Abstract
In fish, amino acids are food-related important olfactory cues to elicit an attractive response. However, the neural circuit underlying this olfactory behavior is not fully elucidated. In the present study, we applied the Tol2 transposon-mediated gene trap method to dissect the zebrafish olfactory system genetically. Four zebrafish lines (SAGFF27A, SAGFF91B, SAGFF179A, and SAGFF228C) were established in which the modified transcription activator Gal4FF was expressed in distinct subsets of olfactory sensory neurons (OSNs). The OSNs in individual lines projected axons to partially overlapping but mostly different glomeruli in the olfactory bulb (OB). In SAGFF27A, Gal4FF was expressed predominantly in microvillous OSNs innervating the lateral glomerular cluster that corresponded to the amino acid-responsive region in the OB. To clarify the olfactory neural pathway mediating the feeding behavior, we genetically expressed tetanus neurotoxin in the Gal4FF lines to block synaptic transmission in distinct populations of glomeruli and examined their behavioral response to amino acids. The attractive response to amino acids was abolished only in SAGFF27A fish carrying the tetanus neurotoxin transgene. These findings clearly demonstrate the functional significance of the microvillous OSNs innervating the lateral glomerular cluster in the amino acid-mediated feeding behavior of zebrafish. Thus, the integrated approach combining genetic, neuroanatomical, and behavioral methods enables us to elucidate the neural circuit mechanism underlying various olfactory behaviors in adult zebrafish.
Keywords: feeding behavior, Gal4/UAS, neural transmission blockade, olfaction
How are various odors represented in the brain and then transformed into specific behavioral outputs? Animals detect a huge variety of odorants in the environment; transmit the information to the brain; and convert it into appropriate output responses, including odor perception and discrimination, emotional change, hormonal release control, and behavioral expression. The olfactory system is equipped with highly organized neural circuits from the olfactory epithelium (OE) to the olfactory bulb (OB). For example, in mice, individual olfactory sensory neurons (OSNs) express only a single type of odorant receptor (OR) gene from a repertoire of ≈1,000 (“one neuron–one receptor rule”) (1–4). The OSNs expressing a given OR converge their axons onto a few specific glomeruli in the OB (“axon convergence to target glomeruli”), which is enabled by the hierarchical and combinatorial actions of multiple axon guidance molecules (5–7). Based on these 2 principles, topographical odor maps are established on the glomerular array of the OB, which depict the internal representations of odor stimuli in the brain (8, 9). However, it is largely unknown how the odor maps on the OB are integrated and transmitted to higher brain centers to elicit various olfactory behaviors.
Fish detect a variety of water-soluble odorants, which evoke different types of fundamental behaviors, such as food finding, alarm response, predator avoidance, social communication, and reproductive activity. The fish OE contains 3 types of morphologically distinct sensory neurons: ciliated OSNs, microvillous OSNs, and crypt cells. Each type of those neurons is supposed to express different classes of chemosensory receptors and signal transduction molecules, project axons to distinct regions of the OB, and mediate different physiological responses (10–12). The microvillous OSNs expressing V2R-type olfactory receptors and transient receptor potential channel C2 innervate the lateral chain glomeruli that respond to amino acids, which are potential feeding cues (11, 13–16). The ciliated OSNs expressing OR-type olfactory receptors, GTP-binding protein Gαolf/s, cyclic nucleotide-gated channel A2 subunit, and olfactory marker protein (OMP) predominantly target the anteromedial glomeruli that respond to bile acids, putative social pheromones (11, 13, 14, 16–18). In contrast, little is known about the molecular constituents, axonal projection, and function of the crypt cells (11, 12, 19, 20), despite their unique properties such as seasonal variability (21). We previously demonstrated that the 2 basic principles, the one neuron–one receptor rule and axon convergence to target glomeruli, are essentially conserved also in the zebrafish olfactory system (22). Although this conservation renders the zebrafish an excellent animal model to analyze the olfactory system (23), it has been difficult to correlate various olfactory behaviors with the OSN types, glomerular identities, and neural circuits.
In addition to general advantages (e.g., external fertilization, large clutch size, rapid development, optical transparency of larvae), the zebrafish is amenable to various techniques of both forward and reverse genetics. Recently, we developed a method for targeted gene expression in zebrafish by combining the Tol2 transposable element with the Gal4/UAS system (24, 25). With this method, a number of fish lines that express the modified yeast transcription activator Gal4FF in specific tissues and cells are created by gene and enhancer trapping. By crossing with effector lines that contain genes of interest downstream of the Gal4 recognition sequence UAS, selective visualization and manipulation of the Gal4FF-expressing cells can be achieved.
Here, we analyzed the zebrafish olfactory circuits responsible for attraction to amino acids, taking advantage of the Tol2-mediated Gal4 gene trap method. Initially, we created transgenic fish lines that expressed Gal4FF in the OE. Then, we performed a detailed anatomical analysis and identified fish lines that expressed Gal4FF in specific subpopulations of OSNs. Finally, we crossed these fish with the UAS:TeTxLC effector fish carrying a gene for tetanus neurotoxin light chain (TeTxLC), which blocks synaptic transmission, and analyzed behavioral phenotypes of the double-transgenic fish. We demonstrate that the attractive behavior of zebrafish to amino acids is mediated through the neural circuitry involving the lateral chain glomeruli in the OB, which receive odor information from the microvillous OSNs. Thus, this is a unique genetic study providing definitive evidence for the selective neural circuit underlying a specific behavior in adult zebrafish.
Results
Identification of Gene Trap Lines with Gal4FF Expression in OSN Subsets Innervating Distinct Glomeruli.
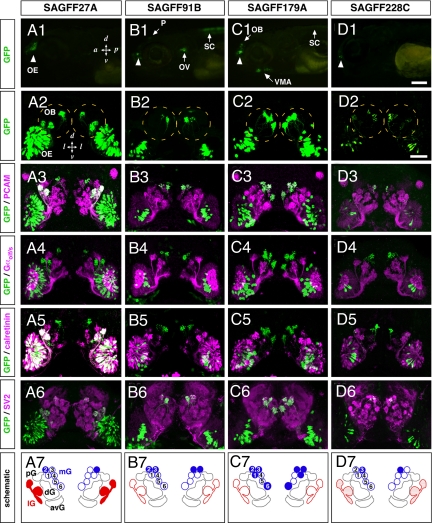
We previously conducted gene trap and enhancer trap screens in zebrafish by using the Tol2 transposon constructs containing the Gal4FF transcription activator and established 185 transgenic lines that expressed Gal4FF in specific tissues and cells (25). We crossed these lines with the UAS:GFP reporter fish and analyzed the double transgenic larvae for GFP expression in the olfactory system. Among 16 lines that showed GFP fluorescence in the OE in a variety of patterns at 5 days after fertilization [supporting information (SI) Fig. S1], we identified 4 gene trap lines (SAGFF27A, SAGFF91B, SAGFF179A, and SAGFF228C) that expressed GFP in distinct subpopulations of OSNs (Fig. 1A1–D1). In these larvae, GFP-expressing OSNs projected their axons to partially overlapping but mostly different glomeruli in the OB. In all 4 lines, the GFP-positive axons differentially targeted one or some of the glomeruli located in the medial region of the OB (Fig. 1 A2–D2). In SAGFF27A, the GFP-positive axons innervated a chain of glomeruli positioned in the lateral OB in addition to the medial glomerulus (Fig. 1A2). GFP expression was restricted to OSNs in SAGFF27A and SAGFF228C (Fig. 1A1 and 1D1), whereas SAGFF91B and SAGFF179A also showed GFP fluorescence in other tissues of larvae: the otic vesicle, pineal gland, and spinal cord in SAGFF91B (Fig. 1B1) and the ventral mandibular arch and spinal cord in SAGFF179A (Fig. 1C1).
Fig. 1.
Gene trap zebrafish lines with Gal4FF expression in OSN subsets innervating distinct glomeruli. GFP expression patterns were examined in 5-day-old larvae of the 4 gene trap Gal4FF-expressing lines (A1–A7, SAGFF27A; B1–B7, SAGFF91B; C1–C7, SAGFF179A; and D1–D7, SAGFF228C) crossed with the UAS:GFP reporter strain. (A1–D1) Lateral views of transgenic larval heads with GAL4FF-driven GFP fluorescence in the OE (arrowheads). In SAGFF91B and SAGFF179A, additional fluorescent signals are observed. (B1) The otic vesicle (OV), pineal (P), and spinal cord (SC) in SAGFF91B. (C1) The ventral mandibular arch (VMA) and SC in SAGFF179A. (A2–D2) Frontal views of larvae showing GFP fluorescence in subsets of OSNs innervating the OB (dashed circles) with different patterns. (A3–D6) Whole-mount double-immunofluorescence labeling with antibodies against GFP (A3–D6; green) and PCAM (A3–D3; magenta), Gαolf/s (A4–D4; magenta), calretinin (A5–D5; magenta), or SV2 (A6–D6; magenta). (A7–D7) Schematic diagrams illustrating different patterns of glomerular innervation by Gal4FF-expressing OSNs in the gene trap lines. The 4 lines show different combinations of glomerular targeting in the medial cluster (mG1–6, blue). SAGFF27A and SAGFF228C also show strong and weak GFP expression, respectively, in OSNs innervating the lateral glomeruli (lG, red). avG, anteroventral glomeruli; dG, dorsal glomeruli; mG, medial glomeruli; pG, posterior glomeruli. [Scale bars: (in D1), A1–D1, 200 μm; (in D2), A2–D6, 50 μm.]
To examine detailed axonal trajectories of the GFP-expressing OSNs, we carried out whole-mount immunohistochemistry of 5-day-old larvae using several antibodies. The cell adhesion molecule (PCAM) is a marker for all OSN axons (26, 27) (Fig. S2). GFP-positive axons in all 4 lines were PCAM-positive, confirming that they were the OSN axons (Fig. 1 A3–D3). These axons terminated in glomeruli of the OB, where synaptic vesicle protein SV2 was highly accumulated (Fig. 1 A6–D6). The SV2 immunostaining revealed spatial segregation of 5 major glomerular clusters in the larval OB. We designated these clusters as the anteroventral, dorsal, posterior, lateral, and medial glomerular clusters, with a slight modification of the previous designations for 3.5-day-old larvae (28). In all 4 lines, the GFP-positive axons were completely negative for Gαolf/s immunoreactivity, suggesting that they were not ciliated OSNs (Fig. 1 A4–D4). The calcium-binding protein, calretinin, is expressed mainly by microvillous OSNs innervating the lateral glomerular cluster and also by a small population of ciliated OSNs innervating the dorsal and posterior glomerular clusters (27). Double-labeling with anti-calretinin antibody revealed that the majority of GFP-expressing axons in SAGFF27A were positive for calretinin, innervating the lateral glomerular cluster. This result indicates that SAGFF27A fish expressed Gal4FF predominantly in the microvillous OSNs (Fig. 1A5). In contrast, there was no overlap between GFP and calretinin signals in SAGFF91B and SAGFF179A and only a slight overlap in SAGFF228C (Fig. 1 B5–D5). We also noticed that in SAGFF91B, SAGFF179A, and SAGFF228C, the Gal4FF-expressing OSNs were confined to the central region of the OE, although calretinin-positive microvillous OSNs were distributed broadly in the whole OE (Fig. 1 B5–D5). Thus, the Gal4FF-expressing OSNs in this central region of the OE predominantly innervated the medial glomerular cluster in the OB, suggesting the topographical organization of OSN axon projection.
The medial glomerular cluster is unique in that it is innervated by neither Gαolf/s-positive ciliated OSNs nor calretinin-positive microvillous OSNs (16). The SV2 immunostaining clearly delineated 6 glomeruli (mG1–6) in this cluster (Fig. 1 A6–D6). The GFP-positive axons in 4 transgenic lines differentially targeted one or some of the 6 glomeruli in the medial cluster (i.e., SAGFF27A, mG2; SAGFF91B, mG2 and mG3; SAGFF179A, mG1, mG2, mG3, and mG6; SAGFF228C, mG3). The patterns of glomerular innervation in individual lines are schematically depicted in Fig. 1 A7–D7. In the adult SAGFF179A, we observed many GFP-positive OSNs that were located in the most apical layer of the OE, displayed an ovoid cell shape, and expressed calcium-binding protein S100 (data not shown). These anatomical, morphological, and molecular features are characteristic of the third type of fish OSNs, crypt cells (11, 12, 19, 20), suggesting that SAGFF179A may express Gal4FF in the crypt cells, possibly innervating some of the glomeruli in the medial cluster.
Glomerular Innervation of Gal4FF-Expressing OSNs in Adult Zebrafish.
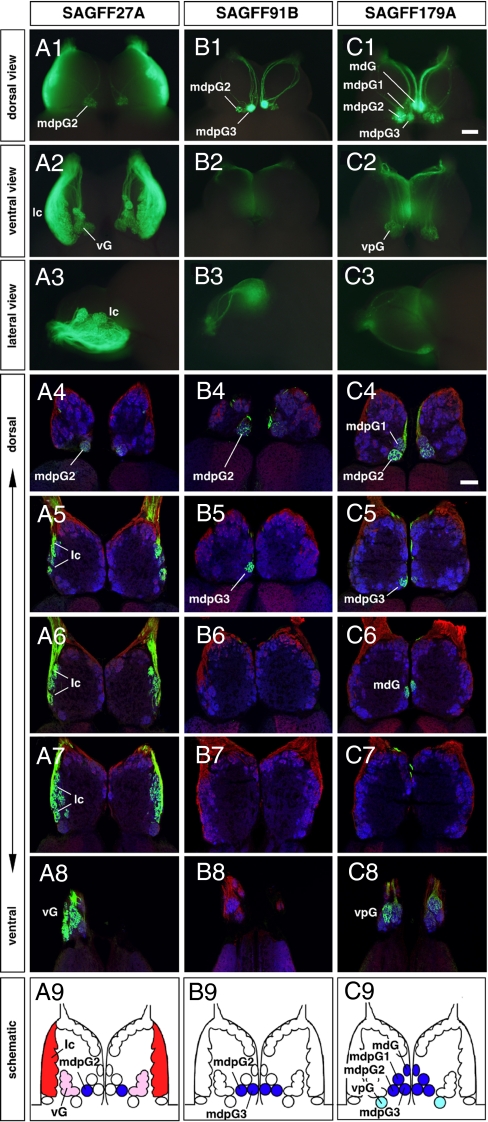
We next asked whether the characteristic Gal4FF expression patterns in the transgenic zebrafish larvae are also observed in the adult fish. Intense GFP expression was detected in whole-mount preparations and sections of the OE and OB from the 3 transgenic lines (SAGFF27A, SAGFF91B, and SAGFF179A) crossed with UAS:GFP reporter fish (Fig. 2A1–C3 and Figs. S3 and S4). For SAGFF228C, however, no GFP signal was observed in the adult fish (data not shown), suggesting transient Gal4FF expression only at early developmental stages in this line.
Fig. 2.
Conserved patterns of differential glomerular innervation of GFP-positive olfactory axons in 3 gene trap lines in adult zebrafish: (A1–A9) SAGFF27A, (B1–B9) SAGFF91B, and (C1–C9) SAGFF179A. (A1–C3) Whole-mount OBs with GFP fluorescence viewed from dorsal (A1–C1), ventral (A2–C2), and lateral (A3–C3) sides. (A4–C8) Horizontal sections of OBs triple-labeled with antibodies against GFP (green), PCAM (red), and SV2 (blue). For individual lines, 5 sections are shown from the dorsal-most (A4–C4) to ventral-most (A8–C8) levels. (A9–C9) Schematic diagrams illustrating different patterns of glomerular innervation by GAL4FF-expressing OSNs in the adult gene trap lines. In SAGFF27A, GFP-positive olfactory axons innervate the lateral chain (lc, red), the ventral glomeruli (vG, pink), and 1 of the mediodorsal posterior glomeruli (mdpG2, blue). In SAGFF91B, GFP-positive olfactory axons innervate 2 of the mediodorsal posterior glomeruli (mdpG2 and mdpG3, blue). In SAGFF179A, GFP-positive olfactory axons innervate the mdpG1-3 (blue), the mediodorsal glomerulus (mdG, blue), and the ventroposterior glomerulus (vpG, light blue). (Scale bars: 100 μm.)
Detailed glomerular innervation patterns of Gal4FF-expressing OSNs in the adult SAGFF27A, SAGFF91B, and SAGFF179A were analyzed on OB horizontal sections triple-labeled with anti-GFP, anti-PCAM, and anti-SV2 antibodies (Fig. 2 A4–C8). We essentially used the nomenclature of individual glomeruli in the adult OB designated by Baier and Korsching (29). For SAGFF27A, strong GFP fluorescence was observed mainly in the lateral region of the OB (Fig. 2 A1–A3). A careful examination of OB sections revealed that these GFP-positive OSN axons predominantly innervate the lateral chain of glomeruli and the ventral glomeruli (vG) (Fig. 2 A4–A9).
Consistent with the distinctive projections of OSN axons onto the medial glomerular cluster observed in larvae, the 3 transgenic lines also maintained similar patterns of glomerular innervation in adults. Among the 3 mediodorsal posterior glomeruli in the adult OB, mdpG2 was positive for GFP in all 3 lines, mdpG3 was positive for GFP in 2 lines (SAGFF91B and SAGFF179A), and mdpG1 was positive for GFP only in 1 line (SAGFF179A) (Fig. 2 A4–C5 and A9–C9). These discrete labeling patterns indicate that the adult mdpG1, mdpG2, and mdpG3 may correspond to the larval mG1, mG2, and mG3, respectively. The mediodorsal glomerulus (mdG) showed GFP immunoreactivity only in SAGFF179A (Fig. 2 C6 and C9). Based on a comparison of relative glomerular positions along the dorsoventral axis of the OB, the adult mdG is likely to be derived from the larval mG6. Thus, a clear correlation was observed between the larval and adult expression patterns in the 3 transgenic lines.
On the other hand, another GFP-positive glomerulus (vpG) was observed in the most ventroposterior region of the OB, distant and isolated from other GFP-positive glomeruli, in SAGFF179A (Fig. 2 C8 and C9). Because the GFP-positive vpG was detected in the adult fish but not in the 5-day-old larvae, it is likely that this glomerulus was formed at some later stage during development.
Genetic Behavioral Analysis of Attractive Response to Amino Acids.
Because the unique and restricted patterns of Gal4FF expression were maintained in the adult transgenic fish, we reasoned that it was feasible to take an integrated approach combining the genetic manipulation of neural transmission with the olfactory behavioral assays to specific odor stimuli. In particular, we were interested in SAGFF27A, because the Gal4FF-expressing OSNs predominantly innervated the lateral chain of glomeruli that had been shown to respond to amino acids in zebrafish and implied to mediate feeding behavior in goldfish and crucian carp (11, 13, 17, 30–32).
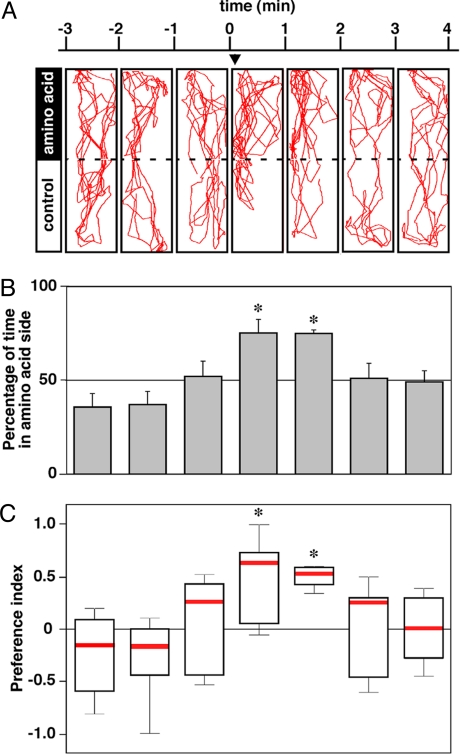
To analyze the odor preference behavior in zebrafish, we developed an assay system to monitor fish movement in response to odorants. Fig. 3A shows an example of the swimming path of a WT zebrafish, which clearly displayed an attractive response to the amino acid mix (also see Movie S1). During the prestimulating period, WT fish used the entire space of a tank for swimming. However, on the introduction of an amino acid mix into a corner of the tank (Fig. 3A, arrowhead), fish preferentially stayed in the area where the amino acid mix was applied (amino acid side) rather than on the opposite side. To quantify the degree of attraction, we measured the percentage of time spent on the amino acid side for every 1 min (Fig. 3B) and calculated the preference index (PI; Fig. 3C). A significant preference for the amino acid side was observed for 2 min after the application (Fig. 3 B and C).
Fig. 3.
Attractive response of zebrafish to amino acids. (A) Representative swimming paths of zebrafish on amino acid application. Results are presented for every 1 min from 3 min before (Left) to 4 min after (Right) the application of the amino acid mix. The arrowhead indicates the corner of amino acid application. (B) Percentage of time spent on the amino acid side by zebrafish (n = 7). Data are shown as mean ± SEM (*P < 0.05, compared with 1-min period before the application of the amino acid mix, 2-tailed Student's t test). (C) The degrees of attraction to amino acids are represented by the PI. Box plots represent the median (red horizontal line), 25 to 75% quantiles (boxes), and ranges (whiskers) of data. Each time bin was analyzed for a significant deviation from 0 using the Wilcoxon sign-rank test (*P < 0.05).
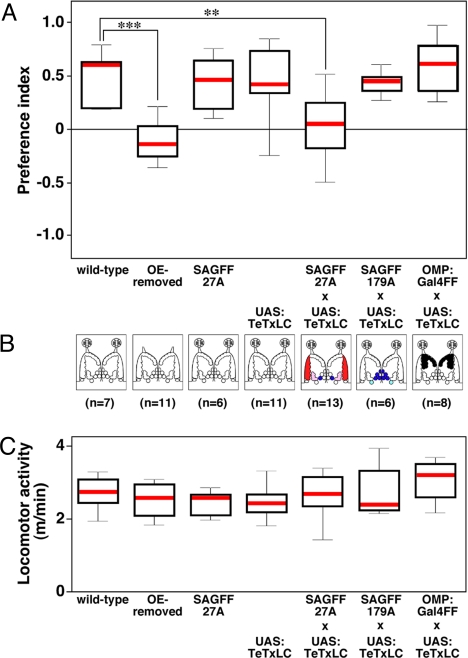
To determine whether this attractive response is dependent on the olfaction, we surgically removed the OE bilaterally and carried out the same behavioral experiment. The OE-removed zebrafish showed no preference for the amino acid mix (Fig. 4A). This result indicates that the olfactory system is required for the attractive response to amino acids in zebrafish, although we cannot exclude the possibility of some contribution of other chemosensory systems for the detection of amino acids.
Fig. 4.
Attractive response to amino acids is abolished in SAGFF27A expressing tetanus neurotoxin. (A) The attractive responses to amino acids for individual genotypes are represented by the PI. The OE-removed fish and the SAGFF27A;UAS:TeTxLC fish show no preference for amino acids, whereas other genotypes (WT, SAGFF27A, UAS:TeTxLC, SAGFF179A;UAS:TeTxLC, and OMP:Gal4FF;UAS:TeTxLC) show normal attractive responses to amino acids. Significance was assessed by Mann-Whitney U tests (***P < 0.001, **P < 0.01 compared with WT fish). (B) Schematic diagrams showing surgical removal of the OE (second panel from Left) and genetic manipulations of OSNs (3 panels on Right). Glomeruli innervated by TeTxLC-expressing OSNs are shown in colors. (C) All the genotypes show normal locomotor activities.
To identify olfactory neural circuitry mediating the attraction to amino acids, we genetically expressed TeTxLC under the control of a Gal4/UAS system (25). TeTxLC proteolytically cleaves a synaptic vesicle protein, synaptobrevin, leading to defective exocytosis and neural transmission blockade (33). We used 3 transgenic lines, SAGFF27A, SAGFF179A, and OMP:Gal4FF, which express Gal4FF in subsets of OSNs innervating mostly different regions of the OB (SAGFF27A: lateral, SAGFF179A: posteromedial, OMP:Gal4FF: anteromedial) (Fig. 4B and Figs. S3 and S4). These Gal4FF-expressing lines were crossed with UAS:TeTxLC fish to block synaptic transmission within selective glomerular clusters, and a behavioral analysis was performed. A normal attractive response to the amino acid mix was observed in SAGFF179A;UAS:TeTxLC and OMP:Gal4FF;UAS:TeTxLC double-transgenic adult fish, whereas the attractive response was completely abolished in SAGFF27A;UAS:TeTxLC double-transgenic fish (Fig. 4A and Movie S2). Either SAGFF27A or UAS:TeTxLC single-transgenic fish showed an attractive response similar to that of WT fish, suggesting that the neural transmission blockade of Gal4FF-expressing OSNs in SAGFF27A is the cause of the behavioral abnormality regarding amino acids. In contrast, the attractive response to taurocholic acid, a putative social pheromone, was abrogated only in OMP:Gal4FF;UAS:TeTxLC double-transgenic fish (Fig. S5), which is consistent with previous reports on the roles of ciliated OSNs and anteromedial glomeruli in the bile acid response (11, 17). The locomotor activity of all single- and double-transgenic fish was indistinguishable from that of WT fish (Fig. 4C). These results suggest that the functional neural circuit involving the lateral chain glomeruli is absolutely required for the attractive response to amino acids in adult zebrafish.
Discussion
The zebrafish has become one of the most useful and important model organisms in biology because of its general experimental advantages as well as its amenability to both forward and reverse genetic techniques (23). Here, we have succeeded in genetic and functional dissection of the olfactory neural circuits in zebrafish by combining (i) a Tol2 transposon-mediated gene trap approach, (ii) a Gal4/UAS-based targeted gene expression system, (iii) visualization of axon projection patterns with genetically engineered fluorescent proteins in both the larva and adult, (iv) TeTxLC-mediated neural transmission blockade of selective OSNs, and (v) a simple behavioral assay for specific olfactory responses in adult zebrafish. Until recently, such a multidisciplinary strategy for anatomical and functional analyses of selective neural circuits was possible only in a few invertebrate species such as Caenorhabditis elegans and Drosophila melanogaster. However, rapid advances of various genetic techniques in the zebrafish have rendered it as a useful model organism comparable to these 2 invertebrates. This study provides several lines of evidence that reinforce the usefulness of zebrafish in integrative genetic analyses and opens up a unique avenue for future research on the functional organization of the vertebrate neural circuits. In particular, we showed that Gal4FF-mediated TeTxLC expression in distinct subsets of OSNs sufficiently generates an unambiguous behavioral phenotype in adult zebrafish.
One of the most intriguing findings obtained from our gene trap screens is the diversity of Gal4FF expression patterns in the OE. In particular, the Gal4FF expression in several lines was confined to small subpopulations of OSNs in particular regions of the OE, which project axons to distinct subsets of glomeruli in the OB. For instance, in SAGFF91B, SAGFF179A, and SAGFF228C, the Gal4FF-expressing OSNs were located in the central region of the OE and predominantly innervated the medial glomerular cluster in the OB. Such a topographical organization of the OSN axon projection from the OE to the OB in zebrafish is reminiscent of the zonal organization in the rodent olfactory system (34), suggesting the phylogenic conservation from fishes to mammals. Furthermore, these results suggest that OSNs are heterogeneous with respect to their spatial distribution and gene expression, both of which presumably correlate with functional aspects of distinct OSN subpopulations.
In SAGFF27A, the majority of Gal4FF-expressing cells were the microvillous OSNs innervating the lateral chain glomeruli in the OB, although weak Gal4FF expression was also observed in OSNs innervating one of the mediodorsal posterior glomeruli, mdpG2. Neural transmission blockade of these OSNs by TeTxLC expression in the double-transgenic zebrafish (SAGFF27A;UAS:TeTxLC) resulted in the absence of an attractive response to amino acids. In contrast, 2 other lines (SAGFF179A and OMP:Gal4FF) expressing Gal4FF in largely nonoverlapping subsets of OSNs showed normal attractive responses to amino acids after being crossed with UAS:TeTxLC fish. Because SAGFF27A and SAGFF179A share mdpG2 as a common target glomerulus of the Gal4FF-expressing OSNs, it is unlikely that mdpG2 is involved in the amino acid-evoked attractive response. Although we cannot exclude a possible involvement of another Gal4FF-positive glomerular cluster vG, the present findings strongly suggest that the microvillous OSNs innervating the lateral chain glomeruli are important to the preference for amino acids, probably leading to feeding behaviors of zebrafish.
The present results are consistent with several previous reports on amino acid responses of various fish as follows: (i) amino acids bind to V2R-type olfactory receptors on the microvillous OSNs in goldfish and zebrafish (15, 35, 36) and activate these neurons in zebrafish (37); (ii) zebrafish microvillous OSNs mainly innervate the lateral chain glomeruli (16); (iii) lateral chain glomeruli and the nearby mitral cells (the output neurons in the OB) in channel catfish (38) and zebrafish (13, 17, 39) are activated by amino acid stimuli; (iv) in freely swimming cod, feeding behaviors are elicited by electrical stimulation of the lateral olfactory tract that contains mitral cell axons originating from the lateral region of the OB (40); and (v) feeding behaviors are impaired by surgical transection of the lateral olfactory tract in crucian carp (32, 41). By combining genetics, neuroanatomy, and behavioral analysis, the present results integrate these previous circumstantial findings into convincing evidence to prove that the selective neural pathway involving the lateral chain glomeruli mediates the amino acid-induced attractive behavior in zebrafish. The next important step will be to elucidate how the amino acid information is transmitted beyond the OB and decoded in higher olfactory centers for execution of the attractive behavior.
Materials and Methods
Zebrafish.
Zebrafish larvae were obtained in natural crosses and staged as previously described (42, 43). Further details are provided in SI Text.
To generate OMP:Gal4FF transgenic zebrafish, the hsp70 promoter of T2KhspGFF plasmid containing a Gal4 DNA binding domain, 2 VP16 transactivation modules, and 2 Tol2 transposon elements (25) was replaced with the 5′-flanking 2-kb sequence of the zebrafish OMP gene. The plasmid DNA was coinjected with Tol2 transposase mRNA into one-cell–staged fertilized eggs (24). The resultant fish were crossed with homozygous UAS:GFP reporter fish. F1 larvae with the brightest GFP expression in ciliated OSNs were raised to establish the OMP:Gal4FF transgenic line.
Gene trap lines (SAGFF), enhancer trap lines (hspGFF and hspGGFF), UAS:GFP fish, and UAS:TeTxLC fish were the same as described previously (25). After several outcrosses, the fish lines with single insertions of SAGFF27A, SAGFF91B, and SAGFF179A were established. Details of integration sites of SAGFF are described in SI Text.
To prepare zebrafish without the OE, the olfactory rosettes were surgically removed under anesthesia with 0.016% tricaine (ethyl-m-aminobenzoate methanesulfonate; Nacalai Tesque). For OE-removed fish, the behavioral analysis was performed 1 week after the surgery.
Immunohistochemistry.
Immunohistochemistry was performed for whole-mount larvae and adult brain sections essentially as described previously (27). Further details are provided in SI Text and Table S1.
Behavioral Assay.
Five- to 7-month-old fish (body length ≈4 cm) were assayed for responses to a mixture of 8 amino acids (Ala, Cys, His, Lys, Met, Phe, Trp, and Val; Sigma). Each fish was transferred to an experimental tank (6 × 25 × 17 cm filled with 600 mL of water) and allowed to acclimate to the environment for at least 30 min twice within a week before the experiments. Fish were starved for 24 h before the experiments. A single fish was put in a tank, and its movement was recorded for 12–16 min in each trial. After the prestimulation period (6–8 min), 0.6 mL of amino acid mix (0.1 mM each) was delivered into a corner of the tank through a peristaltic pump (1.5 mL/min). The swimming paths were analyzed using an automated videotracking system (Videotrack; ViewPoint Life Sciences). The same behavioral assay system was used to analyze zebrafish responses to taurocholic acid (Fig. S5).
Data Analyses.
A PI was calculated from the following equation: PI = (TA − TC)/(TA + TC). TA and TC denote periods of time (min) for which fish stayed on the amino acid side or control side, respectively. PI values for every 1 min during 7 min of observation were plotted in Fig. 3C, whereas PI values for 2 min after the amino acid application are shown in Fig. 4A. Further details are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Susanne C. Hoyer for help in statistical analysis and critical reading of the manuscript, Dr. Rumiko Mizuguchi for helpful comments on the manuscript, and members of Yoshihara laboratory for fish care and discussion. We also thank Dr. Keiko Abe (University of Tokyo) for helpful discussion. This work was supported in part by a Grant-in-Aid for Scientific Research (C) (to T.K.); a Grant-in-Aid for Scientific Research (B) and on Priority Area (Cellular Sensor) (to Y.Y.); a Grant-in-Aid for Scientific Research (S) and on Priority Areas (to K.K.); the National BioResource Project (K.K. and Y.Y.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and a postdoctoral fellowship from the Japan Society for the Promotion of Science (to K.A.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900470106/DCSupplemental.
References
- 1.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 2.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 3.Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci. 1999;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- 4.Serizawa S, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- 6.Imai T, Sakano H. Roles of odorant receptors in projecting axons in the mouse olfactory system. Curr Opin Neurobiol. 2007;17:507–515. doi: 10.1016/j.conb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Mori K, Nagao H, Yoshihara Y. The olfactory bulb: Coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- 9.Mori K, Takahashi YK, Igarashi KM, Yamaguchi M. Maps of odorant molecular features in the mammalian olfactory bulb. Physiol Rev. 2006;86:409–433. doi: 10.1152/physrev.00021.2005. [DOI] [PubMed] [Google Scholar]
- 10.Morita Y, Finger TE. Differential projections of ciliated and microvillous olfactory receptor cells in the catfish, Ictalurus punctatus. J Comp Neurol. 1998;398:539–550. doi: 10.1002/(sici)1096-9861(19980907)398:4<539::aid-cne6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Hansen A, et al. Correlation between olfactory receptor cell type and function in the channel catfish. J Neurosci. 2003;23:9328–9339. doi: 10.1523/JNEUROSCI.23-28-09328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen A, Anderson KT, Finger TE. Differential distribution of olfactory receptor neurons in goldfish: Structural and molecular correlates. J Comp Neurol. 2004;477:347–359. doi: 10.1002/cne.20202. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich RW, Korsching SI. Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron. 1997;18:737–752. doi: 10.1016/s0896-6273(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Oh BC, Stryer L. Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc Natl Acad Sci USA. 1998;95:11987–11992. doi: 10.1073/pnas.95.20.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speca DJ, et al. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich RW, Korsching SI. Chemotopic, combinatorial, and noncombinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J Neurosci. 1998;18:9977–9988. doi: 10.1523/JNEUROSCI.18-23-09977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellstrom T, Døving KB. Chemoreception of taurocholate in anosmic and sham-operated cod, Gadus morhua. Behav Brain Res. 1986;21:155–162. doi: 10.1016/0166-4328(86)90093-8. [DOI] [PubMed] [Google Scholar]
- 19.Hansen A, Finger TE. Phyletic distribution of crypt-type olfactory receptor neurons in fishes. Brain Behav Evol. 2000;55:100–110. doi: 10.1159/000006645. [DOI] [PubMed] [Google Scholar]
- 20.Germana A, et al. S100 protein-like immunoreactivity in the crypt olfactory neurons of the adult zebrafish. Neurosci Lett. 2004;371:196–198. doi: 10.1016/j.neulet.2004.08.077. [DOI] [PubMed] [Google Scholar]
- 21.Hamdani el H, Lastein S, Gregersen F, Døving KB. Seasonal variations in olfactory sensory neurons-fish sensitivity to sex pheromones explained? Chem Senses. 2008;33:119–123. doi: 10.1093/chemse/bjm072. [DOI] [PubMed] [Google Scholar]
- 22.Sato Y, Miyasaka N, Yoshihara Y. Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci. 2007;27:1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara Y. In: Chemosensory Systems in Mammals, Fishes and Insects. Meyerhof W, Korsching S, editors. Heidelberg: Springer; 2009. in press. [PubMed] [Google Scholar]
- 24.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Asakawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno T, et al. Molecular diversity in zebrafish NCAM family: Three members with different VASE usage and distinct localization. Mol Cell Neurosci. 2001;18:119–130. doi: 10.1006/mcne.2001.1007. [DOI] [PubMed] [Google Scholar]
- 27.Miyasaka N, et al. Robo2 is required for establishment of a precise glomerular map in the zebrafish olfactory system. Development. 2005;132:1283–1293. doi: 10.1242/dev.01698. [DOI] [PubMed] [Google Scholar]
- 28.Dynes JL, Ngai J. Pathfinding of olfactory neuron axons to stereotyped glomerular targets revealed by dynamic imaging in living zebrafish embryos. Neuron. 1998;20:1081–1091. doi: 10.1016/s0896-6273(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 29.Baier H, Korsching S. Olfactory glomeruli in the zebrafish form an invariant pattern and are identifiable across animals. J Neurosci. 1994;14:219–230. doi: 10.1523/JNEUROSCI.14-01-00219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Rekowski C, Zippel HP. In goldfish the qualitative discriminative ability for odors rapidly returns after bilateral nerve axotomy and lateral olfactory tract transection. Brain Res. 1993;618:338–340. doi: 10.1016/0006-8993(93)91287-3. [DOI] [PubMed] [Google Scholar]
- 31.Hamdani EH, Alexander G, Døving KB. Projection of sensory neurons with microvilli to the lateral olfactory tract indicates their participation in feeding behaviour in crucian carp. Chem Senses. 2001;26:1139–1144. doi: 10.1093/chemse/26.9.1139. [DOI] [PubMed] [Google Scholar]
- 32.Hamdani EH, Kasumyan A, Døving KB. Is feeding behaviour in crucian carp mediated by the lateral olfactory tract? Chem Senses. 2001;26:1133–1138. doi: 10.1093/chemse/26.9.1133. [DOI] [PubMed] [Google Scholar]
- 33.Schiavo G, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 34.Mori K, von Campenhause H, Yoshihara Y. Zonal organization of the mammalian main and accessory olfactory systems. Philos Trans R Soc Lond B. 2000;355:1801–1812. doi: 10.1098/rstb.2000.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luu P, Acher F, Bertrand HO, Fan J, Ngai J. Molecular determinants of ligand selectivity in a vertebrate odorant receptor. J Neurosci. 2004;24:10128–10137. doi: 10.1523/JNEUROSCI.3117-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: Candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipschitz DL, Michel WC. Amino acid odorants stimulate microvillar sensory neurons. Chem Senses. 2002;27:277–286. doi: 10.1093/chemse/27.3.277. [DOI] [PubMed] [Google Scholar]
- 38.Nikonov AA, Caprio J. Electrophysiological evidence for a chemotopy of biologically relevant odors in the olfactory bulb of the channel catfish. J Neurophysiol. 2001;86:1869–1876. doi: 10.1152/jn.2001.86.4.1869. [DOI] [PubMed] [Google Scholar]
- 39.Yaksi E JB, Friedrich RW. Topological reorganization of odor representations in the olfactory bulb. PLoS Biol. 2007;5:e178. doi: 10.1371/journal.pbio.0050178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Døving KB, Selset R. Behavior patterns in cod released by electrical stimulation of olfactory tract bundlets. Science. 1980;207:559–560. doi: 10.1126/science.7352272. [DOI] [PubMed] [Google Scholar]
- 41.Hamdani el H, Døving KB. The functional organization of the fish olfactory system. Prog Neurobiol. 2007;82:80–86. doi: 10.1016/j.pneurobio.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 43.Westerfield M. The Zebrafish Book. Eugene, OR: Univ of Oregon Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.