Abstract
The structural integrity of the ubiquitous enzyme superoxide dismutase (SOD1) relies critically on the correct coordination of Cu and Zn. Loss of these cofactors not only promotes SOD1 aggregation in vitro but also seems to be a key prerequisite for pathogenic misfolding in the neurodegenerative disease amyotrophic lateral sclerosis (ALS). We examine here the consequences of Zn2+ loss by selectively removing the Zn site, which has been implicated as the main modulator of SOD1 stability and disease competence. After Zn-site removal, the remaining Cu ligands can coordinate a nonnative Zn2+ ion with μM affinity in the denatured state, and then retain this ion throughout the folding reaction. Without the restriction of a metallated Zn site, however, the Cu ligands fail to correctly coordinate the nonnative Zn2+ ion: Trapping of a water molecule causes H48 to change rotamer and swing outwards. The misligation is sterically incompatible with the native structure. As a consequence, SOD1 unfolds locally and interacts with neighboring molecules in the crystal lattice. The findings point to a critical role for the native Zn site in controlling SOD1 misfolding, and show that even subtle changes of the metal-loading sequence can render the wild-type protein the same structural properties as ALS-provoking mutations. This frustrated character of the SOD1 molecule seems to arise from a compromise between optimization of functional and structural features.
Keywords: functional evolution, protein disease, protein misfolding
The mechanisms by which protein misfolding and aggregation induce cytotoxicity in neurodegenerative disorders are not yet established. For several of these disorders, like e.g., Alzheimer's and Huntington's disease, the precursor proteins are structurally flexible and thus directly free to adopt pathogenic structures (1). For the motor-neuron disease ALS, however, the precursor protein SOD1 is a stable and perfectly soluble metalloenzyme that needs first to be locally or globally unfolded to acquire pathologic function (Fig. 1). Consistent with this, a notable property of ALS-provoking SOD1 mutations is decreased protein stability (2, 3). Although these mutations are only responsible for a small fraction of the ALS cases, accumulating evidence suggests that the more common, sporadic, cases are caused by an analogous mechanism involving the wild-type protein (4–6). This seemingly inherent susceptibility of SOD1 to provoke neural damage has drawn attention to the role of the Cu and Zn cofactors. Under normal conditions, the Cu and Zn ions orchestrate the catalytic degradation of superoxide radicals (3), and their removal substantially destabilizes the protein structure (7). Somewhat unexpectedly, the largest part of this effect has been assigned to the removal of the locally coordinated Zn2+ ion, with a stability loss that matches, or even exceeds, that of the most extreme ALS-mutations (8): The free-energy penalty of Zn removal exceeds 8 kcal/mol (5, 9), whereas the corresponding value for the ALS mutation A4V is <5 kcal/mol (2). At a structural level, Zn depletion is observed to cause distortion of the dimer interface (10), partial loop unfolding and increased solvent accessibility of the Cu2+ ion (11). More indirectly, metallation of the Zn site also seems to determine the ensemble composition of immature, and potentially disease provoking, SOD1 species in vivo by being a prerequisite for chaperone-assisted loading of Cu (12). Finally, selective removal of Zn from wild-type protein induced apoptosis in cultured cells (4). The remaining SOD1 containing just a bound Cu2+ ion did not offer any protection against the observed cytotoxicity, but was rather found to boost it by rendering anomalous catalytic properties to SOD1 (4). In this study, we examine the role of Zn2+ binding in folding of the SOD1 monomer. This folding step is the first, critical event in the maturation pathway of the native enzyme and is the point where cytotoxic misfolding is most likely to occur. The results show that, in the absence of native Zn ligands, Zn2+ ions bind readily but incorrectly to the SOD1 Cu ligands. As a result the protein is prevented from attaining its correctly folded structure: Exclusion of H48 from the inner ligand sphere obstructs the backbone packing and causes local unfolding of the active-site loops.
Fig. 1.
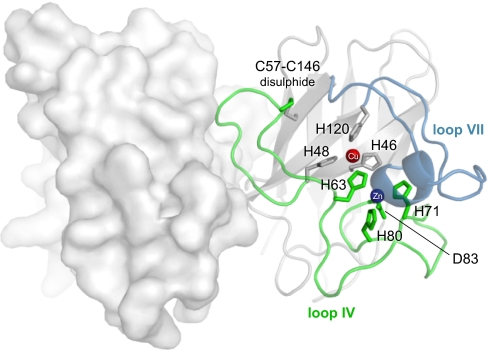
The functional features of the SOD1 homodimer (PDB entry 1SPD) are all contained by the long loops IV (green) and VII (blue) that protrude from either side of the central Ig-like scaffold. These loops not only coordinate and encapsulate the Cu and Zn ions but form also the substrate channel and part of the dimer interface. In this study we analyze a monomeric variant of SOD1 where H63, H71, H80, and D83 have been mutated to S, to mimic a protein that has lost, or fails to coordinate, the native Zn2+ ion.
Results and Discussion
Zn Ligands Compromise the Structural Stability of the apo Monomer.
Similarly to other Ig-like proteins, the apo SOD1 monomer is observed to acquire its structure in a cooperative 2-state transition over a single free-energy barrier (5, 13) (Fig. 2). This barrier is also the rate-limiting step for dimer formation (5, 13) and the feature that safeguards the integrity of the native structure. Partly structured, and potentially sticky, intermediates are prevented from populating because they are unstable (14). The most apparent deviation from this cooperative folding pattern is seen in the functional loops IV (residues 48–85) and VII (residues 120–144), which display considerable backbone dynamics in the absence of coordinated metals (15). To probe the extent of 3-dimensional structure of loop IV in the apo monomer, we mutated the Zn ligands H63, H71, H80, and D83 to S (SOD1mutapo). Despite the extensive mutations, the SOD1mutapo structure remains wild-type-like, indicated by the unperturbed folding m values and HSQC NMR spectrum (Fig. 2, SI Text, and Fig. S1). Even so, the mutations have a clear effect on protein stability: The unfolding rate constant (ku) decreases and the transition midpoint shifts to higher urea concentrations (Fig. 2 and Table 1). This specific response to mutation indicates that loop IV is not completely unfolded in the apo monomer but retains some degree of ordered structure. Moreover, the unaffected refolding rate constant (kf) suggests that this structure is only present in the folded monomer and ruptures relatively early in the unfolding process: The net value of φ = Δlogkf/(Δlogku − Δlogkf) for the mutations is ≈0 (Table 1) (16, 17). A similarly low φ value is observed for the mutation C111A in the adjacent loop VI (5). The most striking effect of the Zn-site mutations, however, is that they stabilize the folded apo structure by ≈0.5 kcal/mol (Table 1). The origin of this stability gain could be that the native Zn ligands yield unfavorable desolvation terms and/or loss of conformational entropy upon transfer to the protein interior in the absence of Zn2+. From the perspective of the folding-energy landscape (18), such structural penalties give rise to energetic frustration that increases the occupancy of high-energy states in which the Zn-binding loop is fully disordered. Without the Zn site, the occupancy of these intermediates would go down by a factor of 5/2, as crudely estimated from the stability gain upon mutation shown in Table 1. The Zn site of the SOD1 structure seems thus associated with a drawback in the form of a more labile apo state. Such a conflict between the functional and structural properties of enzymes is not unique for SOD1, but seems to be a general theme in protein evolution (19–21).
Fig. 2.
The effects of Zn-site mutation and metallation on folding and stability of the SOD1 monomer. SOD1pWTapo (black), SOD1mutapo (red) and SOD1mutZn (blue). (A) Chevron plots of logkobs = log(kf+ku) vs. [urea] (Eq. 2). (B) Equilibrium unfolding transitions at 0 and 500 μM [Zn] (Eq. 1). Measurements at 50 and 500 μM [Zn] yield indistinguishable results. (C) The corresponding folding free-energy profiles, showing the unfolded state (U), the transition state (‡) and the folded monomer (F). The folded state becomes more solvent accessible upon Zn binding to the Cu site, indicated by decreased value of mu (Eqs. 1 and 2 and Table 1).
Table 1.
Kinetic and thermodynamic parameters
| Protein | logkfH2O* | logkuH2 O* | mf* | mu* | mU/F† | MPeq†, M | ΔG‡, kcal/mol |
|---|---|---|---|---|---|---|---|
| SOD1pWTapo | −1.12 ± 0.1 | −3.9 ± 0.05 | −1.07 ± 0.1 | 0.46 ± 0.01 | 1.17 ± 0.10 | 2.12 ± 0.06 | 3.6 ± 0.2 |
| SOD1mutapo | −1.25 ± 0.05 | −4.41 ± 0.08 | −0.96 ± 0.04 | 0.48 ± 0.01 | 1.13 ± 0.09 | 2.54 ± 0.04 | 4.1 ± 0.2 |
| SOD1mutZn | 0.76 ± 0.02 | −4.7 ± 0.12 | −0.81 ± 0.01 | 0.22 ± 0.01 | 0.84 ± 0.05 | 5.85 ± 0.05 | 7.2 ± 0.2 |
*Derived from kinetic data by Eq. 2. The rate constants for SOD1pWTapo are slightly different from those published in refs. 5 and 14, which are measured at higher ionic strength. mf of the apo species are slightly overestimated due to a upward kink of logkf at low [urea]. The effect seems due to shifts of the folding reaction to parallel pathways and is unlikely to affect the conclusions of the current study.
†Derived from equilibrium data by Eq 1.
‡Average of ΔGkin = −2.3RT(logkuH2 O − logkfH2 O) and ΔGeq = − 2.3RT(mU/FxMPeq).
Effects of Metal Loading.
After overexpression and purification, SOD1mut contains ≈25% Cu (Fig. S2), consistent with the observation that a filled Zn site is required for efficient Cu loading by the coexpressed chaperone CCS (12). In the remaining fraction of the molecules the Cu site contains a Zn2+ ion. This selective metallation of the Cu site has 2 characteristic effects on the SOD1 monomer (Fig. 2). It increases protein stability and alters the structure of the folded state. Looking at the chevron data, the latter is indicated by an anomalous reduction of the unfolding m value (mu) (Eq. 2). Demetallated protein reconstituted exclusively with Zn2+ (SOD1mutZn) yields indistinguishable results. Molecules that carry Cu2+ must then have properties very similar to those of the Zn2+ loaded protein. Consistently, preliminary studies of the completely Cu2+ loaded protein yield logku values that essentially overlap with those of the Zn2+ species. Also, the stability and chevron data of SOD1mutZn are insensitive to the mutation H63S, indicating that this native Cu ligand is not part of the coordination sphere in the absence of a filled Zn site (Fig. S2). The gain of protein stability upon Zn2+ binding to the Cu site is kinetically seen as an increase of kf, accompanied by a decrease of ku, leading to a shift of the transition midpoint (MP) to higher urea concentrations (Fig. 2). The increased kf observed for the Zn2+-bound protein, however, shows that the binding of metal to the Cu ligands occurs early in the folding process and lowers the folding barrier. The associated value of φ = Δlogkf/(Δlogku − Δlogkf) = 0.9 (at 0M urea) indicates that the coordination geometry precedes the formation of the folding nucleus (17). An energetic view of what happens with the folded structure upon coordination of the metal is provided by the selective reduction of mu. In keeping with basic mass-action criteria, this diminished sensitivity to [urea] suggests that the folded monomer has become more denatured-like, i.e., its solvent-accessible surface area has increased (18).
Cu-Ligands Coordinate Metals Already in the Denatured State.
To examine the likelihood of metals coordinating directly to unfolded SOD1, we titrated denatured SOD1mutapo with Zn2+ using ITC. The result shows that unfolded SOD1mutapo binds Zn2+ with an affinity of KUZn = 180 × 10−9 M, in the presence of 7 M urea (Fig. S3). Analogous experiments with the more promiscuous Cu2+ ions, however, failed to give reproducible results, possibly because of Cu2+-promoted aggregation or competing ligation by other side chains. The Cu ligands of unfolded SOD1 seem thus to have the capacity to pick up Zn2+ at physiologically relevant concentrations and to retain this ion coordinated throughout the folding process. Similar coordination behavior was recently reported for binding of Zn2+ to the native Zn ligands (8). The findings indicate not only how SOD1 avoids Cu loss during natural structural fluctuations, but also raises questions about the actual levels of metal-free protein in vivo. Taken to the extreme, it can even be envisaged that immature SOD1 functions as a ubiquitous Zn2+ reservoir from which the active protein is recruited through metal replacement by the Cu-loading chaperone CCS.
Formal Relationship Between Metal-Binding and Protein Stability.
The effect of metal binding on protein stability is ultimately determined by mass-action: The addition of metal shifts the folding equilibrium toward the species with the highest metal-binding affinity (22). Here, this species is the folded SOD1 monomer. A general formalism for the link between metal binding and protein stability is presented in Fig. S4. The first thing to be noted is that the stability change of SOD1mut shows key agreement with the ITC data: Above 10 μM, where both the folded and unfolded states are fully metallated, the folding equilibrium becomes insensitive to further additions of Zn2+ (Fig. S4). Second, the gain in protein stability, i.e., 3.1 ± 0.2 kcal/mol (Table 1), indicates that the Zn2+ affinity increases 100-fold upon acquisition of the folded state. The dissociation constant is expected to change from KUZn = 180 nM in the unfolded state to KFZn = 1.8 nM in the folded monomer. Similar, folding-induced changes in Zn2+ affinity have been reported for azurin (22) and, more generally, for mutational insertion of His pairs to probe folding transition states (23). As a control of Zn2+ binding to folded SOD1mut we measured KFZn directly by ITC, yielding a value of ≈13 nM (Fig. S3). Considering that this measurement is at the detection limit of the ITC instrument, the agreement with the kinetic estimate is quite reasonable. For further discussion of metal binding to SOD1, see Fig. S3.
Crystal Structure Reveals Obstructive Misligation of the Cu Site.
The crystal structure of SOD1mutZn was determined to 2.2Å resolution (Fig. 3). With respect to the central β strands, the structure aligns well with that of monomeric holo SOD1 (24), having an rms deviation of 0.67 Å for 112 equivalent Cα positions. However, SOD1mutZn displays an unprecedented degree of disorder of loops IV and VII, which are invisible in the X-ray data. In addition, a Zn2+ ion is coordinated tetrahedrally at the Cu site by H46, H120 and 2 water molecules (Fig. 3). Notably, the loop disorder of SOD1mutZn accounts precisely for the increased solvent-accessible surface area of the folded state implicated by the folding data in Fig. 2. The calculated contribution of loop unfolding to the mD-N value is 30% (16492Å2 → 11522Å2, Fig. S5), which is in good accord with the measured 26% change upon metallation (mD-N decreases from 1.13 to 0.84) (Table 1). The good agreement between folding and crystallographic data are a strong indication that the structure reports on the solution properties of the SOD1 monomer.
Fig. 3.
Misligation of Zn2+ to the Cu site of the SOD1 monomer. (A) Structural superposition of the misligated SOD1 monomer (orange) and the monomeric holo SOD1 (gray). The misligated Zn ion and water molecules in the Cu-binding site are shown in blue and red, respectively. Interpretable electron densities for loops IV and VII are missing in the misligated protein. (B) Close-up of the Cu site showing that Zn binding to the Cu site of the SOD1 mutant causes H48 to change rotamer. This altered orientation of H48 is sterically incompatible with the native arrangement of loop IV. Electron densities are shown as green mesh.
Strikingly, we also observe that H48 has changed rotamer and is now oriented away from the Zn2+ ion, which is instead coordinated by a water molecule (Fig. 3). The new orientation of H48 is further sterically incompatible with the native conformation of G61 and P62 in loop IV, explaining why the active site ruptures. Since the folding m values indicate that solvent-accessible surface area of SOD1mutapo is smaller than for SOD1mutZn, we conclude that the rupture of loop IV is induced by metallation rather than preexisting to any large extent in the apo state, i.e., the loop is mainly folded in the apo state and pushed out by the misligation. Due to this rearrangement, the metal ion at the Cu site becomes more solvent accessible than in the correctly folded holo-monomer [1MFM (24)], i.e., 12.6 Å2 vs. 1.0 Å2. Also, the catalytically important R143 is rotated away from its canonical position and adopts a conformation that would clash with the native conformation of A60 in loop IV. Judging from the structure of Zn-site mutated dimeric SOD1 (11), the native H48 rotamer is restored upon dimerization, together with most of the loop IV topology (11). Even so, this restoration leaves the Cu site slightly more solvent accessible than in the fully metallated protein due to a widened access channel. This increased accessibility of the catalytic Cu to small substrates has been coupled to the gain of anomalous redox activity: The Cu ion becomes more rapidly reduced and reacts with NO to catalyze adverse tyrosine nitration (11, 25). It is conceivable that the susceptibility to undergo such noxious side reactions is even higher in the more extensively opened-up, Zn-depleted monomers.
From the perspective of protein-aggregation disease, a second interesting effect of the misligation is that residues 128–131 of loop VII stretch out to interact with the β-barrel of the neighboring molecule in the crystal lattice. Local unfolding leads to a gain of interaction between the monomers that buries ≈560 Å2 of solvent-accessible surface area per chain. This pattern of protein–protein contacts is essentially the same as that observed in the dimeric ALS mutants apo-S134N and apo-H46R (26) (Fig. 4) in the crystal structure of the Zn2+-free SOD1 dimer (11). The fact that the interaction is almost identical in these 4 mutants, representing monomeric and dimeric Zn-deficient forms, which have crystallized with 4 different crystal symmetries, implies that loop VII, unless securely tied down by Zn2+, possesses an intrinsic propensity to interact erroneously with the edge strands of other SOD1 molecules. If so, failure to correctly coordinate Zn2+ would promote both of the suggested causes of ALS, namely, erroneous catalytic function (25) and protein aggregation (3).
Fig. 4.
Loop VII makes amyloid-filament-like interactions with neighboring molecules in the crystals of 4 different SOD1 mutants. (A) Close-up of the interaction between loop VII and 2 edge β-strands of the neighboring molecule. (B) Overview of the crystallographic 2-fold interaction seen in the present work. (C) Superposition of the intermolecular interaction seen in the present work (orange) with the interactions seen in the dimeric structures of the ALS mutants H46R (1OZT, green) and S134N (1OZU, cyan), and the dimeric Zn-free mutant H80S/D83S (2R27, blue). (D) The fibril-like arrangement of protein seen in crystals of the dimeric Zn-deficient SOD1 mutants (Right) and the situation in monomeric Zn-misligated SOD1 (Left).
Loop Dynamics Confirmed by Solution NMR.
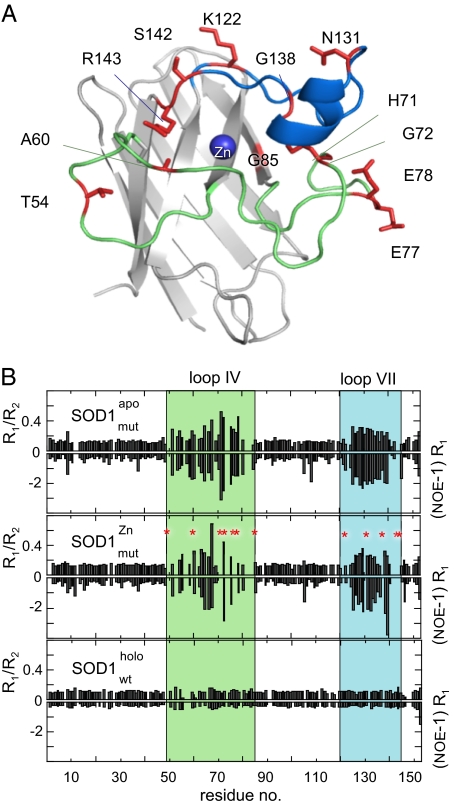
As a final test, we investigated by NMR how the crystallographic data relate to the solution structure of the SOD1 monomer. Comparison of the 1H,15N-HSQC spectra of SOD1pWTapo and SOD1mutapo lacking the zinc ligands reveals no significant structural difference induced by the mutations (SI Text and Fig. S1). Moreover, NMR relaxation measurements can be used to obtain specific information about molecular motion and dynamics. The global rotational correlation time and conformational dynamics of local structural exchange can be estimated from the ratio of the relaxation rates R1/R2 (27). Very fast dynamics however are related to the NOE phenomenon and may be estimated from the product (NOE-1)R1 (27). The results show that SOD1mutapo exhibits high mobility and ns dynamics in loops IV and VII, similar to what is observed for SOD1pWTapo (15) (Fig. 5). According to the folding data these dynamic motions would still occur within a native-like context with restricted access of solvent molecules. Loading of Zn2+ to the Cu site of SOD1mutapo changes the chemical shifts mainly in the loop regions and in the flanking β-sheet (Fig. 5). Consistent with the crystal structure, these chemical-shift changes suggest a structural reorganization of the loops IV and VII and local changes close to the copper-binding site. Whether the chemical-shift changes in the β-strands are due to structural alterations or are just a secondary reflection of the altered loops is not yet clear; by comparison the signals are relatively small. In addition to the chemical-shift changes, several loop resonances of SOD1mutZn display significant line broadening, i.e., the resonance peaks disappear, indicating structural exchange on an intermediate NMR time scale, namely microseconds to milliseconds (Fig. 5). Strikingly, the fast dynamics of the remaining loop resonances remain unaffected by the Zn2+ binding. Taken together these results show that the Zn2+ ion does not suppress the dynamics of loops IV and VII but changes its character. This additional dynamic complexity upon Zn2+ binding to the Cu site is in sharp contrast to the effect of native metal ligation. In holo SOD1 the loop dynamics are severely diminished rendering the loops as rigid as the β-barrel (Fig. 5). Although the precise nature of the loop alterations in Fig. 5 cannot be deduced from the current NMR data, the results show that loops IV and VII are disordered in the solution structure of SOD1mutZn; thus the folding analysis, X-ray structure and NMR data are in full agreement.
Fig. 5.
NMR shows that loop IV and loop VII of the misligated SOD1 monomer exhibit a high degree of backbone dynamics at both fast and intermediate time scales. (A). Structural representation of the SOD1 monomer showing the residues (red) in loops IV and VII that display increased dynamic complexity upon misligation of Zn. The rest of loops IV and VII retain apo-like dynamics. (B) Increased dynamics in the intermediate time scale is indicated by increased values of R1/R2 and increased fast dynamics as increased negative values of (NOE-1)R1. Apo SOD1 with mutated Zn ligands (SOD1mutapo) shows a dynamic pattern that is very similar to that of the apo state of the wild-type monomer. Upon misligation of Zn to the Cu site (SOD1mutZn) the resonances from some residues in loops IV and VII becomes line-broadened beyond detection (red). In the fully metallated wild-type monomer (SOD1pwtholo) (35) loops IV and VII are as fixed as the rest of the protein.
Compromise Between Folding and Function.
The evolution of biological function sometimes occurs at the expense of structural properties. The functional residues could, for example, decrease protein stability or lead to competing interactions in the folding-energy landscape (19, 20). Because of this built-in compromise between function and folding, the functional features are typically developed in connection to loop regions where the energetic and structural penalties of mutations are relatively small (28). From this perspective, SOD1 stands out as a masterpiece. The entire catalytic site is composed of long loops that protrude from either end of the protein to pinch the Cu and Zn ions against one side of the β-scaffold (Fig. 1). Moreover, the detailed geometry of these loops controls the dimer-interface strength (11, 29), allows intersubunit communication (30) and orchestrates metal loading by CCS (12). Function seems to be entirely contained within one piece that is separate from the Ig-like framework. Even so, the high degree of conservation of the “nonfunctional” parts of the SOD1 sequence (31) indicates that the development has come at a price. In this study we verify the existence of such a conflict between function and structure in the SOD1 molecule and uncover some of its details. Most strikingly, the ligands of the Cu site have the conflicting ability to pick up Zn2+ in the denatured state, which leads to misfolding of the SOD1 monomer. It is apparent that one role of the native Zn site is to compensate for such misfolding by restricting the active-site loops. However, the very acquisition of the native Zn site seems associated with a second level of structural penalty: Its ligands destabilize the apo structure. In this context, it is notable that the native Zn2+ is not required for enzymatic activity (32). The primary role of the Zn site seems to be to maintain the structural integrity of the SOD1 Cu site (11). Even if the implication of this functional frustration for ALS is not yet known, it is interesting to note that the protein that is linked to the cause of this disease has such conspicuous tendencies.
Materials
Protein Preparation.
The mutants were constructed on a background of C6A/C111A combined with the dimer-splitting substitutions F50E/G51E (24), coexpressed with yCCS, 3 mM CuSO4 and 30 mM ZnSO4 (33), and purified as described in ref. 5. Apo protein was prepared as in (2) and buffers were 10 mM Mes or Bis-Tris (SIGMA) at pH 6.3. Zn loading was achieved by incubating 50 or 500 μM ZnCl2 with the native protein for 3 h.
Equilibrium and Kinetic Measurements.
Equilibrium unfolding and slow kinetics (logkobs < −2.5) were done on a Varian Cary Eclipse spectrophotometer (Varian) with excitation wavelength 280 nm and emission at 360 nm and stopped-flow measurements as described in ref. 5. SOD1 concentration in the detection volume was in all cases 4 μM (monomer), and all measurements were done at 25 °C, using urea as denaturant (ultra PURE, MP Biomedicals, Inc.). Data analysis was with the software Kalaidagraph. The SOD1 monomer was assumed to fold by a 2-state transition KU/F = [U]/[F] = ku/kf, where KU/F is the equilibrium constant KU/F, and ku and kf are the unfolding and refolding rate constants, respectively (5, 16). Protein stability, ΔGU/F = −2.3RTlogKU/F, was assumed to depend linearly on [urea], yielding logKU/F = logKH2O + mU/F[urea], logkf = logkfH2 O +mf[urea] and logku = logkuH2 O + mu[urea]. The transition midpoint (MP) and mU/F were derived from equilibrium data (Fig. 2) according to
where Fobs is the measured fluorescence, and a and b are sloping base lines (16). The kinetic data (Fig. 2) were fitted to the standard 2-state expression (5)
where kfH2 O and kuH2 O are the rate constants at 0 M urea, and mf and mu the slopes of the refolding and unfolding limbs, respectively.
ITC Measurements.
Isothermal titration calorimetry (ITC) experiments were performed on a MicroCalorimeter (MicroCal) with a sample cell volume of 1.43 mL at 25 °C. The injected volume was 10 μL during 10 s, at intervals of 10 min, and baselines were estimated by Zn-free control. Injection enthalpies and Zn-binding transitions were analyzed by the software MicroCal/Origin (OriginLab) software.
NMR.
Standard 1H,15N-HSQC, T1, T2 and steady state heteronuclear NOE experiments were performed on Bruker 700 and 500-MHz spectrometers (Bruker) equipped with a cryogenically cooled triple resonance probe. All experiments were performed at 25 °C and pH 7.2. Spectra were transformed using nmrPipe and analyzed using the program Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, UCsf). In the T1 and T2 experiments the signal attenuation, from 10 different relaxation delays, was fitted to a single exponential decay using MATLAB (Mathworks).
Crystallization and Data Collection.
Crystallization was at 20 °C by hanging drop vapor diffusion. Drops with 1 μL of protein in water mixed with 1 μL of a reservoir solution containing 16% PEG 6000, 10% Jeffamine M-600 (pH 7.0), 0.1 M Tris·HCl (pH 8.0), and 0.1 M ZnCl2 were equilibrated against 1 mL of reservoir. Crystals grew to a final size of ≈0.1 × 0.1 × 0.1 mm in 10 days. They diffracted to 2.2 Å resolution and belong to space group P3121, with 1 molecule in the asymmetric unit and a solvent content of 30.6%. Further the crystals were soaked with 1 mM methyl mercury chloride for 26 h before data collection to label the potentially reduced Cys-57-Cys-146 disulfide bond. No traces of mercury are found in the structure indicating that the Cys-57-Cys-146 bond is formed (Table S1). The crystals were cryoprotected using 15% glycerol in the mother liquor. All crystallographic data were collected at 100 K at the I911–3 station of MAX-lab, Lund, Sweden. Structure determination was as described in Table S2 and the coordinates have been deposited in PDB with ID code 3HFF.
Supplementary Material
Acknowledgments.
This work was supported by the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Bertil Hållsten Foundation, and Hjärnfonden.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HFF).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812046106/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Lindberg MJ, Byström R, Boknäs N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci USA. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw BF, Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Estevez AG, et al. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 5.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rumfeldt JA, Lepock JR, Meiering EM. Unfolding and folding kinetics of amyotrophic lateral sclerosis-associated mutant Cu,Zn superoxide dismutases. J Mol Biol. 2009;385:278–298. doi: 10.1016/j.jmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Kayatekin C, Zitzewitz JA, Matthews CR. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J Mol Biol. 2008;384:540–555. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stroppolo ME, Malvezzi-Campeggi F, Mei G, Rosato N, Desideri A. Role of the tertiary and quaternary structures in the stability of dimeric copper, zinc superoxide dismutases. Arch Biochem Biophys. 2000;377:215–218. doi: 10.1006/abbi.2000.1780. [DOI] [PubMed] [Google Scholar]
- 10.Mulligan VK, Kerman A, Ho S, Chakrabartty A. Denaturational stress induces formation of zinc-deficient monomers of Cu,Zn superoxide dismutase: Implications for pathogenesis in amyotrophic lateral sclerosis. J Mol Biol. 2008;383:424–436. doi: 10.1016/j.jmb.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Roberts BR, et al. Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J Mol Biol. 2007;373:877–890. doi: 10.1016/j.jmb.2007.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb AL, Torres AS, O'Halloran TV, Rosenzweig AC. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat Struct Biol. 2001;8:751–755. doi: 10.1038/nsb0901-751. [DOI] [PubMed] [Google Scholar]
- 13.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. J Mol Biol. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg M, Tångrot J, Oliveberg M. Complete change of the protein folding transition state upon circular permutation. Nat Struct Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- 15.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu,Zn superoxide dismutase: Role of metal ions in protein folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 16.Fersht AR. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: WH Freeman; 1999. [Google Scholar]
- 17.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: Parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 19.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. Localizing frustration in native proteins and protein assemblies. Proc Natl Acad Sci USA. 2007;104:19819–19824. doi: 10.1073/pnas.0709915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosavi S, Whitford PC, Jennings PA, Onuchic JN. Extracting function from a beta-trefoil folding motif. Proc Natl Acad Sci USA. 2008;105:10384–10389. doi: 10.1073/pnas.0801343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friel CT, Smith AD, Vendruscolo M, Gsponer G, Radford SE. The mechanism of folding of Im7 reveals competition between functional and kinetic evolutionary constraints. Nat Struct Mol Biol. 2009;16:318–324. doi: 10.1038/nsmb.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson CJ, Apiyo D, Wittung-Stafshede P. Role of cofactors in metalloprotein folding. Q Rev Biophys. 2004;37:285–314. doi: 10.1017/S003358350500404X. [DOI] [PubMed] [Google Scholar]
- 23.Pandit AD, Krantz BA, Dothager RS, Sosnick TR. Characterizing protein folding transition States using Psi-analysis. Methods Mol Biol. 2007;350:83–104. doi: 10.1385/1-59745-189-4:83. [DOI] [PubMed] [Google Scholar]
- 24.Ferraroni M, et al. The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited. J Mol Biol. 1999;288:413–426. doi: 10.1006/jmbi.1999.2681. [DOI] [PubMed] [Google Scholar]
- 25.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 26.Elam JS, et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 27.Mäler L, Kowalewski J. Nuclear Spin Relaxations in Liquids. Theory, Experimnets and Applications. New York: Taylor and Francis; 2006. [Google Scholar]
- 28.Brändén CI, Tooze J. Introduction to Protein Structure. New York: Garland; 1991. [Google Scholar]
- 29.Hornberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365:333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Potter SZ, et al. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 31.Sandelin E, Nordlund A, Andersen PM, Marklund SS, Oliveberg M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J Biol Chem. 2007;282:21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- 32.Valentine JS, Pantoliano MW, McDonnell PJ, Burger AR, Lippard SJ. pH-dependent migration of copper(II) to the vacant zinc-binding site of zinc-free bovine erythrocyte superoxide dismutase. Proc Natl Acad Sci USA. 1979;76:4245–4249. doi: 10.1073/pnas.76.9.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahl IM, Lindberg MJ, Tibell LA. Coexpression of yeast copper chaperone (yCCS) and CuZn-superoxide dismutases in Escherichia coli yields protein with high copper contents. Protein Expr Purif. 2004;37:311–319. doi: 10.1016/j.pep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Oliveberg M, Vuilleumier S, Fersht AR. Thermodynamic study of the acid denaturation of barnase and its dependence on ionic strength: Evidence for residual electrostatic interactions in the acid/thermally denatured state. Biochemistry. 1994;33:8826–8832. doi: 10.1021/bi00195a026. [DOI] [PubMed] [Google Scholar]
- 35.Banci L, Bertini I, Cantini F, D'Onofrio M, Viezzoli MS. Structure and dynamics of copper-free SOD: The protein before binding copper. Protein Sci. 2002;11:2479–2492. doi: 10.1110/ps.0210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.