Introduction
In spite of their varied appearances, insects have in common a body plan that consists of head, thorax, and abdomen, with different numbers and types of segments. Insects generally possess a head region made up of 6-7 segments, a thorax of three segments, and an abdomen of 8-11 segments. This format is laid out during embryogenesis when patterning genes specify the body plan. Virtually all insects start development in a syncytial environment where nuclei divide without cell membranes to separate cells. Patterning factors may thus diffuse and have direct access to nuclei to provide patterning information. We currently have a high molecular resolution map in Drosophila of how an embryo patterns its segments. However, Drosophila has a highly derived type of embryogenesis in which virtually all segments are patterned simultaneously at the syncytial stage. Therefore, the study of other insects is invaluable for piecing together how the ancestor of all insects established its segmented body plan, and how this process can be plastic during evolution. Though we only have extensive molecular understanding of the development of very few non-drosophilid insects, these data show that, while the paths taken to achieve similar adult body plans share common features, they are often very different.
This review will discuss the evolution of Antero-Posterior (A-P) patterning mechanisms in insects. It will first describe the two distinct modes of insect development, long and short germ development, and how these two modes of patterning are achieved. We will then summarize how A-P patterning occurs in the long-germ Drosophila, where most of our knowledge comes from, and in the well-studied short-germ insect, Tribolium. Examples drawn from other insects will highlight differences in patterns of expression, but also foci of evolutionary change.
Germ types
G. Krause is credited with the designations now in use to distinguish among the different developmental strategies used by insects ([1]; Figure 1). He observed that some insects produce eggs in which the antecedents (“anlagen”) of all the future segments of the embryo are represented and patterned during the syncytial blastoderm stage. Since the embryo (“germ”) fills up the majority of the egg and all segments of the germ anlage are present before gastrulation, with a relatively small portion designated for extraembryonic tissue, he called these embryos “large germ”. These embryos were also called “long germ” referring in part to the number of segments specified in the germ anlage by the time gastrulation occurred ([1, 2]; Figure 1A). Embryos that fall into this category include the higher Diptera Drosophila melanogaster and Musca domestica ([3-5]), as well as the parasitic wasp Nasonia vitripennis (Hymenoptera) ([6]). In contrast, G. Krause noted that other insects produce eggs in which most of the material is specified as extraembryonic tissue, with the germ tissues largely restricted to the posterior-ventral side of the egg. In these insects, only the anteriormost embryonic structures are patterned at the syncytial stages. He called these insects “short germ”, since the germ anlage represents only a few (the anteriormost) segments of the embryo, while the remaining (posterior) segments are generated via a posterior ‘growth zone’ after cellularization. Many were also “small germ”, in reference to the relative small portion of the egg that serves as germ material (Figure 1B). Short germ insects include the well-studied flour beetle, Tribolium castaneum (Coleoptera), the milkweed bug Oncopeltus fasciatus (Hemiptera), the cricket Gryllus bimaculatus (Orthoptera), and the grasshopper Schistocerca americana (Orthoptera). Differences between short and long germ insects may arise in part in the ovary where maternal information originates. It has been suggested that ovaries that possess nurse cells (meroistic ovaries), in contrast to the more ancestral panoistic ovaries, which lack them, may have been a critical intermediate in the progression from short germ development to long germ. The reliance of long germ embryogenesis on nurse cells to supply nutrients and determinants is largely supported ([7])(The reverse is not necessarily true, as several short germ insects also possess meroistic ovaries.)
Figure 1. Schematic representation of long germ and short germ embryos.
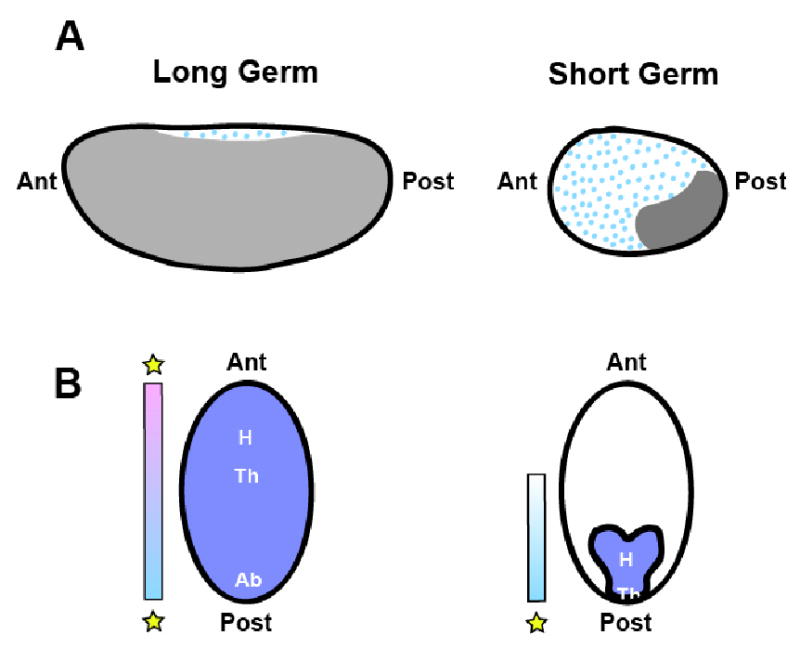
(A) Lateral view. In long germ embryos, relatively little of the egg is allocated for extraembryonic tissues (blue dots), while the germ occupies the majority. The germ rudiment of short germ insects is restricted to the ventral posterior of the blastoderm, while the remainder of the egg is occupied by extraembryonic tissues. (B) Ventral view. In long germ insects, all of the future segments (head [H], thorax [T] and abdomen [A]) will be patterned in the precellular blastoderm by two patterning centers (indicated by stars). Short germ insects pattern only head and thorax before gastrulation, from a single patterning center at the posterior.
As there are orders of insects, such as Coleoptera, which possess members of both long and short germ types, this suggests that long germ embryogenesis was either lost in some short germ insects, or that it arose independently several times. This latter view is currently favored ([8], [9]). How did the transition(s) from short germ to long germ embryogenesis occur? The most informative approach to answering this question is to describe the long and short germ modes of development, and then revisit those aspects that have been transformed. We will begin with the best-described program: that of the long germ Drosophila.
Long Germ Embryogenesis: The Drosophila Paradigm
Drosophila embryogenesis begins in the meroistic ovaries of the female (reviewed in [10]). The Drosophila oocyte is specified as one of 16 sister cells, the 15 others becoming nurse cells that will transcribe maternal messages encoding morphogens, as well as provide cellular machinery and mitochondria for the oocyte to develop rapidly. These are transferred into the oocyte via ring canals that connect the nurse cells and the oocyte. The nurse cells dump their contents into the oocyte, including mRNA's for several maternal factors that become localized to the poles of the oocyte. After fertilization, these mRNA's are translated and act as transcriptional or translational regulators. Syncytial nuclear divisions in the blastoderm embryo continue for 13 cycles, with the onset of the earliest zygotic transcription occurring around cycle 10, when the nuclei migrate out to the periphery of the embryo, with another wave during cycle 14 at the maternal to zygotic transition (MZT), when the nuclei become separated by cell membranes and the embryo becomes cellularized. By the end of cycle 14, the specification of the segments of the Drosophila embryo has largely been achieved.
The segmented Drosophila body plan is specified by a cascade of transcription factors that subdivide the embryo into increasingly small domains. In the late 1970's, large-scale saturation mutagenesis screens were carried out by Nüsslein-Volhard, Wieschaus, Schupbach and others to identify genes involved in segmentation ([11, 12]). The mutants identified in the screens were grouped according to the patterns of defects observed: genes required maternally for embryogenesis (maternal effect genes) that affect large regions of the body, like head, thorax, abdomen, or the non-segmented termini; genes that cause defects in groups of adjacent segments (gap genes); genes that affect alternating segments (pair-rule genes), and genes that affect every segment (segment polarity genes). These genes clearly act sequentially, patterning finer and finer regions of the embryo. The maternal effect genes, or coordinate genes, establish the initial polarity of the embryo, and initiate expression and position of the first zygotic genes, the gap genes, which act regionally in the embryo to regulate the downstream pair-rule genes. These, in turn, combinatorially regulate segment polarity genes in every segment, establishing the final number and boundaries of the segments. The identification of developmental genes allowed us to obtain one of the most precise descriptions of any complex biological system, the Drosophila embryo. It also provided the template to study how patterning of other animals is initiated, since the genes identified in Drosophila are among the first inroads made into the understanding of the embryogenesis of other insect (and vertebrate) models. A schematic representation of the patterns of expression of genes described in this section is provided in Figure 2.
Figure 2. Schematic representation of segmentation gene expression patterns in long and short germ insects.
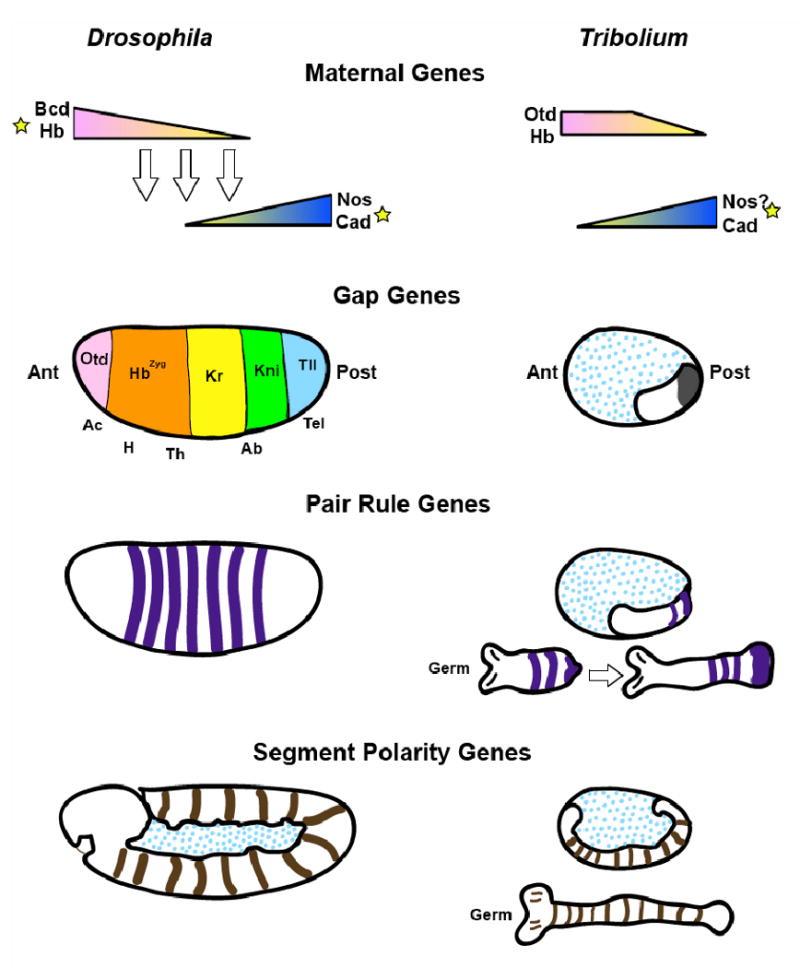
Patterning centers are indicated by stars and non-germ tissue by blue dots. Stripes indicate approximate domains of gene expression, triangles indicate morphogen gradients. Abbreviations: Ac- acron; H-head; Th- thorax; Ab- abdomen; Tel- telson.
Maternal genes
Most maternal effect mutations affect genes whose messages are loaded by the mother into the oocyte to establish polarity in the early embryo (reviewed in [13], [14], [15]). These coordinate genes produce, for instance, the anterior determinant Bicoid (Bcd) [16] and the posterior determinant Nanos [17]. In early experiments in other Diptera, embryos were pricked at the anterior and the cytoplasm was permitted to leak out, resulting in embryos lacking the anterior structures- this indicated that instructive factors are contained in the cytoplasm at the poles of the oocyte of these species ([18]). Indeed, the bcd and nanos mRNAs are localized to their respective poles using elements in their 3′ UTRs that allow the mRNA to be loaded onto cytoskeletal motors and transported to or trapped at the poles. The 3′UTRs also mediate translational repression where protein product activity must be suppressed ([19, 20]). Both bcd in the anterior and nos in the posterior form strongly localized mRNA sources in the oocyte, which result in protein gradients during early embryogenesis. The localization of maternal messages is a critical step in establishing A-P polarity in embryos (see below). In contrast, mRNA of other maternal genes such as hunchback (hb) or caudal (cad) is loaded and evenly distributed throughout the early embryo [21](see below).
bcd mutant mothers give rise to embryos that lack all anterior structures (head, thorax, part of the abdomen) and have a duplicated posterior telson. However, the remaining abdominal segments retain proper polarity (reviewed in [14]). Bcd is a transcription factor that promotes anterior identity through the activation of zygotic hb expression as well as the expression of a number of other target genes that respond to the different concentrations of Bcd. This is the instructive function of Bcd. Maternal Hb cooperates with Bcd for the activation of head gap genes ([22]). Bcd also has a permissive function in repressing posterior and trunk identity, which it achieves through the activation of the repressor, giant ([23]). It also acts as a translation factor that binds to 3′UTR of uniformly distributed cad mRNA and prevents the anterior translation of this posterior determinant.
hb, which is an anterior gene, should inhibit abdominal identity at the posterior. However, posterior hb is not translated because of a Nanos response element (NRE) in its 3′UTR that is bound by Pumilio and Nanos proteins ([24], [25], [26]), allowing maintenance of posterior identity in the embryo. Lack of nanos causes loss of abdominal segments due to ectopic maternal Hb protein at the posterior ([27], [28], [17]). However, embryos from mothers double mutant for hb and nanos are viable ([27, 28], [21, 29]), suggesting that the only segmentation role of Nanos is to repress maternal hb at the posterior, and that this maternal hb function is not essential. Nanos is also required for the specification of pole cells ([17]). Maternal cad, which is translationally suppressed in the anterior, is active in the posterior, activating posterior gap genes knirps (kni) and gt and patterning abdominal segments.
One separate class of maternal genes affects the termini (reviewed in [30]). These genes include maternal Torso, torso-like, and trunk. Unlike the anterior and posterior systems, the terminal system is not associated with a transplantable factor localized within the cytoplasm at the ends of the embryo ([31]). The activities of the terminal genes, which comprise a receptor tyrosine kinase cascade with Torso as the receptor, inhibit activity of the segmentation genes to set apart the termini from the rest of the body axis.
In summary, maternal Bcd and Hb cooperate to promote anterior identity through transcription control, while Bcd translationally inhibits cad; cad that is translated promotes posterior identity, in cooperation with Nos, which permits posterior identity through the inhibition of Hb, thus establishing the initial A-P polarity in Drosophila.
Segmentation genes
The mutant phenotype of gap genes (hb, gt, Krüppel (Kr), kni) includes “gaps” in the segmented pattern of the larval cuticle resulting from loss of contiguous segments. Gap genes encode a series of transcription factors that are expressed regionally in the embryo. orthodenticle (otd), empty spiracles (ems) and buttonhead (btd) are head gap genes activated by Bcd at the most anterior tip of the embryo but act independently from the rest of the segmentation cascade. Tailless and huckebein are gap genes that act downstream of Torso, cooperating to carry out its function at the termini ([32]).
Mutations in Kr lead to deletion of the thorax and the first five abdominal segments ([33]). kni mutants exhibit deletions of adjacent abdominal segments, leaving only the termini and last abdominal segment intact ([34]). Flies lacking gt expression exhibit labial-T1 fusions and defects in abdominal segments 5-7 ([35], [36]). Several gap gene products act as morphogens (e.g. Kr and Hb;[27, 37, 38]). For instance, high concentrations of Hb repress anterior Kr expression, while lower concentrations activate it. The products of gap genes refine each other's borders of expression. For instance, Kr and gt exhibit mutually repressive interactions ([39]) while gt also negatively regulates kni, leading indirectly to the regulation of kni by Kr ([40]). This repressive network establishes overlapping domains within the trunk of the embryo that then lead to regulation of the downstream pair rule genes.
Pair-rule mutant phenotypes are characterized by the loss of alternating segments or parts of segments. hairy, even skipped (eve), and runt are expressed as seven or eight stripes that each results directly or indirectly from a combination of activation and repression by gap genes. They represent a transcriptional readout of gap gene activity at each position along the A-P axis. The eve promoter, a case study in modular gene regulation, contains 5 discrete elements that control expression of individual or pairs of stripes in response to maternal and gap genes ([41, 42]): the eve stripe 2 element, for instance, is controlled by bcd, hb, Kr, and gt [42, 43]. hairy exhibits similar modular control ([44, 45]). In contrast, the seven stripes of secondary pair-rule genes odd-skipped, runt, paired, sloppy paired and fushi-tarazu (ftz) appear to respond to the primary pair-rule genes. The seven ftz stripes are controlled by a single “zebra element” whose regulation is still poorly understood ([46]). Most pair-rule genes are also expressed in segmental stripes later during gastrulation, which may arise either de novo, as in the case of eve or runt, or by splitting of the primary stripes, e.g. paired (reviewed in [13]). Finally, each segment has its own intrinsic polarity/patterning, which is established through regulation by the pair rule genes of the segment polarity genes, such as engrailed (en) and wingless (wg). wg is expressed at the anterior border of the compartment boundary while en is expressed in the entire posterior compartment. By this stage, the embryo is no longer a syncytium and regulation involves signaling molecules such as wingless or hedgehog.
Long germ segmentation thus involves the coordinated actions of several tiers of genes that pattern all segments essentially simultaneously. Pair rule gene regulation, exemplified by the modularity of the eve promoter, make this possible, by interpreting the gradient and boundary information of the gap genes, as well as the repressive effects of other pair rule genes, and translating this into individual stripes of expression which predict segments. This cascade of events is well suited to the germ, which is completely represented in the blastoderm stages. Other long germ band insects share some but not all aspects of this program. Recent studies in the long germ wasp Nasonia provide an excellent context for addressing which conserved aspects are requisite for evolution of long germ development, and which represent merely one way of “inventing” the long germ mode.
An Alternate Mode of Long Germ Segmentation: Nasonia
Nasonia is a wasp (Hymenoptera) that diverged from Diptera more than 200 million years ago. Its embryogenesis begins like Drosophila, except that some of the earliest events in embryogenesis- namely the loading and localization of maternal messages in the egg- are significantly different. Bcd is unique to higher Diptera and is absent from the genomes of Nasonia and all other insects. Instead, the ortholog of the fly head gap gene otd ([47]) is maternally contributed and localized, like bcd mRNA, to the anterior in Nasonia ([9, 48]). The Otd homeodomain has the ability to recognize the same DNA sequence as that of Bcd through its K50 homeodomain, which establishes similar contacts with DNA[49]. It functions in combination with maternal hb and likely maternal gt ([50]) to pattern the anterior. Surprisingly, otd is also localized to the posterior pole in Nasonia which it also patterns by interaction with other genes, likely cad.
In Drosophila, maternal cad is uniformly distributed as mRNA but is translationally repressed in the anterior by Bcd, leading to the formation of a Cad protein gradient. In Nasonia, in the absence of Bcd to regulate its translation, maternal cad has invented a different strategy to limit its function to the posterior: maternal cad mRNA is itself localized to the posterior, creating a gradient of mRNA ([51]). Loss of cad activity in Nasonia results in embryos that lack most abdominal segments, placing cad atop the cascade of segmentation genes, while its role in Drosophila is more restricted. otd and cad cooperate to activate and position posterior gap gene expression in Nasonia.
Thus, in long germ development, one key feature that appears to be widely conserved is the use of two patterning centers: an anterior morphogenetic center, which utilizes a combination of Hb and another factor (Bcd in flies, Otd in Nasonia) to achieve widespread gene activation and anterior identity, and a posterior center, using Cad (and likely the terminal system in flies; Cad works with Otd in Nasonia). However, while the activity (and targets) of the anterior morphogen is conserved, its identity is not. This theme of conserved activity with interchangeable factors is discussed in the context of the anterior morphogenetic center in the final section below. In addition, the relative positions of gap and maternal genes in regulation of the segmentation hierarchy are labile. Otd is a gap gene in Drosophila and an important maternal gene as well in Nasonia (and in Tribolium, discussed below). otd temporal and spatial shift of expression correlates with its additional regulatory responsibilities.
One other noteworthy difference between Nasonia and Drosophila is the heavy early reliance of Nasonia on zygotic as well as maternal gene expression. A screen for patterning mutants in Nasonia revealed several zygotic mutations whose phenotypes affected adjacent segments, mimicking the Drosophila gap gene phenotypes [52]. The zygotic headless/hb mutation exhibits a more severe phenotype than hb in flies, losing most head structures and thorax in addition to several anterior abdominal segments. This phenotype mimics loss of both maternal and zygotic hb combined with reduction of bcd in Drosophila. The zygotic cad mutant phenotype, called head only [51], exhibits loss of posterior structures; head en stripes are intact while all trunk en stripes are lost ([52]). This is comparable to the phenotype of maternal and zygotic cad mutants in Drosophila, where head and thorax are unaffected, but the posterior segments are reduced ([53]). The significance of the inability of maternal genes in Nasonia to rescue some of the early zygotic defects is discussed below.
Comparison of these two long germ insects provides information about which viable solutions of the short-to-long germ transition are flexible, and which aspects are constrained: the use of an anterior patterning center seems to be essential, while the use of bcd in that center is not. In contrast, short germ insects utilize a very different strategy, in which posterior segments of the germ form in a reiterative, progressive fashion, and are patterned primarily from a posterior center. This type of development has been studied in the milkweed bug, Oncopeltus (Hemiptera), crickets and grasshoppers of Orthoptera, and has been most deeply explored in the beetle, Tribolium castaneum (Coleoptera).
Short Germ Segmentation: The Tribolium paradigm
The best studied short germ insect is the flour beetle Tribolium, where genetic screens have been successfully carried out to identify genes involved in segmentation ([54],[55]). Although these screens identified classes of mutants similar to those of Drosophila, an exciting outcome was the identification of new classes of mutants unknown in Drosophila.
Unlike Drosophila, short germ insects produce embryos that only fill a small portion of the total egg. Tribolium females possess meroistic ovaries but provide a different cohort of factors to the developing embryo. The Tribolium syncytial embryo is differently shaped than Drosophila and undergoes a transition in which nuclei aggregate in the posterior on the ventral side, forming a cell layer that will become the germ anlage. As gastrulation begins, the primitive pit forms at the future caudal end of the embryo, which then dives into the yolk. The germ anlage undergoes a process similar to convergent extension as cells aggregate at the posterior and the germ band narrows and is extended anteriorly along the ventral surface of the embryo. As the head forms, segments are added at the posterior end through cell addition in the growth zone, until the germ band is fully extended along the ventral side of the embryo, and the caudal end of the ectoderm sits on the dorsal surface of the embryo near the head. At full extension, the head lobes are formed at the anterior and the germ band retracts. The germ band of Tribolium becomes overtly segmented in an anterior-posterior fashion well after gastrulation has begun. ([56]). It is important to note several key differences from Drosophila: first, at cellularization, only the future head segments and thorax are represented in the germ anlage toward the posterior of the Tribolium embryo. Abdominal segments arise later from a posterior growth zone in Tribolium, whereas abdominal segment primordia are present in the precellularization blastoderm in Drosophila and Nasonia. A schematic representation of A-P patterning gene expression in short germ insects, as exemplified by Tribolium, is given in Figure 2.
Gene function in Tribolium can be disrupted using parental RNAi (pRNAi) ([57, 58]). RNAi of genes like Torso which prevent addition of segments from the growth zone phase of development results in embryos whose head and anterior thorax are unaffected, but which lack all structures that develop during post-blastodermal growth ([59]). This means that there are two phases or programs at play in Tribolium development and likely all short germ insects: one controlling patterning of the extraembryonic membranes and anterior germ anlage in the precellular blastoderm, and another controlling generation and patterning of posterior (abdominal) segments in a cellular environment.
Like Nasonia, Tribolium lacks Bcd. Bcd is widely thought to have originated from a duplication of the related, nearby gene, Zerknullt/Hox3 (Zen), which is involved in specification of amnioserosa in Drosophila. zen is conserved in Tribolium, both in sequence and in function ([60]). The ancestral anterior patterning configuration utilizing maternal otd and hb was first discovered in Tribolium ([61, 62]). Schröder ([62]) used parental RNAi to knockdown otd in Tribolium embryos: These embryos lack all head structures. pRNAi for hb, which is also provided maternally in Tribolium, leads to embryos that lack maxillary, labial, and thoracic structures, but whose head segments are intact. Embryos from otd and hb double pRNAi females result in embryos that phenocopy a severe bcd phenotype indicating that Otd and Hb work together in Tribolium to carry out the function that Bcd and Hb serve in Drosophila. cad pRNAi in Tribolium results in arrest of axis elongation, and as in Nasonia, causes severe truncations of the embryo ([63], [51]). Thus, otd, hb, and cad serve as coordinate genes in Tribolium.
Orthologues of Drosophila gap genes were studied in Tribolium through reverse genetics (pRNAi). Tribolium Kr is expressed at the posterior pole of the blastoderm embryo, which is anterior to most of the future abdominal segments, meaning that its position relative to the primordia of the larval segments is conserved. Knockdown of Kr in Tribolium results in embryos whose head segments are intact, but whose thoracic segments are transformed into a reiterated series of gnathal segments, followed by a deletion of several abdominal segments. The mutant jaws, later identified as Tribolium Krüppel [54, 64], causes both a homeotic transformation in the thorax and first abdominal segment, as well as a deletion of most of the abdominal segments. Thus, Kr seems to behave differently in Tribolium than in flies: it is expressed in a more anterior domain, and its loss of function causes a disorder in segmentation that is propagated in all segments that are formed posterior to its expression domain, underscoring the two programs of segmentation utilized by short germ insects. Tribolium gt, like Drosophila gt, is expressed in two domains: one in the head and one in the trunk, coincident with the primitive pit. Like Kr, the posterior domain of Tribolium gt is more anteriorly located than its Drosophila counterpart. pRNAi of Tribolium gt also causes segmentation defects affecting all segments (both thoracic and abdominal) including those posterior to its expression domain[65].
An EST expression screen in embryos identified a novel gap gene in Tribolium called Millepattes (Mlpt). Mlpt is a polycistronic RNA encoding several peptides that is expressed in the head lobes, as well as at the onset of primitive pit formation. It is then expressed in a stripe at the posterior of the mandibular segment, which expands during elongation before a new posterior domain is formed. Embryos mutant for mlpt are shortened, but possess additional pairs of legs, indicating a transformation of abdominal segments into leg-bearing thoracic ones. ([66]). The homologous Drosophila gene, Tarsal-less or polished rice, ([67]) has a lethal phenotype that includes an array of defects but expression of gap and homeotic genes is unaffected. The identification of this “novel” segmentation gene provides an important reminder that the Drosophila paradigm is unlikely to be representative of the entire genetic repertoire utilized across taxa to generate segments, and rather, underscores the importance of unbiased approaches to the study of the developmental programs of other insects. Additional gap genes have been identified by other forward genetic means, including krusty (kry) and bollig (bol), whose molecular lesions have not yet been mapped ([55]). kry mutants exhibit absent labial and T3 segments and deletions of adjacent abdominal segments. Bol mutants have intact head, pregnathal and gnathal regions but lack posterior thoracic and the first abdominal segments ([55]). Each gene affects contiguous segments of which some are formed in a cellularized environment and some before cellularization, indicating that gap genes still specify regional identity through function in different mechanistic contexts.
Pair rule gene expression in Tribolium, and indeed in all short-germ insects, is different from Drosophila. This is due in part to the altered expression domains and apparent longer-term effects of the gap genes, but also reflects the role of the posterior growth zone in generating the abdominal segments of the embryo. Because most abdominal segments are made after cellularization, a short germ gap gene must either be expressed before pair rule stripe formation and anterior to its final location or be expressed in the growth zone in waves concomitant with the formation and patterning of the abdominal segments to affect stripe expression. The organization of pair rule gene promoters in short germ insects should therefore reflect this different strategy. While regulation of pair-rule genes has not been extensively described in Tribolium, study of the genomic region upstream of Tribolium hairy identified a fragment that largely recapitulates the endogenous expression pattern in embryos. A subfragment of the initial 8.8kb gave expression in a subset of stripes, but these fragments could not easily be reduced to identify individual stripe-specific enhancers, suggesting that individual stripes of expression in Tribolium are not controlled by compact modular elements like their Drosophila counterparts ([68]).
Orthologs of the pair rule genes eve, hairy, ftz, odd-skipped, sloppy-paired, runt and paired have been studied in Tribolium ([69],[70],[71], [72], [73]). These genes are all expressed in pair rule stripes in two waves: one in the early blastoderm, followed by a second wave of expression in the growth-zone during posterior growth. Surprisingly, pRNAi of odd-skipped, eve, or runt produces truncated embryos lacking virtually all segments, resulting from cessation of germ elongation, while pRNAi knock down of sloppy-paired or paired in Tribolium results in a phenotype which more closely resembles the canonical pair-rule phenotype. Interestingly, despite its expression in stripes, a mutation affecting expression of ftz does not cause a pair-rule phenotype, suggesting that ftz does not function as a pair-rule gene in Tribolium([74]). Similarly, pRNAi for hairy only causes anterior defects in head segments ([71]), suggesting that hairy also does not play a major role in segmentation in Tribolium. Two mutants with pair-rule phenotypes were isolated in a genetic screen for segmentation mutants (icy and scy; [55]). These mutants phenocopy the RNAi phenotypes of Tribolium slp and prd, though the lesions have not been mapped ([75]).
Thus, while homologs of Drosophila pair rule genes are conserved across species at the sequence and often expression levels, their utilization during A-P patterning has changed during evolution from short to long germ. This is reflected in the variable expression timing and patterns of these genes across species studied to date. Clearly, the shift of genes from one level of the hierarchy to another by evolution (gain or loss) of cis-regulatory sites or by altering timing of expression is an important way that A-P patterning has evolved. This type of change has been extensively documented and discussed elsewhere in several excellent reviews on segmentation in insects (see [5],[76, 77], [78]; and many others) and will not be discussed further here.
Short and long germ insects progress through embryogenesis differently. In an embryo that fills the oocyte, having two patterning centers (one at each pole- indicated by stars in Figure 2) creates a sufficient complexity of gradient information to pattern all segments. However, in a short germ insect, whose head anlage is positioned near the posterior of the embryo, an anterior determinant at the anterior pole of the egg would not have enough gradient information to reach the head anlage. A posterior patterning center could play a significant role in the precellular blastoderm of these insects patterning anterior (early) segments, since posterior segments do not arise at all until after cellularization, when simple diffusion can no longer suffice to deliver gradient information. Subsequent patterning would have to be differently controlled in the growth zone to ensure that emergent posterior segments receive the appropriate instruction at the appropriate time, to generate the correct number and types of segments. Comparison of the three insects described so far shows that in the evolution of long germ development from ancestral short germ development, specific activities (but not specific factors) have been coopted in the creation of an anterior patterning center, so that what is localized and how it is localized may vary among species while the outcome of their activities are the same. We will now discuss these transition mechanisms of mRNA localization and maternal vs. zygotic expression in the context of their potential to contribute to germ type evolution, and how short-to-long germ transitions may have occurred.
Variations on a Theme: mRNA Localization
Localization of maternal factors is a common strategy among animals for establishing axial polarity [79]. The localization of maternal bicoid and nanos mRNAs provides positional information to the Drosophila embryo, and this maternal prepattern likely correlates with its extremely rapid early development. While comparatively very little is known about the significance of maternally localized factors in patterning the embryos of other short germ insects, some recent results as well as classic embryological experiments suggest how the use of the localization strategy is employed and changed in evolution.
Short germ insects
In short germ insects, though an anterior patterning center would have little consequence, a posterior determinant is required since the primordium of the embryo proper arises near the posterior of the egg, ([2], [77]). Consistent with this model, classic studies using embryological manipulations of short germ embryos provided strong evidence for a posteriorly localized patterning center, but weak or no evidence for an anterior center. Of particular interest are the fragmentation experiments of Sander using embryos of the leafhopper, Euscelis [80]. In these experiments, posterior pole material was moved by gentle mechanical pressure toward the anterior of the egg, followed by separation of the anterior and posterior halves (by ligation). The results of these experiments demonstrated that the posterior pole material possesses a strong posteriorizing activity, which appears to act in a graded, morphogen-like fashion ([80, 81]). In addition, ligature experiments in the cricket Gryllus showed that a posterior localized factor is required for the establishment of the germ anlage ([81]) while no anterior morphogenetic center could be found.
In the molecular age, more evidence has mounted that posteriorly localized patterning information is important in short germ embryos. As mentioned previously, patterning of Tribolium embryo depends on gradients of Hb and Otd1 activity, both of which are provided maternally as ubiquitous mRNA ([62]). Anterior to posterior gradients of both proteins appear to form as a result of translational repression activity emanating from the posterior. As both Tribolium otd1 and hb mRNAs possess NREs in their 3′UTRs, it is tempting to conclude that Nanos is responsible for creating the two transcription factor gradients that together pattern the AP axis at the blastoderm stage, although this has never been formally demonstrated. Thus, posterior nanos may provide all of the positional information required to specify the head and thoracic segments at the blastoderm stage ([62]).
Further evidence that posteriorly localized nanos mRNA has a conserved role in establishing AP polarity comes from the short germ grasshopper Schistocerca. In this organism, nanos mRNA is expressed maternally and is restricted to the posterior cortex of the oocyte. In the embryo, the posterior localization persists into the germband stage, and the region of nanos localization corresponds to regions where Hb protein is absent. This pattern, along with the fact that Schistocerca hb mRNA contains an NRE, indicates that Nanos has a conserved role in regulating the formation of the AP axis throughout the insect clade ([82]).
It may be worthwhile to revisit the classical embryological experiments with a new molecular perspective and techniques (such as in situ hybridization and RNAi). For example, it should be possible to test the hypothesis that the posterior pole material of Euscelis contains nanos mRNA, and that loss of nanos message results in the loss of the posteriorizing potency of this material in translocation experiments similar to those of Sander.
Long germ insects
Before the molecular identification of bicoid and nanos, there was already evidence from manipulations of the embryos of dipterans, the honeybee Apis, and of long germ beetles (e.g. Callosobruchus) for both a posterior and an anterior patterning center in long germ insects. Particularly interesting were experiments performed using chyronomid midges (lower Diptera) wherein embryos were centrifuged, and gave rise to double-headed or double-tailed larvae, depending on the direction and timing of the centrifugation ([83]), suggesting the existence of two morphogenetic centers.
The wasp Nasonia has emerged as a long germ model alternative to Drosophila that is highly amenable to functional analyses, permitting a detailed understanding of the strategies for localizing maternal factors that pattern the embryo. In Nasonia, as in Drosophila, maternal factors localized to both poles are important for patterning the AP axis. In fact, Nasonia seems to utilize mRNA localization much more extensively and in different ways than Drosophila.
Nasonia otd1 was originally examined as a strong candidate to replace the anterior patterning function of a missing bicoid ortholog. In Tribolium, otd1 patterns the anterior of the short germ insect but its activity is regulated from the posterior morphogenetic center. Nasonia otd1 functions in place of bcd, and even more interestingly, mRNA for this gene is maternally localized at the anterior pole of the egg. Thus, in two long germ insects, mRNA for a transcription factor is maternally localized at the embryo's anterior and used as an information source for anterior patterning. That this strategy has arisen independently in two distantly diverged insects indicates that anterior localization may be a common tool to solve the problem of patterning a long germ embryo ([9]).
Surprisingly, Nasonia otd1 mRNA is also localized to the posterior end of the egg, in a structure corresponding to the wasp germ plasm (the oosome), and this also has a patterning function, but of the posterior fates, and with different target genes ([9]). The difference in the ability of the two gradients of Nasonia Otd1 to activate specific targets was attributed to its synergy with maternal Nasonia Hb at the anterior (which is anteriorly restricted by translational repression by Nanos, whose mRNA is also localized to the oosome ([84]; JAL and CD, manuscript in preparation), and with Cad at the posterior (see below). In Drosophila, there is no posteriorly localized mRNA for a transcription factor with a function equivalent to that of posterior Nasonia otd1, and patterning of the posterior might rely on the terminal system in flies [27]. This indicates that the use of localized mRNAs can be quite labile in evolution.
Two other localized factors in Nasonia further demonstrate the great potential for the use of maternal localization in the evolution of long germ embryogenesis. The Nasonia gap gene giant is maternally expressed and localized at the anterior pole. Unlike Nasonia otd1, which remains tightly localized, gt mRNA becomes delocalized and diffuses through the anterior part of the embryo in the early stages of embryogenesis, where it has an important role in repressing Nasonia Kr expression, thus allowing zygotic hunchback expression and anterior patterning (Kr is a negative regulator of hb transcription). This represents another novel variation on maternal localization in Nasonia: in Drosophila the repression of Kr at the anterior depends on the combination of Bcd (probably through its target gt) and terminal system activity ([50]).
The second localized mRNA is caudal, whose function was described in an earlier section. Nasonia caudal is initially incorporated into the oosome at the posterior of the embryo. Then, in the early stages of embryogenesis, cad mRNA is released from the oosome and diffuses in the posterior part of the embryo, generating a gradient of cad mRNA ([51]). This is different from what is seen for posterior otd1 and nanos, where most of the mRNA remains tightly associated with the oosome until the formation of the pole cells ([9], JAL and CD, in preparation). This mechanism of cad regulation is different from what is seen in Drosophila, and cad may be controlled in other insects by still other mechanisms that have yet to be explored experimentally.
Thus, the Nasonia system, along with Drosophila and earlier work with long germ embryos, demonstrate a much greater dependence of long germ patterning on maternal localization. This also shows that the development of a strong anterior source of positional information is required for this type of embryogenesis, in addition to the posterior center: Long germ embryos must establish many more cell fates at the blastoderm stage and utilize the entire length of the embryo for patterning, making a single patterning center at only one pole (the posterior) insufficient. Studies in additional, independently-evolved long germ insects (such as long germ beetles), as well in lower dipterans that are long germ but do not use bcd, will provide valuable information about the various ways that mRNA localization may be used to pattern embryos.
Evolution of localization mechanisms
The broad conservation of at least a localized posterior patterning center in insects points to an ancestral role for mRNA localization in establishment of AP polarity. One widely conserved component of the ancestral posterior patterning center is the use of nanos, a gene with a highly conserved role in the germline throughout metazoa. Despite its broad conservation, there is evidence that the use of germ plasm to localize nanos and establish AP polarity is still a potential target for evolution, since several different variations on this mechanism have already been found. One example comes from Drosophila, where a maternal gene, oskar, is absolutely critical for the assembly and proper localization of the germ plasm ([85, 86]). Despite this critical role, it appears that oskar is a unique invention within Diptera, as orthologs of this gene cannot be found in the fully sequenced genomes of the silkmoth, beetle, bee or wasp (JAL, personal observation). The study of mechanisms of germ plasm assembly in non-dipteran insects will allow the understanding of the ancestral mode of AP axis formation. Another interesting case comes from the honeybee, where the germline does not appear to be specified maternally, but rather is induced late in embryogenesis. Despite this, nanos mRNA is posteriorly localized in a way that is consistent with a role in posterior patterning, indicating that the two functions of nanos can be separated depending on mechanisms of germline determination ([87]).
Analyses in Drosophila have shown that intact and properly polarized actin and microtubule cytoskeletons are critical for proper localization of maternal mRNAs ([88]). In this system, oocyte microtubules are initially assembled at an organizing center (MTOC) located at the posterior pole of the oocyte. After signaling events between the posteriorly located oocyte nucleus and the follicle cells overlying the posterior pole of the oocyte, the posteriorly organized microtubule cytoskeleton disassembles and is replaced by one in which microtubules are nucleated primarily from the anterior cortex of the oocyte. This reorganization is also correlated with the migration of the oocyte nucleus to the anterior dorsal region of the oocyte. These events are absolutely critical for the proper localization of bicoid mRNA to the anterior pole, and the formation of the germ plasm (along with localization of nanos mRNA) at the posterior ([89-91]). This also serves to determine the dorso-ventral axis.
At the moment, how broadly the Drosophila mechanisms for mRNA localization are conserved is unknown. However, some evidence obtained in Nasonia indicates that some of these mechanisms are conserved at least in these two species. The pattern of otd1 mRNA localization first gave a clue that a reorganization of the cytoskeleton occurred in Nasonia: otd1 is first seen to accumulate at the posterior pole, then, as the follicle matures, the first evidence of anterior otd1 localization can be seen.
Functional studies using drugs disrupting the actin and microtubule cytoskeletons as well as RNAi against genes involved in maintaining microtubule polarity gave further insight into the role of theses structures in localizing mRNAs in the Nasonia oocyte. Most localized mRNAs in Nasonia (anterior giant and otd1, and caudal at the posterior) depend on the presence and proper polarity of microtubules for their localization. In contrast, localization of Nasonia nanos and the posterior portion of otd1 mRNA are unaffected by colchicine treatment, and instead rely on the actin cytoskeleton for proper anchoring to the posterior pole ([84]). It has been shown in Drosophila that localization of some cargos of cytoskeletal motors involves retention of the motor with the cargo (dynein is retained at the apical cortex to anchor ectopic, injected pair rule RNA, while actin is not required; [92]). If anterior otd1 in Nasonia (like Drosophila bcd) requires continual transport using dynein for anterior localization ([93]), then separating motors might result in differential localization of subpopulations of the same message. This separation might also reflect additional requirements for maintenance or timing of localization, and may also be necessary to avoid depletion of resources required for continual transport. If the delay in anterior localization of Nasonia otd1 instead reflects a dependence on a rearrangement of cytoskeletal polarity, then motors may also be sequentially utilized according to their response to the sequence or timing of cytoskeletal rearrangements that are to take place. Further analysis of the Nasonia system, as well as expansion of analyses of oogenesis to other insects, should provide deeper insights into how mechanisms of localization can be used and changed during evolution.
Maternal vs. zygotic gene expression
Another critical parameter that has been modulated during evolution is the maternal provision vs. zygotic transcription of patterning determinants. The significance of the maternal contribution can be seen in cases where both maternal and zygotic expressions are utilized: for example, hb mRNA is maternally deposited in the Drosophila oocyte. While the maternal component is not required for embryogenesis, elimination of both maternal and zygotic hb expression results in a phenotype that is much more severe than eliminating zygotic expression alone ([22]). Thus, timing of activity might be important. Evolution could affect messages of this type by altering the degree of continuity of transcripts expressed both maternally and zygotically, by shifting from purely zygotic to addition of maternal expression (or vice versa), or may allocate discrete functions by temporally or spatially separating domains of gene expression. Continuity of maternal and zygotic expression would be modulated at the maternal zygotic transition (MZT), which is very tightly regulated: Approximately 30% of maternally contributed mRNAs in Drosophila decrease dramatically in abundance at MZT, including a subset of genes that require the degradation of the maternal transcripts to permit zygotic transcription ([94]). A variety of mechanisms to ensure clearance of maternal messages at MZT are represented among that group, including use of several types of 3′ UTR motifs known to mediate mRNA instability ([94]). Transcription of a different subset of zygotic genes is required for degradation of maternal messages, a mechanism that likely includes transcription of microRNAs. Consistent with this model, at least one microRNA, miR-430, was recently identified in zebrafish as an important regulator of MZT ([95]). These mechanisms of regulation of maternal messages comprise one aspect of control over this transition.
Interestingly, promoters of purely zygotic transcripts are enriched for the heptamer sites called TAGteam ([48, 94]) Presence of these sites is not sufficient for transcriptional activation, but the sequence is overrepresented in promoters enriched for Dorsal and Bcd binding sites, suggesting cooperation with those factors. These intriguing data suggest a coordinated mechanism for activation of early, purely zygotic transcripts. If this mechanism is indeed widespread, then other species, such as Nasonia, which lack Bcd but possess the TAGteam factor, may depend on other maternally encoded proteins, such as Otd, to cooperate in the activation of zygotic targets. In this model, a gene that was shifting from purely maternal to early zygotic expression would be under selection for TAGteam site enrichment. Several gatekeepers of MZT including regulators of maternal mRNA stability and early activators of zygotic transcription may thus be important potentiators of evolution of patterning.
The (dis)continuity of maternal and zygotic expression of specific genes may also be important to consider. Although redundancy provided by temporal overlap of expression ensures that essential functions will not be neglected, zygotic activity instead of maternal control provides for spatial control that would not be obtainable from diffusing maternal product, or may permit divergence of functions. In this way, zygotic vs. maternal gene expression is an aspect of the developmental program that may be a powerful focus of change. For instance, in Drosophila, maternal and zygotic hb have different promoters controlling the expression of the same protein ([96]). Maternal control is ceded at the onset of zygotic transcription, when maternal hb is degraded, making the expression of hb essentially continuous. Nasonia still extensively utilizes the activity of hb in patterning of the early embryo, but maternal and zygotic expressions are not continuous. This means that no rescue of zygotic function can be achieved by perdurance of maternal message. The consequence of this non-overlap is that the phenotype resulting from zygotic loss of hb function in Nasonia is more severe than loss of zygotic hb function in Drosophila, resembling the loss of both maternal and zygotic hb and one copy of bcd ([97],[98]). This is consistent with a model in which, in ancestral insects, hb had a critical zygotic function. Since Nasonia spends far more time in the syncytial blastoderm stage than Drosophila, despite the approximate equivalence of the time required for post-gastrulation events [52], extensive reliance on critical zygotic gene functions may be a more general trend in Nasonia, and may be similar in other long germ insects.
In some cases, zygotic function is overtly divergent from maternal gene function, as well. For instance, the short germ grasshopper Schistocerca exhibits an apparent discontinuity in maternal/zygotic hb expression, where axial patterning is carried out by zygotic hb which appears to form a gradient, while maternal hb is thought to play a separate role in setting aside the germ primordium from the extraembryonic rudiment. Unlike Drosophila, zygotic hb expression in Schistocerca does not occur in a syncytial environment, but achieves a stepped graded pattern of expression by zygotic transcriptional control ([99]). It is the zygotic promoter that permits spatial control in a cellular environment that could not be achieved by the maternal cohort. Perhaps the separate control of non-overlapping domains of gene expression was permissive of additional functional divergence.
In summary, what is transcribed maternally and how maternal and zygotic transcripts are regulated become important potential contributors to evolutionary plasticity. The changes that have occurred in the independent evolution of long germ from short germ have included shifts both in localization of mRNA's and their maternal vs. zygotic regulation and function. How these changes have come together is summarized below.
Evolution of long germ mode of development
During evolution, the germ has undergone several transitions. Short germ insects are often small germ as well (i.e. only a small proportion of the oocyte is allocated for the embryo rudiment), and pattern few segments early. To evolve long germ embryogenesis from a short germ mode, at least three changes have to occur: the expansion of the proportion of the oocyte dedicated to embryo formation, the positioning of a patterning center at the anterior to pattern a compact head and thorax in the anterior portion of the egg, and early positioning of posterior gap genes to permit abdominal segment patterning at the syncytial stage.
The first step in transitioning from short to long germ would be an increase in the proportion of the oocyte allocated for the germ. An example of this type of transition in process may resemble the Oncopeltus embryo, which is large germ, occupying virtually all of the oocyte, yet only the head and thoracic segments are patterned from this material during the blastoderm stage. Like other basal insects, there is no bcd (or, in general, anterior morphogenetic center) to position head and thoracic segments within the germ, but like other insects, it does possess zen orthologs, which pattern the most anterior regions of the egg as extraembryonic membranes. pRNAi of Oncopeltus zen results in a complex phenotype, including an everted embryo and failed katatrepsis ([100]). However, pRNAi of Tribolium zen1 results in a shift in the amnion to cover the dorsal side of the embryo and expansion of the germ anlage anteriorly, which is largely tolerated and compensated for later in development [60]. This suggests that zen could play a significant role in changing the size of the germ.
Acquisition of an anterior patterning center is an important second step in transitioning from short to long germ. Bcd has been studied for its dynamics in the early embryo, since it is a maternally provided morphogen with two important early functions: it activates expression of head and thoracic gap genes, and it regulates translation of cad, whose resulting posterior-to-anterior gradient permits posterior abdominal segments to form ([101-103]). Embryos with 6 copies of bcd exhibit an overly large head, and a posterior shift of the fate map, resulting in patterning defects from compression of the remaining posterior segments into a small proportion of the embryo anlage ([104]). This suggests that the overexpression of bcd, leading to the activation farther posteriorly of hb and the restriction of the domain of Cad expression, affects the ability of the embryo to proportion itself appropriately. Thus, Bcd is an essential maternal signal for allocating space in the fly embryo for the head and designating boundaries for positioning of the remaining segments, through regulation of hb and cad.
While long germ Drosophila and Nasonia have solved the anterior patterning center problem by localizing bcd and otd to the anterior, Drosophila may have also coopted components of the Torso terminal system to create an anterior morphogenetic center. Activation of Torso signaling in the anterior of the oocyte is required for formation of anterior head structures, although this can be bypassed by increased concentrations of bcd, suggesting that these two pathways act on common targets ([105],[106]). Subsequent addition of a localized activator at the anterior also serves to move the Kr domain more toward the middle of the germ; in Nasonia, this is accomplished by anteriorly localized gt [50]. Lastly, posterior gap gene expression must be established for patterning of abdominal segments. This likely involves cad, which is required for abdominal segment formation and gap gene activation, but does not act as a morphogen.
Unlike the Drosophila paradigm, which relies on bcd, both short germ and other long germ insects (like Nasonia) heavily utilize the posterior gradient of cad to position hb and Kr correctly to proportionally allocate the embryo. In Nasonia, cad is maternally deposited and posteriorly localized in the ooctye, and forms an RNA gradient ([51]). RNAi of cad results in reduction of Kr, gt, and kni expression and a consequent posterior shift in hb expression; the cuticles of cad-RNAi embryos exhibit a head-only phenotype. Though this parceling of the germ anlage in non-higher Diptera is heavily reliant on cad for general activation of gap genes, its anterior spread must nonetheless be limited to leave room for head and thoracic structures. As Bcd, which regulates cad translation in the anterior of Drosophila, is not present in these insects, and Otd cannot carry out Bcd's translation repression function, other mechanisms must be invoked. Nasonia has uniquely achieved this through localization of cad mRNA. One alternative in other insects would be to use different translational regulators, such as orthologs of the C.elegans gene mex-3 which inhibits translation of the cad-like gene, pal-1, in C.elegans ([107]). That Nasonia and Tribolium require cad for formation of at least some anterior segments in addition to posterior segments argues strongly for the importance of cad regulation in evolution of patterning. The molecule that translationally regulates cad, then, would be an important capacitor of the change from short to long germ embryogenesis, as a modulator of the allocation of the germ anlage via placement of the hb and Kr boundaries.
Conclusion
During evolution, insects have found a number of different ways to use the genetic tools present in ancestral insects to fashion a developmental program that culminates in a segmented body plan. Different solutions have incorporated conserved developmental modules and modified these over time with acquisition of novel gene products (e.g. Bcd) and concurrent changes in germ proportions, resulting in new patterning regimes, functionalities, and transitions. The points discussed in this review represent only some of the ways in which these transitions may have occurred. Further experimentation in new insect models will provide answers as to how evolution has shaped development.
Figure 3. Model for transition from short germ to long germ mode of embryogenesis.
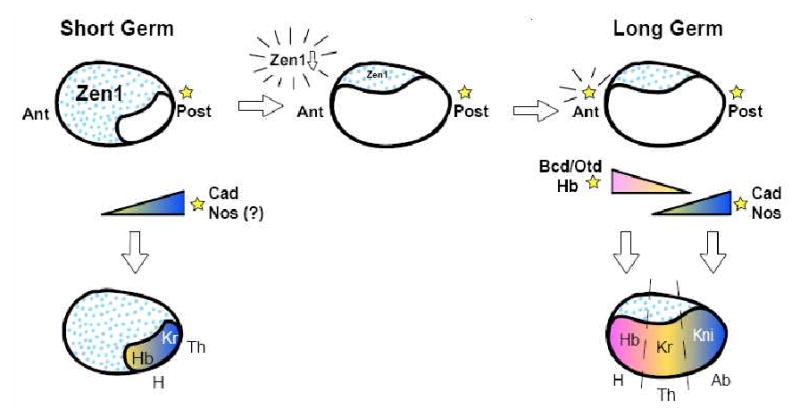
In the first step, the germ is expanded anteriorly, for instance, through reduction of Zen expression. Second, an anterior patterning center is acquired to localize morphogens near the future head. Lastly, gap gene boundaries, such as Kr, shift anteriorly, leaving room for patterning of abdominal segments from the remaining blastoderm. Patterning centers are indicated by stars, and extraembryonic tissues by blue dots. Abbreviations: H- head; Th-thorax; Ab- abdomen.
Acknowledgments
The authors would like to thank Steve Small, Chris Rushlow, and Anita Fernandez for critical reading of the manuscript and helpful comments and discussions. They also apologize for all the papers that have not been properly referred to, due to lack of space.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krause G. Die Eitypen der Insekten. Biologischen Zentralblatt. 1939;59:495–536. [Google Scholar]
- 2.Davis GK, Patel NH. Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu Rev Entomol. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- 3.Liu PZ, Kaufman TC. Kruppel is a gap gene in the intermediate germband insect Oncopeltus fasciatus and is required for development of both blastoderm and germband-derived segments. Development. 2004;131:4567–4579. doi: 10.1242/dev.01311. [DOI] [PubMed] [Google Scholar]
- 4.Sommer R, Tautz D. Segmentation gene expression in the housefly Musca domestica. Development. 1991;113:419–430. doi: 10.1242/dev.113.2.419. [DOI] [PubMed] [Google Scholar]
- 5.Liu PZ, Kaufman TC. Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol Dev. 2005;7:629–646. doi: 10.1111/j.1525-142X.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- 6.Pultz MA, Leaf DS. The jewel wasp Nasonia: querying the genome with haplo-diploid genetics. Genesis. 2003;35:185–191. doi: 10.1002/gene.10189. [DOI] [PubMed] [Google Scholar]
- 7.Bier K, Kunz K, Ribbert D. Structure and function of oocyte chromosomes nucleoli and as well as the extra DNA during oogenesis in panoistic and meroistic insects. Chromosoma. 1967;23:214–254. doi: 10.1007/BF00331114. [DOI] [PubMed] [Google Scholar]
- 8.Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, Tettelin H, Lercher MJ. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature. 2006;439:728–732. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- 10.Spradling AC. Developmental Genetics of Oogenesis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. pp. 1–70. [Google Scholar]
- 11.Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- 12.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 13.Pankratz MJ, Jackle H. Blastoderm segmentation. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. [Google Scholar]
- 14.Driever W. Maternal control of anterior development in the Drosophila embryo. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. [Google Scholar]
- 15.St Johnston D. Pole Plasm and the Posterior Group Genes. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. [Google Scholar]
- 16.Nusslein-Volhard C, Frohnhofer HG, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt O, Zissler D, Sander K, Kalthoff K. Switch in pattern formation after puncturing the anterior pole of Smittia eggs (Chironomidae, Diptera) Dev Biol. 1975;46:216–221. doi: 10.1016/0012-1606(75)90099-8. [DOI] [PubMed] [Google Scholar]
- 19.Schnorrer F, Bohmann K, Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 20.Arn EA, Cha BJ, Theurkauf WE, Macdonald PM. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell. 2003;4:41–51. doi: 10.1016/s1534-5807(02)00397-0. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann R, Nusslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- 22.Simpson-Brose M, Treisman J, Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78:855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- 23.Kraut R, Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991;111:601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- 24.Gamberi C, Peterson DS, He L, Gottlieb E. An anterior function for the Drosophila posterior determinant Pumilio. Development. 2002;129:2699–2710. doi: 10.1242/dev.129.11.2699. [DOI] [PubMed] [Google Scholar]
- 25.Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 26.Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Struhl G. Differing strategies for organizing anterior and posterior body pattern in Drosophila embryos. Nature. 1989;338:741–744. doi: 10.1038/338741a0. [DOI] [PubMed] [Google Scholar]
- 28.Hulskamp M, Schroder C, Pfeifle C, Jackle H, Tautz D. Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature. 1989;338:629–632. doi: 10.1038/338629a0. [DOI] [PubMed] [Google Scholar]
- 29.Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- 30.Sprenger F, Nusslein-Volhard C. The terminal system of axis determination in the Drosophila embryo. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, New york: 1993. [Google Scholar]
- 31.Klingler M, Erdelyi M, Szabad J, Nusslein-Volhard C. Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature. 1988;335:275–277. doi: 10.1038/335275a0. [DOI] [PubMed] [Google Scholar]
- 32.Bronner G, Jackle H. Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech Dev. 1991;35:205–211. doi: 10.1016/0925-4773(91)90019-3. [DOI] [PubMed] [Google Scholar]
- 33.Gloor H. Schadigungsmuster eines Letalfaktors (Kr) von Drosophila melanogaster. Arch Jul Klauds Stiftung. 1950;25:38–44. [Google Scholar]
- 34.Nauber U, Pankratz MJ, Kienlin A, Seifert E, Klemm U, Jackle H. Abdominal segmentation of the Drosophila embryo requires a hormone receptor-like protein encoded by the gap gene knirps. Nature. 1988;336:489–492. doi: 10.1038/336489a0. [DOI] [PubMed] [Google Scholar]
- 35.Petschek JP, Perrimon N, Mahowald AP. Region-specific defects in l(1)giant embryos of Drosophila melanogaster. Dev Biol. 1987;119:175–189. doi: 10.1016/0012-1606(87)90219-3. [DOI] [PubMed] [Google Scholar]
- 36.Sanders LR, Patel M, Mahaffey JW. The Drosophila gap gene giant has an anterior segment identity function mediated through disconnected and teashirt. Genetics. 2008;179:441–453. doi: 10.1534/genetics.107.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hulskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Kruppel and knirps in the early Drosophila embryo. Nature. 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- 38.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 39.Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Kruppel define middle body regions of the Drosophila embryo. Development. 1991;111:611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 40.Capovilla M, Eldon ED, Pirrotta V. The giant gene of Drosophila encodes a b-ZIP DNA-binding protein that regulates the expression of other segmentation gap genes. Development. 1992;114:99–112. doi: 10.1242/dev.114.1.99. [DOI] [PubMed] [Google Scholar]
- 41.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 43.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 44.Howard KR, Struhl G. Decoding positional information: regulation of the pair-rule gene hairy. Development. 1990;110:1223–1231. doi: 10.1242/dev.110.4.1223. [DOI] [PubMed] [Google Scholar]
- 45.Riddihough G, Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 1991;5:840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- 46.Dearolf CR, Topol J, Parker CS. Transcriptional control of Drosophila fushi tarazu zebra stripe expression. Genes Dev. 1989;3:384–398. doi: 10.1101/gad.3.3.384. [DOI] [PubMed] [Google Scholar]
- 47.Finkelstein R, Perrimon N. The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature. 1990;346:485–488. doi: 10.1038/346485a0. [DOI] [PubMed] [Google Scholar]
- 48.ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 49.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 50.Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science. 2007;315:1841–1843. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- 51.Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, Desplan C. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development. 2006;133:3973–3982. doi: 10.1242/dev.02576. [DOI] [PubMed] [Google Scholar]
- 52.Pultz MA, Pitt JN, Alto NM. Extensive zygotic control of the anteroposterior axis in the wasp Nasonia vitripennis. Development. 1999;126:701–710. doi: 10.1242/dev.126.4.701. [DOI] [PubMed] [Google Scholar]
- 53.Macdonald PM, Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 54.Sulston IA, Anderson KV. Embryonic patterning mutants of Tribolium castaneum. Development. 1996;122:805–814. doi: 10.1242/dev.122.3.805. [DOI] [PubMed] [Google Scholar]
- 55.Maderspacher F, Bucher G, Klingler M. Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev Genes Evol. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- 56.Brown SJ, Parrish JK, Denell RE, Beeman RW. Genetic control of early embryogenesis in the red flour beetle, Tribolium castaneum. Am Zool. 1994;34:343–352. doi: 10.1093/icb/34.3.343. [DOI] [PubMed] [Google Scholar]
- 57.Brown SJ, Mahaffey JP, Lorenzen MD, Denell RE, Mahaffey JW. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 58.Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12:R85–86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 59.Schoppmeier M, Schroder R. Maternal torso signaling controls body axis elongation in a short germ insect. Curr Biol. 2005;15:2131–2136. doi: 10.1016/j.cub.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 60.van der Zee M, Berns N, Roth S. Distinct functions of the Tribolium zerknullt genes in serosa specification and dorsal closure. Curr Biol. 2005;15:624–636. doi: 10.1016/j.cub.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 61.Schulz C, Schroder R, Hausdorf B, Wolff C, Tautz D. A caudal homologue in the short germ band beetle Tribolium shows similarities to both, the Drosophila and the vertebrate caudal expression patterns. Dev Genes Evol. 1998;208:283–289. doi: 10.1007/s004270050183. [DOI] [PubMed] [Google Scholar]
- 62.Schroder R. The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature. 2003;422:621–625. doi: 10.1038/nature01536. [DOI] [PubMed] [Google Scholar]
- 63.Copf T, Schroder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc Natl Acad Sci U S A. 2004;101:17711–17715. doi: 10.1073/pnas.0407327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cerny AC, Bucher G, Schroder R, Klingler M. Breakdown of abdominal patterning in the Tribolium Kruppel mutant jaws. Development. 2005;132:5353–5363. doi: 10.1242/dev.02154. [DOI] [PubMed] [Google Scholar]
- 65.Bucher G, Klingler M. Divergent segmentation mechanism in the short germ insect Tribolium revealed by giant expression and function. Development. 2004;131:1729–1740. doi: 10.1242/dev.01073. [DOI] [PubMed] [Google Scholar]
- 66.Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 67.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 68.Eckert C, Aranda M, Wolff C, Tautz D. Separable stripe enhancer elements for the pair-rule gene hairy in the beetle Tribolium. EMBO Rep. 2004;5:638–642. doi: 10.1038/sj.embor.7400148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown SJ, Parrish JK, Beeman RW, Denell RE. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech Dev. 1997;61:165–173. doi: 10.1016/s0925-4773(96)00642-9. [DOI] [PubMed] [Google Scholar]
- 70.Patel NH, Condron BG, Zinn K. Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- 71.Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci U S A. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sommer RJ, Tautz D. Involvement of an orthologue of the Drosophila pair-rule gene hairy in segment formation of the short germ-band embryo of Tribolium (Coleoptera) Nature. 1993;361:448–450. doi: 10.1038/361448a0. [DOI] [PubMed] [Google Scholar]
- 73.Brown SJ, Hilgenfeld RB, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc Natl Acad Sci U S A. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stuart JJ, Brown SJ, Beeman RW, Denell RE. A deficiency of the homeotic complex of the beetle Tribolium. Nature. 1991;350:72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- 75.Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peel AD, Chipman AD, Akam M. Arthropod segmentation: beyond the Drosophila paradigm. Nat Rev Genet. 2005;6:905–916. doi: 10.1038/nrg1724. [DOI] [PubMed] [Google Scholar]
- 77.Lynch J, Desplan C. Evolution of development: beyond bicoid. Curr Biol. 2003;13:R557–559. doi: 10.1016/s0960-9822(03)00472-x. [DOI] [PubMed] [Google Scholar]
- 78.Nasiadka A, Dietrich BH, Krause G. Anterior-posterior patterning in the Drosophila embryo. Advances in Developmental Biology and Biochemistry. 2002;12:156–204. [Google Scholar]
- 79.Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- 80.Sander K. Pattern specification in the insect embryo. Ciba Found Symp. 1975;0:241–263. doi: 10.1002/9780470720110.ch12. [DOI] [PubMed] [Google Scholar]
- 81.Sander K. Specification of the basic body pattern in insect embryogenesis. In: Treherne J, Berridge M, Wigglesworth V, editors. Advances in Insect Physiology. Vol. 12. Academic Press; 1969. pp. 125–235. [Google Scholar]
- 82.Lall S, Ludwig MZ, Patel NH. Nanos plays a conserved role in axial patterning outside of the Diptera. Curr Biol. 2003;13:224–229. doi: 10.1016/s0960-9822(03)00045-9. [DOI] [PubMed] [Google Scholar]
- 83.Counce S. The Causal Analysis of Insect Embryogenesis. In: Counce S, Waddington C, editors. Developmental Systems: Insects. Vol. 2. 1973. pp. 2–156. [Google Scholar]
- 84.Olesnicky EC, Desplan C. Distinct mechanisms for mRNA localization during embryonic axis specification in the wasp Nasonia. Dev Biol. 2007;306:134–142. doi: 10.1016/j.ydbio.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 86.Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 87.Dearden PK. Germ cell development in the Honeybee (Apis mellifera); vasa and nanos expression. BMC Dev Biol. 2006;6:6. doi: 10.1186/1471-213X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev. 2001;11:374–383. doi: 10.1016/s0959-437x(00)00207-0. [DOI] [PubMed] [Google Scholar]
- 89.Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- 90.Gonzalez-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature. 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 91.Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 92.Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 93.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 94.De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 96.Schroder C, Tautz D, Seifert E, Jackle H. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 1988;7:2881–2887. doi: 10.1002/j.1460-2075.1988.tb03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pultz MA, Westendorf L, Gale SD, Hawkins K, Lynch J, Pitt JN, Reeves NL, Yao JC, Small S, Desplan C, Leaf DS. A major role for zygotic hunchback in patterning the Nasonia embryo. Development. 2005;132:3705–3715. doi: 10.1242/dev.01939. [DOI] [PubMed] [Google Scholar]
- 98.Wimmer EA, Carleton A, Harjes P, Turner T, Desplan C. Bicoid-independent formation of thoracic segments in Drosophila. Science. 2000;287:2476–2479. doi: 10.1126/science.287.5462.2476. [DOI] [PubMed] [Google Scholar]
- 99.Patel NH, Hayward DC, Lall S, Pirkl NR, DiPietro D, Ball EE. Grasshopper hunchback expression reveals conserved and novel aspects of axis formation and segmentation. Development. 2001;128:3459–3472. doi: 10.1242/dev.128.18.3459. [DOI] [PubMed] [Google Scholar]
- 100.Panfilio KA, Liu PZ, Akam M, Kaufman TC. Oncopeltus fasciatus zen is essential for serosal tissue function in katatrepsis. Dev Biol. 2006;292:226–243. doi: 10.1016/j.ydbio.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 101.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 103.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 104.Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 105.Furriols M, Ventura G, Casanova J. Two distinct but convergent groups of cells trigger Torso receptor tyrosine kinase activation by independently expressing torso-like. Proc Natl Acad Sci U S A. 2007;104:11660–11665. doi: 10.1073/pnas.0700991104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schaeffer V, Killian D, Desplan C, Wimmer EA. High bicoid levels render the terminal system dispensable for Drosophila head development. Development. 2000;127:3993–3999. doi: 10.1242/dev.127.18.3993. [DOI] [PubMed] [Google Scholar]
- 107.Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]


