Abstract
Herpes simplex virus type 1 (HSV1) capsids undergo extensive structural changes during maturation and DNA packaging. As a result, they become more stable and competent for nuclear egress. To further elucidate this stabilization process, we used biochemical and nanoindentation approaches to analyze the structural and mechanical properties of scaffold-containing (B), empty (A), and DNA-containing (C) nuclear capsids. Atomic force microscopy experiments revealed that A and C capsids were mechanically indistinguishable, indicating that the presence of DNA does not account for changes in mechanical properties during capsid maturation. Despite having the same rigidity, the scaffold-containing B capsids broke at significantly lower forces than A and C capsids. An extraction of pentons with guanidine hydrochloride (GuHCl) increased the flexibility of all capsids. Surprisingly, the breaking forces of the modified A and C capsids dropped to similar values as those of the GuHCl-treated B capsids, indicating that mechanical reinforcement occurs at the vertices. Nonetheless, it also showed that HSV1 capsids possess a remarkable structural integrity that was preserved after removal of pentons. We suggest that HSV1 capsids are stabilized after removal of the scaffold proteins, and that this stabilization is triggered by the packaging of DNA, but independent of the actual presence of DNA.
Keywords: atomic force microscopy, nanoindentation, penton, viral structure, virus mechanics
Herpes simplex virus type 1 (HSV1) virions encapsidate their 152 kbp double-stranded DNA genome in an icosahedral capsid that is surrounded by an amorphous protein layer, called the tegument, and a lipid-containing envelope. Assembly of herpesviruses is initiated in the nucleus where procapsids self-assemble around a protein scaffold and subsequently mature (1–4). This transformation is characterized by massive conformational changes of the ≈200 MDa shell, resulting in stable, mature capsids, and is in many aspects analogous to maturation processes in bacteriophages (5, 6). The scaffold is proteolytically cleaved and removed, whereas the outer shell transforms from a spherical into an icosahedral shape (7, 8). The mature capsid has an outer diameter of 125 nm with an overall shell thickness of ≈15 nm (9). Capsomeres converge at their proximal ends, forming a contiguous shell of ≈4 nm thickness that is only interrupted by channels passing through all of the 162 capsomeres (10, 11). The capsomeres are hexamers (hexons) and pentamers (pentons) of the major capsid protein VP5 that form a shell with a triangulation number of T = 16. In total, 320 triplexes, each formed by a heterotrimer of 1 VP19c and 2 VP23 molecules, connect these capsomeres (10). One of the 12 capsid vertices is occupied by the pUL6 portal (12). Three particle types can be isolated from the nuclei of infected cells due to their different sedimentation behavior: B capsids, which still have the scaffold inside; the lighter A capsids, which are empty; and the denser C capsids, which contain the DNA genome (13, 14). B and A capsids are considered to be defective particles: B capsids failed to properly initiate (3), and A capsids were unsuccessful in completing (15) DNA packaging (1, 4).
B, A, and C capsids all have mature angularized shells, but biochemical and electron microscopy experiments show that there are differences in their protein composition and their morphology (8, 16). However, it is unclear whether they also differ in terms of mechanical properties (16, 17). Recently developed nanoindentation techniques using atomic force microscopy (AFM) allow for the analysis of mechanical properties of viruses at the single particle level (18–20). Such studies show that DNA packaging increases the mechanical strength of capsids of the minute virus of mice (21) and the bacteriophage λ (22). Studies on the maturation of murine leukemia virus and HIV have revealed a mechanical switch that is linked to viral infectivity (23, 24). Furthermore, it was shown that capsid mutations can alter the mechanical strength of cowpea chlorotic mottle virus and minute virus of mice (25, 26). Nanoindentation techniques have also been used to analyze the morphology of different capsids of one virus (e.g., the T = 3 and T = 4 hepatitis B capsids), which were shown to have mechanically similar shells (27).
Here we studied the mechanical properties of scaffold-containing B, empty A, and DNA-filled C capsids of HSV1. Compared with B, the latter 2 capsid types showed identical increased capsid stability. However, removal of capsid pentons with 2.0 M GuHCl from both empty and DNA-filled capsids reduced their mechanical stability to that of B capsids. Our data suggest that HSV1 capsids are stabilized at their vertices upon removal of the internal scaffold proteins and the initiation of DNA packaging. The structural rearrangements responsible for this capsid enforcement do not require maintaining the DNA genome within the capsids, and they can be reversed by removing the pentons with GuHCl.
Results
To study the mechanical and structural features of HSV1, we isolated B, A, and C capsid fractions from the nuclei of HSV1-infected cells. Following isolation, their morphology and purity were analyzed by electron microscopy after negative contrasting (Fig. 1 and Table 1). The scaffold-containing B capsids were characterized by prominent capsomere morphology on their surface, but displayed no additional internal contrast features (Fig. 1A). The majority of the capsids in the A fraction displayed a dark lumen, indicating that the contrasting agent uranyl acetate had penetrated and filled the empty hollow capsid shells (Fig. 1B). The DNA-filled C capsids contained some internal uranyl acetate as indicated by a darker internal rim that excluded a light lumen (Fig. 1C). This suggests that the contrasting agent had penetrated the capsid shell, but could not completely fill it due to the presence of DNA. The C fractions contained 70× to 150× higher amounts of viral genomes than B or A fractions, as detected by quantitative real time-PCR (see Materials and Methods). Furthermore, immunoblot analysis showed similar amounts of the major capsid proteins VP5, VP19c, and VP26 in the different capsid populations (Fig. 2). The B capsid fraction contained in addition large amounts of the scaffold proteins VP21 and VP22a, whereas the A and C capsid fraction had only traces. The minor capsid-associated proteins pUL17 and pUL25 were more abundant in the C capsid fraction, and roughly equal in B and A capsid fractions.
Fig. 1.
Electron microscopy images of negatively contrasted HSV1 nuclear capsids: (A) a scaffold-containing B capsid, (B) an empty A capsid, (C) a DNA-containing C capsid, and (D) a penton-free B, (E) a penton-free A, and (F) a penton-free C capsid. The capsids shown in D–F had been treated with 2.0 M GuHCl. As a result of the extraction of the pentons, these particles were empty and had been filled with uranyl acetate. Depending on the exact focal plane at which the image had been taken, one to several holes in the capsid wall were visible (arrows in D). (Scale bar: 50 nm.)
Table 1.
Characterization of nuclear capsid fractions
| Fraction* | B (n = 1,073) | A (n = 969) | C (n = 715) |
|---|---|---|---|
| B-like | 82% | 2% | 16% |
| A-like | 10% | 73% | 8% |
| C-like | 8% | 14% | 71% |
*A small percentage of the capsids could not be classified unequivocally in the A and C fractions.
Fig. 2.
Protein composition of HSV1 B, A, and C capsids. B, A, and C capsid fractions before (−) and after (+) treatment with 2.0 M GuHCl were analyzed by immunoblot. More GuHCl-treated than -untreated capsids were loaded to demonstrate complete extraction of the proteins VP26, pUL17, pUL25, and the scaffold VP21 and VP22a.
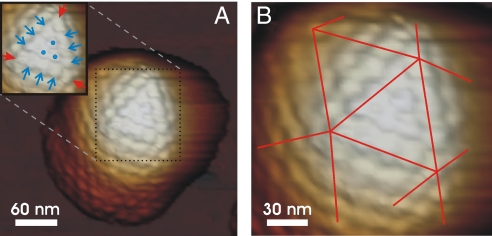
An essential prerequisite for performing nanoindentation experiments was imaging the capsids at high spatial resolution to determine a particle's center. Our atomic force microscopy (AFM) images revealed the 2 different types of capsomeres, hexons and pentons (Fig. 3A), and the triangular faces of the icosahedral capsids (Fig. 3B). Furthermore, we determined the capsid height from these images as 123.6 ± 0.3 nm (n = 37), 123.1 ± 0.5 nm (n = 51), or 122.6 ± 0.3 nm (n = 35) for B, A, or C capsids, respectively. After high-resolution imaging of a capsid, the cantilever tip was directed at its center, and the particle was indented by pushing the tip into the capsid. Fig. 4 shows a particle before (Panel A) and after indentation (Panel B), and the corresponding height profile displayed a hole in the middle of the particle after indentation (Panel C). Zooming in on the capsid center, one can distinguish the individual capsomeres (Fig. 4 D and E). By numbering the capsomeres, we identified those that had been displaced by indentation. Typically, multiple hexons were displaced or removed while the adjacent capsomeres were still in place. It was thus possible to purge adjacent hexons without disrupting the capsid integrity. This showed the robustness of the HSV1 overall capsid structure.
Fig. 3.
AFM images revealing the capsomere organization of icosahedral HSV1 capsids. (A) Image of a B capsid with one triangular face highlighted in the inset. The closed (red) arrows point at the pentons and the open (blue) arrows point at the hexons on the 2-fold axes. The dots mark the 3 central hexons in the middle of the triangular face. (B) Zoom-in on the same particle. The lines mark the borders of the triangular faces of the icosahedral capsid.
Fig. 4.
Capsomere displacement as a result of capsid indentation. (A–C) Image of a B capsid before (A) and after (B) indentation with corresponding height profiles (C). The height profiles were taken along the arrows in A and B. Indentation was performed with a force of 4.0 nN. The upper part of the “Before indentation” curve in C shows the regular pattern of the capsomeres. (D and E) Zoom-in on the central parts of respectively (A and B), with additional numbering of capsomeres. After indentation (E), the hexons with the numbers 8, 9, 12, 13, 14, 17, and 18 were displaced.
The capsids' material properties were determined by analyzing the force indentation (FZ) curves obtained during the indentation experiments (FZ in Fig. 5). The particle spring constant k, indicating the capsids' flexibility, was obtained from the linear part of the FZ curve, taking into account the cantilever's own spring constant (18). Usually, a drop in the force occurred at the end of linearity (Fig. 5A, main panel), but occasionally this occurred at a force which was slightly higher (Fig. 5A, Inset). A drop in the force in the FZ curve is a signature of capsid breakage.
Fig. 5.
Force-indentation (FZ) curves showing the force response of HSV1 capsids upon deformation. To calibrate the system, the reference curve FZ glass was recorded by bending the cantilever on the glass. The indentation curve on the capsid is shown by FZ forward, and the FZ backward curve depicts the retraction of the cantilever. The hysteresis between both curves shows the irreversibility of indentation after breaking. These large-scale disruptions appear permanent because waiting for several hours did not result in reversal to the initial mechanical properties of the capsids. The FZ glass and FZ virus curves were shifted along the x axis to have a coinciding contact point. The experiments were performed on B, A, and C capsids separately and at loading rates of ≈3 nN/sec. (A) Indentation of a penton-containing A capsid showing a clear drop of the force within the linear indentation part. FZ 2 and FZ 3 show the strongly increased flexibility of the particle for additional pushing cycles, indicating that the particle has been broken. (Inset) Indentation of a penton-containing A capsid showing the sometimes-occurring difference between the end of linearity and the breaking of the particle. Whereas the particle in the main panel shows 2 clear breaking events, the particle in the inset shows only one clear breaking event. (B) Indentation of a pentonless B capsid extracted with 2.0 M GuHCl. A qualitatively similar indentation behavior was observed for pentonless and intact capsids.
The mechanical properties of A and C capsids were indistinguishable (Table 2). They had the same spring constant and broke at a comparable force Fbreak of ≈5.7 nN and an indentation of ≈17%. Thus, surprisingly, the presence of the DNA was not noticed while indenting C capsids, despite its dense packing (9). This finding can be related to experiments on phage λ where the mechanical properties of wild-type capsids were compared with that of shorter genome mutants (22). Using a simple geometric formula derived in ref. 28, we can calculate the relative DNA packing density, ρpack, inside HSV1 by ρpack = 0.34 π Nbp/Vcapsid. Here, Nbp denotes the number of base pairs (bp) and Vcapsid the inner volume of the capsid. Assuming a spherical capsid shape with an inner radius of 47.5 nm for HSV1 (11) and a genome length of 152 kbp, we calculated 0.36 to be the relative DNA packing density of HSV1 C capsids. This value is significantly below the packing density of phage λ capsids (ρpack = 0.56; calculated with an inner radius of 27.5 nm) (29), at which the packaged λ DNA became noticeable (22). These findings agree with our data that the presence or absence of the DNA influenced neither the spring constant nor the breaking force.
Table 2.
Mechanical properties of HSV1 nuclear capsids
| Capsid type | Fbreak, nN | k, N/m | Relative indentation* | n |
|---|---|---|---|---|
| B | 3.7 ± 0.2 | 0.33 ± 0.02 | 0.12 ± 0.01 | 37 |
| A | 5.6 ± 0.2 | 0.33 ± 0.02 | 0.18 ± 0.01 | 51 |
| C | 5.8 ± 0.2 | 0.33 ± 0.02 | 0.16 ± 0.01 | 35 |
| B + 2 M GuHCl | 3.1 ± 0.2 | 0.20 ± 0.01 | 0.18 ± 0.02 | 29 |
| A + 2 M GuHCl | 2.4 ± 0.2 | 0.15 ± 0.01 | 0.16 ± 0.02 | 35 |
| C + 2 M GuHCl | 3.6 ± 0.3 | 0.21 ± 0.01 | 0.16 ± 0.01 | 39 |
*Expressed in units of diameter.
B capsids had the same spring constant as A or C capsids, but they broke at much lower forces (Table 2). These findings were surprising, as we did not expect the empty A capsids to be more sturdy than the protein-filled B capsids. Hence, a structural change that strengthens the capsid must take place while the scaffold material leaves the capsids during maturation.
To study the material properties of B, A, and C capsids in more detail, we extracted the pentons by incubating capsids with 2.0 M GuHCl (10, 30). This GuHCl concentration also removes the scaffold proteins from B capsids, the DNA from C capsids, and the triplexes adjacent to the pentons, but not other triplexes (10). We could thus investigate the influence of the vertices on the mechanical characteristics and stability of the icosahedral capsids, and whether DNA packaging into HSV1 capsids lead to capsid reinforcement near the pentons (8, 16).
In electron microscopy (Fig. 1 D–F) and AFM (Fig. 6) images of GuHCl-treated capsids, the holes, where the pentons were lacking, are clearly visible. All GuHCl-treated capsids had a dark lumen after negative contrasting and imaging by electron microscopy, confirming that they had been emptied. Immunoblot analysis confirmed the extraction of the scaffold proteins VP21 and VP22a from B capsids (Fig. 2). Furthermore, VP26, pUL17, and pUL25 were removed from all 3 capsid types.
Fig. 6.
AFM images of a pentonless particle. (A) Top view of a pentonless B capsid with arrows pointing to 3 holes at the position of the missing pentons. Three-dimensional rendered image with shadowing. (B and C) Zoom-in on the central part of the same particle after contrast enhancement. The vertices where the pentons are missing are denoted with a red 5, the peripentonal hexons with P, the edge hexons with E, and the central hexons with C (2, 31). (D) Height profiles of the same image along the 2-fold axes. Curve I shows the total profile, and curve II (Lower Inset) a zoom-in on the top part of this particle. The profiles clearly reveal the P and E hexons as well as the holes where pentons are missing. The lateral distance between the hexons corresponds to ≈16–17 nm, and is consistent with data obtained from cryo-EM reconstructions (2).
The FZ curves display a qualitatively comparable behavior for pentonless capsids as for penton-containing capsids (Fig. 5). The spring constant of all 3 capsid types decreased from 0.33 N/m to ≈0.19 N/m (Table 2). The holes at the penton positions most likely facilitated hexon rearrangements during capsid deformation, leading to a higher flexibility of the capsid shell. The breaking force of the pentonless B capsids was nearly unchanged.* However, that of the penton-free A and C capsids decreased from ≈6 nN to ≈3 nN, and thus approached the breaking force of B capsids.† Therefore, without the pentons, all 3 particle types had similar mechanical properties, and the increased strength of A and C capsids, as compared with B capsids, had its origin at or near the pentons.
Discussion
We have shown that the A and C capsids of HSV1 have similar mechanical characteristics, whereas B capsids break at lower forces. The present data are a strong indication that removing the internal protein scaffold in combination with the packaging process, which most likely occur concomitantly, induces conformational changes in the nuclear capsid. The removal of the scaffold might be linked to a potential local pH reduction as a result of the incoming negative-charged DNA during packaging, as proposed by McClelland et al. (32). Furthermore, stabilizing proteins, which increase the mechanical strength of the capsids, are possibly recruited during this process. A capsids are particles that most likely have initiated packaging, and which have undergone this structural modification to a degree that they became mechanically indistinguishable from C capsids in our AFM experiments. However, A capsids fail to complete packaging and are empty (1, 15). The increased stability of A and C capsids probably occurred through changes near the vertices, as the removal of the pentons by 2.0 M GuHCl made them virtually indistinguishable from B capsids.
Trus et al. (8) reported an electron density, termed CCSC for C-capsid-specific component, indicating (despite its name) the presence of a protein complex near the pentons of A and C capsids. They proposed that an additional maturation step, resulting from DNA packaging, may trigger this complex formation. A capsids contain 7–8 CCSCs per capsid, and this number increases to 27–33 on C capsids. This complex is completely absent on A capsids derived from a HSV1 mutant lacking pUL25, suggesting that the CCSC may be a heterodimer of pUL25 and pUL17. These minor capsid-associated proteins are required for DNA packaging. pUL25 seems to be necessary for stable packaging or retention of DNA (33–35), and in the absence of pUL17, the concatemeric DNA is neither cleaved nor packaged (36, 37). As the pUL17/pUL25 complex is located near the capsid vertices where it spans over the 2 penton-adjacent triplexes, it is also extracted by treatment with GuHCl (8).
Our data, in conjunction with that of Trus et al. (8), indicate that the increased strength of A and C capsids may be initiated, accompanied, or followed by the formation of this CCSC in close proximity to the vertices. Biochemical analyses detected similar concentrations of pUL25 and pUL17 on A and B capsids (Fig. 2) (8, 16, 38). Thus, there may be 2 conformations of pUL25 and pUL17 on the capsids: the CCSC near the vertices and a structurally less-ordered population scattered around other quasi-equivalent sites (8). In addition to extracting pentons and penton-adjacent proteins, the 2.0 M GuHCl treatment also removes VP26, the smallest capsid protein that covers the hexons, but not the pentons (30, 39). Biochemical experiments revealed that B capsids contain slightly less VP26 than A or C capsids (40, 41). However, in cryoelectron microscopy studies, VP26 is already present on all hexamers in B capsids (11, 39). Therefore, it is most likely not responsible for the stability differences of nuclear HSV1 capsids.
Using a continuum mechanics approach to thin shells we can estimate the capsids elastic (or Young's) modulus E by
with the particle radius R, the shell thickness h, and a proportionality factor α (18, 42). For α, a value of 1 is taken, a reasonable approximation for various capsids (18, 25, 43). Even though it may be argued whether HSV1 capsids possess a thin shell (h/R ≪ 1), this equation has also been successfully applied to thicker shells, such as that of cowpea chlorotic mottle virus (25). Calculating the shells Young's modulus with h = 4 nm as the contiguous part of the shell and R = 49.5 nm as the average radius of this contiguous shell (11) yields E ≈ 1.0 GPa for HSV1 capsids. This approximate value, which could for instance be refined by finite element simulations on detailed capsid models (44), is comparable to the Young's modulus of phage λ capsids (1.0 GPa) (22). However, as a result of the thin shell of the phage λ particles of ≈1.8 nm, the empty phage λ capsids have a lower spring constant (k = 0.13 N/m) than HSV1 nuclear capsids (k = 0.33 N/m). As the spring constant is proportional to the square of the particle shell thickness (see Eq. 1), this would be expected. The relative deformation at breakage of phage λ capsids, which is ≈10% to 12.5% of their diameter, is comparable to that of B capsids, which broke at 12%. These measurements were performed with roughly comparable loading rates (22). Interestingly, A and C capsids broke at a higher indentation, namely around 17%. This shows that the mechanical properties of B capsids are similar to that of phage λ particles, but that A and C capsids have undergone stabilizing modifications that result in increased capsid strength.
Our data also reveal a role of the vertex proteins for the mechanical and structural properties of viral icosahedrons. Because removing the pentons resulted in a dramatic decrease of the spring constant for B, A, and C capsids, this corroborates the importance of the vertex capsomeres in maintaining the icosahedrons' rigidity. This decrease in rigidity likely resulted from the holes in the capsid shell, which alter the stress redistribution occurring during particle deformation. Notably, the spring constant of B capsids decreased from 0.33 N/m to 0.20 N/m, whereas their breaking force was only slightly reduced upon penton removal. Hence, the breaking force and capsid flexibility were decoupled in the B capsids of HSV1.
Liashkovich et al. (17) recently reported a spring constant of HSV1 capsids of ≈0.5 N/m that is substantially higher than our spring constant of 0.33 ± 0.02 N/m. This apparent discrepancy might have been due to different buffers. However, control experiments on C capsids at 100 mM NaCl yielded a spring constant of 0.29 ± 0.03 N/m (n = 12), which was not significantly different from our other measurements at 500 mM NaCl. Thus, a 5-fold difference in the concentration of monovalent salts did not influence the capsid rigidity in our system. Whereas we studied the mechanical properties of nuclear capsids, Liashkovich et al. (17) analyzed viral capsids that had been isolated from viral particles secreted from infected cells, and thus most likely contained significant amounts of tegument attached to them (45, 46). The tegument may well increase the particle spring constant, as the latter is proportional to the square of the shell thickness (see Eq. 1). If one assumes that the tegument has a comparable Young's modulus to the capsid, an increase by only 20% in average shell thickness would be sufficient to explain the increased spring constant of tegumented capsids. With a contiguous HSV1 capsid shell thickness of ≈4 nm, the tegument would have to add an additional layer of only ≈0.8 nm to the capsids.
This reasoning is consistent with experiments on surface-attached tegumented capsids that had been treated with 1 M GuHCl (17), which extracts the DNA from C capsids but leaves the pentons in place (47). Liashkovich et al. (17) assumed that the decreased spring constant of viral capsids treated with 1 M GuHCl was due to the loss of the internal DNA, and thus suggested that the DNA of filled capsids is a major contributor to the capsids' mechanical properties. However, treating tegumented capsids with 1 M GuHCl not only removed the DNA but also solubilized tegument proteins (data not shown). This reduction in tegument may result in a thinner shell, and hence capsid weakening. The GuHCl treatment reduces the spring constant of tegumented capsids to 0.36 ± 0.2 N/m (17). This value is not significantly different from the spring constant that we measured for the untegumented, nuclear capsids (Table 2). Thus, the presence or absence of tegument proteins may be responsible for the higher stiffness of viral capsids in comparison with nuclear capsids.
Our data show that capsids that have undergone or initiated DNA packaging are more stable than capsids that have not, indicating that the replacement of the internal protein scaffold by the genomic DNA triggers capsid reinforcement during HSV1 maturation. This capsid stabilization resulted in an increase of the capsid strength, but not in capsid stiffening. Major structural rearrangements seem to occur at the pentons that may be further stabilized and reinforced by the CCSC, which is most likely a heterodimer of pUL17 and pUL25. Further AFM indentation experiments using capsids of HSV1 mutants lacking pUL25 or pUL17 (33, 36) in combination with capsids treated with GuHCl, urea, and trypsin (8, 16, 38) may reveal the contribution of these proteins to capsid stability and mechanical reinforcement.
Materials and Methods
Preparation of Nuclear B, A, and C Capsids.
BHK-21 cells were infected with 0.01 plaque-forming units per cell of HSV1 strain F (ATCC VR-733) for 2–3 days until the cells had detached from the culture flasks. Infected cells were collected, washed once with MNT buffer (30 mM MES, 100 mM NaCl, 20 mM Tris [pH 7.4]), snap frozen, and stored in single-use aliquots at −80 °C. Nuclear capsids were prepared from these cells as described previously (13, 14, 30, 46, 48). The 3 light-scattering bands corresponding to B, A, and C nuclear capsid fractions were harvested from linear 20%–50% (wt/wt) sucrose gradients and analyzed by electron microscopy after negative contrasting.
GuHCl or Urea Extraction of Capsids.
Pentonless capsids were generated from nuclear B, A, or C fractions as described (10, 30). Capsids were treated with 2.0 M GuHCl or 6.0 M urea for 30 min at 4 °C or 1 h at RT, respectively. Pentonless A and C capsids were resuspended in 1× DNase I buffer (Promega) with 10 mM DTT and protease inhibitors and treated with 0.1 U/μL DNase I (M6101; Promega) for 30 min at 37 °C. The capsids were snap frozen in liquid nitrogen after addition of 1/5 volume of 5× Tris NaCl-EDTA (TNE) buffer (2.5 M NaCl, 5 mM EDTA, 100 mM Tris-HCl [pH 7.5]).
Electron Microscopy.
After adsorption onto carbon and Formvar-film coated 400 mesh copper grids (Stork Veco), the samples were washed with PBS and distilled water before negative contrasting at pH 4.4 with 2% uranyl acetate (Merck) in distilled water. Capsids were analyzed with an EM10CR transmission electron microscope (Carl Zeiss AG) at 80 kV.
Real-Time PCR.
The DNA concentration of the HSV1 capsid preparations was quantified by real-time PCR as described (49, 50). B and A capsid fractions contained few genomes (3.1 × 109 ± 2.2 × 109 or 1.4 × 109 ± 1.0 × 109 genomes/fraction respectively), whereas C capsid fractions contained a higher concentration (2.2 × 1011 ± 8.8 × 1010 genomes/fraction). These data corresponded to the concentration of C capsids that we had estimated in the B and A fractions by electron microscopy. The values are mean values of 3 independent capsid preparations analyzed in 1 or 2 independent PCR experiments (error: SEM).
SDS-PAGE and Immunoblot.
Protein samples were solubilized in sample buffer (125 mM Tris-HCl [pH 6.8] with 4% wt/vol SDS, 20% vol/vol glycerol, 120 mM ß-mercaptoethanol, bromophenol blue), heated to 95 °C and separated on large linear 5%–15% polyacrylamide gradient gels (51). After transfer onto nitrocellulose membrane (Pall Corporation), proteins were detected using specific primary antibodies (anti-VP5 NC1, anti-VP19c NC2, anti-VP22a/VP21 NC3/4 [52], anti-VP26 [53], anti-pUL17 203 [54], anti-pUL25 ID1 [55]) and secondary antibodies coupled to horseradish peroxidase (Pierce, Perbio Science) for ECL detection (SuperSignal West Femto Maximum Sensitivity Substrate; Pierce, Perbio Science).
Atomic Force Microscopy.
Glass coverslips were cleaned in a KOH/ethanol bath and treated with hexamethyldisilazane to render them hydrophobic, as before (18). HSV1 capsids were diluted in TNE buffer to a total volume of 100 μL and pipetted onto a hydrophobic glass coverslip. After an incubation of 20 min, 100 μL of TNE buffer was added and the nanoindentation measurements were started. A Nanotec Electronica AFM operated in jumping mode was used, and all experiments were performed at room temperature in liquid. Olympus OMCL-RC800PSA rectangular silicon nitride cantilevers, with a nominal spring constant of 0.05 N/m, were used. The cantilevers were calibrated by the method of Sader et al. (56) and had an averaged spring constant of 0.051 ± 0.002 (SD) N/m. Throughout this text, when the standard deviation is used as error, this is explicitly noted with SD. All other errors are SEM.
Acknowledgments.
We thank Katinka Döhner and Sabine Hübner (Institute of Virology, Hannover Medical School) for performing real-time PCRs. We are grateful to Jutta Milzer (Institute of Virology, Hannover Medical School) for providing electron microscopy reagents, and to Ernst Ungewickell (Institute of Cell Biology, Hannover Medical School) for granting unlimited access to the electron microscope facility. We thank Gary Cohen and Roselyn Eisenberg (University of Pennsylvania), Prashant Desai (Johns Hopkins University), Daniel Tenney (Bristol-Myers Squibb Pharmaceutical Research Institute), and Valerie Preston (Medical Research Council, Glasgow, Scotland) for generously providing antibodies. This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek grants CW-ECHO (to G.J.L.W.) and Rubicon (to W.H.R.), by Deutsche Forschungsgemeinschaft Grant DFG So403/3 (to B.S.), and by DFG Excellence Cluster REBIRTH (“From Regenerative Biology to Reconstructive Therapy”) funding (to B.S.). K.R. received a Ph.D. fellowship from the Hannover Biomedical Research School (Center of Infection Biology, Hannover Medical School).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
We performed control experiments of penton removal by extracting B capsids with 6.0 M urea instead of 2.0 M GuHCl (10). These particles had identical mechanical properties: k = 0.20 ± 0.02 N/m and Fbreak = 2.6 ± 0.3 nN (n = 21).
The GuHCl-treated A were slightly weaker than the B and C capsids, which could be due to incomplete removal of pentons, as there was a slight variation in the number of removed pentons. Nevertheless, our EM and AFM analyses showed that the majority of the pentons was removed.
References
- 1.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Heymann JB, et al. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 3.Steven AC, Heymann JB, Cheng NQ, Trus BL, Conway JF. Virus maturation: Dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines JS, Weller SK. In: Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Catalano CE, editor. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 135–150. [Google Scholar]
- 5.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Newcomb WW, et al. Assembly of the herpes simplex virus capsid: Characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 8.Trus BL, et al. Allosteric signaling and a nuclear exit strategy: Binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booy FP, et al. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes-simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newcomb WW, et al. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 11.Booy FP, et al. Finding a needle in a haystack: Detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newcomb WW, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdue ML, Cohen JC, Kemp MC, Randall CC, Ocallaghan DJ. Characterization of three species of nucleocapsids of equine herpesvirus type-1 (Ehv-1) Virology. 1975;64:187–204. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 14.Perdue ML, Cohen JC, Randall CC, Ocallaghan DJ. Biochemical studies of maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 15.Sherman G, Bachenheimer SL. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 16.Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liashkovich I, et al. Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J Cell Sci. 2008;121:2287–2292. doi: 10.1242/jcs.032284. [DOI] [PubMed] [Google Scholar]
- 18.Ivanovska IL, et al. Bacteriophage capsids: Tough nanoshells with complex elastic properties. Proc Natl Acad Sci USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJL. Viral capsids: Mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci. 2007;64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roos WH, Wuite GJL. Nanoindentation studies reveal material properties of viruses. Adv Mater. 2009;21:1187–1192. [Google Scholar]
- 21.Carrasco C, et al. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci USA. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanovska I, Wuite G, Jonsson B, Evilevitch A. Internal DNA pressure modifies stability of WT phage. Proc Natl Acad Sci USA. 2007;104:9603–9608. doi: 10.1073/pnas.0703166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kol N, et al. Mechanical properties of murine leukemia virus particles: Effect of maturation. Biophys J. 2006;91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kol N, et al. A stiffness switch in human immunodeficiency virus. Biophys J. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel JP, et al. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrasco C, Castellanos M, de Pablo PJ, Mateu MG. Manipulation of the mechanical properties of a virus by protein engineering. Proc Natl Acad Sci USA. 2008;105:4150–4155. doi: 10.1073/pnas.0708017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uetrecht C, et al. High-resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Natl Acad Sci USA. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purohit PK, et al. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- 30.Newcomb WW, Brown JC. Structure of the herpes-simplex virus capsid: Effects of extraction with guanidine-hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steven AC, et al. Hexavalent capsomers of herpes simplex virus type 2: Symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClelland DA, et al. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J Virol. 2002;76:7407–7417. doi: 10.1128/JVI.76.15.7407-7417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNab AR, et al. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stow ND. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J Virol. 2001;75:10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn J, et al. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J Virol. 2008;82:5725–5734. doi: 10.1128/JVI.02441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klupp BG, Granzow H, Karger A, Mettenleiter TC. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J Virol. 2005;79:13442–13453. doi: 10.1128/JVI.79.21.13442-13453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou ZH, et al. Assembly of Vp26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 40.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNabb DS, Courtney RJ. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1992;66:2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau LD, Lifshitz EM. Theory of Elasticity. Oxford: Elsevier; 1986. [Google Scholar]
- 43.Gibbons MM, Klug WS. Nonlinear finite-element analysis of nanoindentation of viral capsids. Phys Rev E. 2007;75 doi: 10.1103/PhysRevE.75.031901. 031901. [DOI] [PubMed] [Google Scholar]
- 44.Gibbons MM, Klug WS. Influence of nonuniform geometry on nanoindentation of viral capsids. Biophys J. 2008;95:3640–3649. doi: 10.1529/biophysj.108.136176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: Reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfstein A, et al. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 47.Newcomb WW, Brown JC. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology. 2007;361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Döhner K, Radtke K, Schmidt S, Sodeik B. Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol. 2006;80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelmann I, et al. Rapid quantitative PCR assays for the simultaneous detection of herpes simplex virus, varicella zoster virus, cytomegalovirus, Epstein-Barr virus, and human herpesvirus 6 DNA in blood and other clinical specimens. J Med Virol. 2008;80:467–477. doi: 10.1002/jmv.21095. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Cohen GH, et al. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai P, DeLuca NA, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 54.Thurlow JK, et al. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol. 2005;79:150–158. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koslowski KM, Shaver PR, Wang XY, Tenney DJ, Pederson NE. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sader JE, Chon JWM, Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev Sci Instrum. 1999;70:3967–3969. [Google Scholar]