immunohistochemistry
For BrdU labeling, wild type, mutant or rescued mutant larvae were dissected in PBS and then incubated in PBS containing 1 mg/ml BrdU (Sigma B-5002) for 30 min at 25°C. After three rinses with PBS, samples were fixed for 30 min in 4% paraformaldehyde followed by washing 3 times in PBST (PBS, 0.1% Triton X-100) and then treatment with 2M HCl for 30 min. After three washes in PBST, samples were incubated with mouse anti-BrdU (1∶100, ZYMED). TRITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) was used with dilution of 1∶100 for 2 hrs at room temperature. Images were taken under a Leica DM6000 confocal microscope. More than 10 brains were examined per genotype. Rabbit anti-phospho histone H3 (Ser 10) antibody (1∶100) for detecting mitotic phase was purchased from Millipore. FITC-conjugated anti-rabbit secondary antibody (1∶100) was from Jackson ImmunoResearch Laboratories. Over 10 discs were analyzed for each genotype.
Fluorescence activated cell sorting
80–100 wing discs of the same genotypes (the mutant or wild type) were dissected in PBS and digested with trypsin-EDTA (Sigma, T-4174). Cells were dissociated for 5 hours by gentle shaking. The dissociated cells were fixed in 75% ethanol, stained with propidium iodide and analyzed with a Becton Dickinson Vantage Fluorescence activated cell sorter. The events of either the wild type or the mutant were 10000. The experiment was repeated 3 times.
DNA content measurement
Genomic DNA from 100 salivary glands of wild type or dRecQ414 mutant larvae (5 days old) was extracted, followed by A260/280 measurement. Two sample independent t-test was used to determine statistical significance.
Chromosome spreads
Brains from wild type Canton S and dRecQ414 homozygous third instar larvae were dissected in 0.7% saline, treated with colchicine and hypotonic solution, fixed in acetic acid/methanol/H2O (11∶11∶2) and stained for 5 min in 0.2 µg/ml DAPI. The preparations were examined under Leica DM6000 fluorescent microscope.
DNA damage sensitivity tests
Four mutagens were used in this experiment (hydroxyurea, methyl methane sulfonate, paraquat and γ-irradiation, see Table 2). All chemicals were purchased from Sigma. Chemical mutagens were added to fly food at final concentrations that have been used for DNA damage assay in Drosophila (see also Table 2). 20 females and 10 males of y w; dRecQ414/TM3, Sb,Kr-GFP flies were put in the vial that contained different mutagens for 6 h before the parents were discarded. At the 6th day, the heterozygous (GFP+) and homozygous (GFP−) mutants were scored. The survival ratio was measured as the number of GFP− to half the number of GFP+. For γ-irradiation, eggs from y w; dRecQ414/TM3, Kr-GFP flies were collected for 5 hrs and allowed to develop for 12 hrs before being exposed to 9 Gy γ-irradiation with a 60Co source. Only gamma irradiation was used to test the sensitivity of the rescued mutants to DNA damaging reagent. The same dosage was applied as above.
DsRed repair assay
DsRed DSB repair model was carried out essentially according to the Preston method [46]. Since nearly all the dRecQ414 homozygous mutants can live to early pupal stage, the relative survival ratio from early instar larvae to early pupae stage was used to indicate the repair efficiency indirectly, assuming the lethality is caused by unrepaired DSBs. The cross was made as follows: Sp P[Rr3] 48C L/CyO; dRecQ414/TM3 Kr-GFP crossed with Sco/CyO P[UIE] 53D; dRecQ414/TM3 Kr-GFP. Offspring genotypes and phenotypes as judged by fluorescence colors are (homozygous TM3 Kr-GFP animals and homozygous CyO animals do not survive to pupae stage, therefore are not included for counting):
(A): Sp P[Rr3]48C L/CyO P[UIE] 53D; dRecQ414 [GFP (−) DsRed (+)]
(B): Sp P[Rr3]48C L/Sco; dRecQ414 and CyO/Sco; dRecQ414 [GFP (−) DsRed (−)]
(C): Sp P[Rr3]48C L/CyO P[UIE] 53D; dRecQ414/TM3, Kr-GFP [GFP (+) DsRed (+)]
(D): Sp P[Rr3]48C L/Sco; dRecQ414/TM3, Kr-GFP and CyO/Sco; dRecQ414/TM3, Kr-GFP [GFP (+) DsRed (−)]
First instar larvae and early pupae representing each categories were counted for calculation of the relative survival ratio.
Acknowledgments
We thank Drs. Dieter Egli, Zhaohui Wang, Xinhua Lin, William R. Engels and the Bloomington stock center for providing us with plasmids and fly stocks and the Jiao lab member for discussions. We are grateful to the anonymous reviewers for their time and constructive suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work has been supported by National Natural Science Foundation of China (NSFC30623005, 90608029 and 30771217), 973 programs (2005CB522804, 2009CB918702) and Chinese Academy of Sciences (KSCX1-YW-R-70). JM acknowledges support from National Institutes of Health and National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietschy T, Shevelev I, Stagljar I. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell Mol Life Sci. 2007;64:796–802. doi: 10.1007/s00018-007-6468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 4.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, et al. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 6.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 7.Wu L. Wrestling off RAD51: a novel role for RecQ helicases. Bioessays. 2008;30:291–295. doi: 10.1002/bies.20735. [DOI] [PubMed] [Google Scholar]
- 8.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao R, Bachrati CZ, Pedrazzi G, Kuster P, Petkovic M, et al. Physical and functional interaction between the Bloom's syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol Cell Biol. 2004;24:4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 11.Jiao R, Harrigan JA, Shevelev I, Dietschy T, Selak N, et al. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2007;26:3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- 12.Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, et al. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem Soc Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 13.Bagherieh-Najjar MB, de Vries OM, Hille J, Dijkwel PP. Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 2005;43:789–798. doi: 10.1111/j.1365-313X.2005.02501.x. [DOI] [PubMed] [Google Scholar]
- 14.Burks LM, Yin J, Plon SE. Nuclear import and retention domains in the amino terminus of RECQL4. Gene. 2007;391:26–38. doi: 10.1016/j.gene.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Fan W, Luo J. RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA). J Biol Chem. 2008;283:29037–29044. doi: 10.1074/jbc.M801928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006;34:4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, et al. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum Mol Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 20.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 23.Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Capp C, Feng L, Hsieh TS. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev Biol. 2008;323:130–142. doi: 10.1016/j.ydbio.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin J, Kwon YT, Varshavsky A, Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum Mol Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
- 26.Werner SR, Prahalad AK, Yang J, Hock JM. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem Biophys Res Commun. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- 27.Jin W, Liu H, Zhang Y, Otta SK, Plon SE, et al. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum Genet. 2008;123:643–653. doi: 10.1007/s00439-008-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum Mol Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- 29.Smith PJ, Paterson MC. Enhanced radiosensitivity and defective DNA repair in cultured fibroblasts derived from Rothmund Thomson syndrome patients. Mutat Res. 1982;94:213–228. doi: 10.1016/0027-5107(82)90183-x. [DOI] [PubMed] [Google Scholar]
- 30.Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 31.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, et al. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 32.Balraj P, Concannon P, Jamal R, Beghini A, Hoe TS, et al. An unusual mutation in RECQ4 gene leading to Rothmund-Thomson syndrome. Mutat Res. 2002;508:99–105. doi: 10.1016/s0027-5107(02)00189-6. [DOI] [PubMed] [Google Scholar]
- 33.Ichikawa K, Noda T, Furuichi Y. [Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases]. Nippon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- 34.Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 35.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 36.Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama M, Yamaguchi SI, Sagisu Y, Sakurai H, Ito F, et al. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair (Amst) 2008 doi: 10.1016/j.dnarep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki K, Maruyama S, Nakayama M, Matsumoto K, Shibata T. Drosophila melanogaster RECQ5/QE DNA helicase: stimulation by GTP binding. Nucleic Acids Res. 2002;30:3682–3691. doi: 10.1093/nar/gkf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusano K, Johnson-Schlitz DM, Engels WR. Sterility of Drosophila with mutations in the Bloom syndrome gene–complementation by Ku70. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 41.Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trowbridge K, McKim K, Brill SJ, Sekelsky J. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 2007;176:1993–2001. doi: 10.1534/genetics.106.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boubriak I, Mason PA, Clancy DJ, Dockray J, Saunders RD, et al. DmWRNexo is a 3′-5′ exonuclease: phenotypic and biochemical characterization of mutants of the Drosophila orthologue of human WRN exonuclease. Biogerontology. 2008 doi: 10.1007/s10522-008-9181-3. [DOI] [PubMed] [Google Scholar]
- 45.Saunders RD, Boubriak I, Clancy DJ, Cox LS. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell. 2008;7:418–425. doi: 10.1111/j.1474-9726.2008.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston CR, Flores CC, Engels WR. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics. 2006;172:1055–1068. doi: 10.1534/genetics.105.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, et al. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egli D, Selvaraj A, Yepiskoposyan H, Zhang B, Hafen E, et al. Knockout of ‘metal-responsive transcription factor’ MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 2003;22:100–108. doi: 10.1093/emboj/cdg012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egli D, Yepiskoposyan H, Selvaraj A, Balamurugan K, Rajaram R, et al. A family knockout of all four Drosophila metallothioneins reveals a central role in copper homeostasis and detoxification. Mol Cell Biol. 2006;26:2286–2296. doi: 10.1128/MCB.26.6.2286-2296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand F, Castorina P, Morant C, Delobel B, Barouk E, et al. [Rothmund-Thomson syndrome, trisomy 8 mosaicism and RECQ4 gene mutation]. Ann Dermatol Venereol. 2002;129:892–895. [PubMed] [Google Scholar]
- 51.Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, et al. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- 53.Cabral RE, Queille S, Bodemer C, de Prost Y, Neto JB, et al. Identification of new RECQL4 mutations in Caucasian Rothmund-Thomson patients and analysis of sensitivity to a wide range of genotoxic agents. Mutat Res. 2008;643:41–47. doi: 10.1016/j.mrfmmm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Kumata Y, Tada S, Yamanada Y, Tsuyama T, Kobayashi T, et al. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim Biophys Acta. 2007;1773:556–564. doi: 10.1016/j.bbamcr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. Embo J. 2009 doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]