Abstract
Expanded triplet repeats have been identified as the genetic basis for a growing number of neurological and skeletal disorders. To examine the contribution of double-strand break repair to CAG·CTG repeat instability in mammalian systems, we developed zinc finger nucleases (ZFNs) that recognize and cleave CAG repeat sequences. Engineered ZFNs use a tandem array of zinc fingers, fused to the FokI DNA cleavage domain, to direct double-strand breaks (DSBs) in a site-specific manner. We first determined that the ZFNs cleave CAG repeats in vitro. Then, using our previously described tissue culture assay for identifying modifiers of CAG repeat instability, we found that transfection of ZFN-expression vectors induced up to a 15-fold increase in changes to the CAG repeat in human and rodent cell lines, and that longer repeats were much more sensitive to cleavage than shorter ones. Analysis of individual colonies arising after treatment revealed a spectrum of events consistent with ZFN-induced DSBs and dominated by repeat contractions. We also found that expressing a dominant-negative form of RAD51 in combination with a ZFN, dramatically reduced the effect of the nuclease, suggesting that DSB-induced repeat instability is mediated, in part, through homology directed repair. These studies identify a ZFN as a useful reagent for characterizing the effects of DSBs on CAG repeats in cells.
Keywords: DNA repair, gene targeting, triplet repeat instability, zinc finger nucleases
Engineered zinc finger proteins that bind specific DNA targets provide the foundation for a new class of technologies that promise significant gains in the development of novel therapeutics and molecular research tools. The C2H2 zinc finger, the most common DNA-binding domain in eukaryotic genomes, is a 30-aa ββα peptide domain that confers recognition to a triplet DNA sequence (1, 2). Individual zinc fingers have now been engineered in vitro to recognize many DNA triplets (3–6). Zinc fingers are modular, enabling their assembly into linear arrays that can be designed to bind a variety of genome sequences (7–9). These custom zinc finger proteins can be attached to other protein domains to direct various functions to specific locations in the genomes of cells and organisms (10). When fused to the nonspecific DNA cleavage domain of the FokI restriction endonuclease, zinc fingers can direct double-strand breaks (DSBs) to disrupt specific genes (11–14) or in combination with a repair template to induce gene correction via homologous recombination (HR) (15–18). Thus, ZFNs show promise for the treatment of monogenic disorders by promoting the knockout or correction of specific genes.
Here, we describe ZFNs that induce DSBs in CAG·CTG repeat sequences (referred to henceforth as CAG repeats) for the dual purpose of identifying mammalian repair pathways that contribute to repeat instability and to explore the therapeutic potential for shrinking expanded, disease-causing CAG repeat tracts. Expanded CAG repeat tracts are the genetic basis for more than a dozen inherited neurological disorders including myotonic dystrophy, Huntington's disease, and several spinocerebellar ataxias (19, 20). Despite the multitude of pathologies underlying these disorders, they all share common etiology: the expansion of CAG repeats from short benign tracts of <30 repeats to longer, pathogenic lengths that can extend for several thousand repeats. Therapeutic strategies that target this common feature—expanded repeats—in an otherwise diverse group of disorders would have broad applicability.
The mechanisms by which triplet repeats expand to pathogenic lengths remain unclear; however, a contributing cause is the ability of disease-causing repeats to form stable secondary structures that can interfere with aspects of DNA metabolism (21). In Escherichia coli and Saccharomyces cerevisiae, almost any process that exposes single strands of DNA can induce triplet repeat instability, including replication, recombination, and several DNA repair pathways (22). In mammalian cells, we have additionally identified DNA methylation, transcription, and components of mismatch repair and nucleotide excision repair as modifiers of repeat instability (23–26). Deciphering how secondary structures interact with these various processes to trigger repeat instability is an area of active study.
In both yeast and mammalian cells, CAG repeats have been shown to be prone to DSBs (27, 28). To examine whether DSB repair contributes to CAG repeat instability, we designed 2 ZFNs to cooperatively bind and cleave CAG repeat tracts. Here, we describe a ZFN that can cleave CAG repeat tracts in plasmid DNA, as well as in the genomes of human and rodent cells. Treatment with this custom nuclease destabilizes CAG repeat tracts in a length dependent manner, triggering a substantial increase in the frequency of large repeat contractions. The ZFN-mediated effect on CAG repeats in mammalian cells provides proof of principle for the idea that CAG-specific nucleases may be capable of shrinking the expanded CAG repeats at disease loci.
Results
Design of Zinc-Finger Nucleases.
We designed 2 ZFNs to bind and cooperate in the cleavage of a CAG repeat tract, as shown in Fig. 1. Modular assemblies of zinc fingers that recognize GNN triplets have been studied more extensively than those of zinc fingers directed at other triplets, and a recent large-scale study reported fairly high failure rates for modularly designed zinc fingers targeted to non-GNN triplets (29). We therefore chose to link 3 identical GCA-binding domains together with the FokI nuclease domain in 1 nuclease, the zfGCA nuclease, and 3 identical GCT-binding domains with FokI in the other, the zfGCT nuclease. Although a 6-nt separation of the binding sites has been described as optimal, this choice of finger-recognition sites necessitated a spacing of 4, 7, or 10 nt between the zfGCA and zfGCT nucleases. Based on previous results, we chose an amino acid spacer between the zinc fingers and the FokI domain that could accommodate a 7-nt separation of the binding sites and yield a reasonable efficiency of cleavage (15, 30).
Fig. 1.
Schematic for DNA binding by CAG-specific ZFNs. ZFN zfGCA includes a DNA-binding domain consisting of 3 zinc fingers that recognize a triplet GCA sequence, whereas zfGCT recognizes the triplet GCT sequence. The complete binding site is shown in red and can be generalized as: 5′–AGCAGCAGC NNNNNNN GCAGCAGCA–3′, making any CAG repeat of at least 9 copies a target for the nucleases. Each DNA binding domain is fused to a FokI nuclease domain (blue oval) that is inactive until it forms a dimer with a second FokI domain.
Previous in vitro characterization of binding by individual zinc-finger domains that recognize GCT and GCA sequences showed that GCT fingers can sometimes recognize GCA triplets and that GCA fingers can sometimes recognize GCT triplets (3, 5). These results indicated that a homodimer configuration might be sufficient for recognizing and cleaving the repeat tract. Thus, we tested all combinations of nucleases: the heterodimer of zfGCA and zfGCT, and the homodimers of zfGCA and zfGCT.
Cleavage of CAG Repeats in Plasmid DNA.
We constructed plasmid substrates that contain either no CAG repeat tract or a single CAG tract of 38 repeat units, which is greater than the minimum 9 repeats necessary to form a proper recognition site for the zfGCA and zfGCT nuclease pair. To determine whether zfGCA and zfGCT, together or separately, were able to cleave a CAG tract, we incubated the nucleases in various combinations and concentrations with each linearized plasmid. The zfGCA and zfGCT nucleases, singly and in combination, cut the repeat-containing plasmid into fragments consistent with cleavage at the CAG repeat tract (Fig. 2A) but did not cleave the plasmid lacking the repeat (Fig. 2B). Some nonspecific activity at the highest concentration of the nucleases was detected, as indicated by the loss of intensity of some of the bands. Collectively, these results indicate that zfGCA and zfGCT—acting as homodimers—can induce DSBs within CAG repeat tracts, although zfGCT appears to cleave the plasmid somewhat better than zfGCA. We are not aware of another published report of a ZFN that can homodimerize to cleave a target sequence of nonidentical left and right half-sites. However, the result is not completely unexpected given the cross-reactivity of GCA fingers for GCT sequences, and vice versa (3, 5).
Fig. 2.
Cleavage of CAG repeat tracts by ZFNs. The zfGCA and zfGCT nucleases were tested for their ability to cleave CAG repeat tracts, together or separately, at total amounts of 440 ng (1×), 220 ng (0.5×), or 44 ng (0.01x) in 20 μL. The nucleases were incubated with a linearized vector that (A) contains a CAG tract of 38 repeats (4.3 kb), or (B) a vector without a CAG repeat tract (3.9 kb). A 1-kb ladder (L) is shown in the first lane. The second lane (U) contained a reaction with the linearized vector, but no nuclease. Cleavage at the CAG repeat is expected to generate products of 2.5 and 1.8 kb.
Cleavage of CAG Repeats in Mammalian Cells.
To evaluate the mutagenic consequences of inducing DSBs in CAG repeat tracts in mammalian cells, we used 2 previously described selection assays: one uses the APRT gene in CHO cells, and the other uses the HPRT minigene in human cells. In both assays, the reporter genes were initially inactivated by the presence of a (CAG)95 repeat tract within an intron, which interferes with proper splicing and blocks gene expression (24, 31). If the repeat contracts to ≤38 copies in human cells, or ≤34 copies in CHO cells, a sufficient quantity of normal HPRT or APRT mRNA is produced, which restores the activity of the reporter gene and allows cells to survive selection for HPRT+ or APRT+ function (Fig. 3). Counting the surviving colonies provides an assessment of the magnitude of the effect on the repeat tract.
Fig. 3.
Selection assays for CAG repeat instability in human and CHO cells. Long CAG tracts prevent proper splicing, thereby disrupting the production of functional reporter protein. Short CAG tracts are efficiently spliced, allowing sufficient normal reporter protein to be produced. Selection for (A) HPRT+ or (B) APRT+ phenotypes allows for recovery of cells that have undergone a change in the CAG tract.
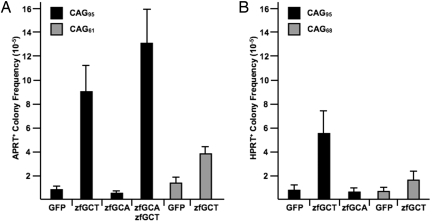
We measured the effect of the zfGCA and zfGCT nucleases on CAG repeat instability by transfecting expression plasmids for the nucleases, or a control plasmid that expresses EGFP, into HPRT(CAG)95 and APRT(CAG)95 cell lines. After 72 h, cells were replated under selection for HPRT+ or APRT+ cells. Expression of EGFP did not affect cell viability or the background frequency of HPRT+ or APRT+ colony formation, nor did expression of either nuclease reduce cell viability (see Materials and Methods). Treatment with the zfGCT nuclease alone increased the frequency of APRT+ colonies 10-fold (P = 0.0004) in CHO cells and increased the frequency of HPRT+ colonies 7-fold (P = 0.0009) in human cells (Fig. 4). By contrast, treatment with the zfGCA nuclease did not increase the frequency of colony formation in human or CHO cells. The combination of zfGCT and zfGCA generated a 15-fold increase in APRT+ colonies, which is a statistically significant increase over zfGCT alone (P = 0.03). These results indicate that the zfGCT nuclease can alter CAG repeat stability in mammalian cells. By contrast, the zfGCA nuclease has no detectable activity by itself and only slightly enhances cleavage in combination with the zfGCT nuclease. The relative ineffectiveness of the zfGCA nuclease in cells is unlikely due to poor expression, because real-time RT-PCR detected ≈85–90% as much zfGCA mRNA as zfGCT mRNA in transfected cells (SI Methods). The contrast between good zfGCA cleavage in vitro and poor cleavage in cells suggests that plasmid digestion may not be a reliable predictor of nuclease activity in cells, as noted by others (9).
Fig. 4.
Induction of CAG repeat instability induced by ZFNs. The ZFNs were evaluated in (A) CHO cells and (B) human cells. The frequencies represent the averages of at least 6 independent experiments, with the exception of the zfGCA+GFP treatments for which there were at least 3 independent experiments. The standard deviations are indicated by the vertical bars. The values (×10−6) for each condition in CHO APRT(CAG)95 cells are: GFP (8.9 ± 2.6), zfGCT+GFP (90 ± 23), zfGCA+GFP (5.0 ± 1.6), and zfGCA+zfGCT (130 ± 31). For CHO APRT(CAG)61 cells, the values are: GFP (15 ± 5.7) and zfGCT+GFP (39 ± 6.6). For the human HPRT(CAG)95 cells, the value were: GFP (8.0 ± 4.5), zfGCT+GFP (55 ± 23), and zfGCA+GFP (4.6 ± 3.3). For the human HPRT(CAG)68 cells, the value are: GFP (8.0 ± 4.1) and zfGCT+GFP (17 ± 7.7).
Length Dependence of Nuclease-Induced Repeat Instability in Mammalian Cells.
Length-dependent modifiers of repeat instability are common (32–35). To explore whether ZFN-induced instability depends on the length of the repeat tract, we transfected the zfGCT nuclease into APRT(CAG)61 CHO cells and human HPRT(CAG)68 cells. Treatment with the zfGCT nuclease induced a 2.6-fold increase in APRT+ colony formation in APRT(CAG)61 cells (Fig. 4A) and a 2-fold increase in HPRT+ colony formation in HPRT(CAG)68 cells (Fig. 4B). Each of these increases is significantly less (P = 0.006 for APRT+ colonies, P = 0.002 for HPRT+ colonies) than observed in the corresponding cells with 95 copies of the CAG repeat. Thus, the zfGCT nuclease destabilizes longer CAG repeats more effectively than shorter ones. This length dependence of the zfGCT nuclease on repeat stability in CHO and human cells is reminiscent of a similar length-dependent response to gamma radiation, which we described previously with these same CHO cell lines (31).
Characterization of Mutation Events.
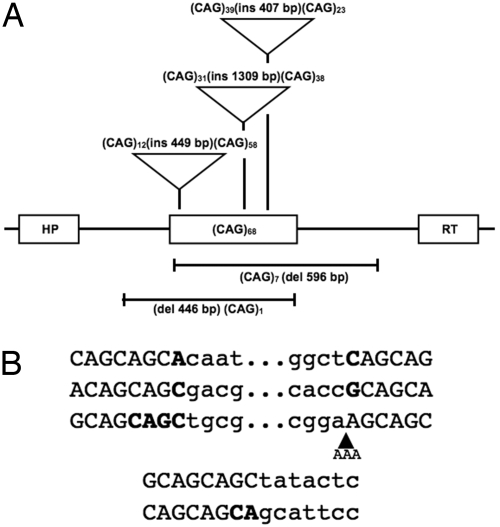
To characterize the changes to the CAG repeats, we sequenced the repeat tracts and flanking DNA in APRT+ and HPRT+ colonies that arose from treatment with the zfGCT nuclease. Previous analyses of positive colonies that arose spontaneously revealed 2 types of event: frequent contractions of the CAG tracts to shorter lengths and occasional deletions that included the repeat sequences and one or both flanks (24, 31). As summarized in Table 1, zfGCT-induced APRT+ and HPRT+ colonies generated contractions (56%) and deletions (20%); however, they also gave rise to a unique class that carried extra DNA inserted into the repeat tract (24%). The insertion and deletion events for HPRT(CAG)68 are illustrated in Fig. 5A. In all 10 characterized insertions, the source of the inserted nucleotides was transfected plasmid DNA. We presume that the inserted sequences permit colony formation by interfering with the ability of the CAG repeats to be spliced into mRNA. We have observed analogous plasmid-derived insertions at DSBs generated by ISceI (36, 37). The deletions and insertions at the CAG repeat tract have junctions that are typical of nonhomologous end joining (NHEJ) (Fig. 5B). The insertion events, in particular, confirm that the zfGCT nuclease induces CAG repeat instability through the production of internal DSBs.
Table 1.
Molecular analysis of zfGCT-induced changes to the CAG repeat tract
| Cell line | Contractions* | Deletions† | Insertions‡ | Total |
|---|---|---|---|---|
| APRT(CAG)95 | 8 | 2 | 4 | 14 |
| HPRT(CAG)95 | 8 | 4 | 3 | 15 |
| HPRT(CAG)68 | 7 | 2 | 3 | 12 |
| Total | 23 (56%) | 8 (20%) | 10 (24%) | 41 (100%) |
*Contractions are defined as shorter, uninterrupted repetitions of pure CAG repeats.
†Deletions are defined as a loss of CAG repeat sequences along with flanking DNA sequences from one or both ends of the repeat.
‡Insertions are defined as an addition of novel DNA sequences at the CAG repeat tract.
Fig. 5.
Detection of zfGCT-induced DSBs within CAG repeats in cells. Deletion and insertion events for the CAG repeat tracts in HPRT+ colonies arising from HPRT(CAG)68 cells are shown. (A) Diagram of deletion and insertion events. (B) The junction sequences for the events shown in A, arranged in the same order from top to bottom. The bold characters indicate microhomologies at the junctions. In one case, an insertion of 3 nt at a repaired junction is denoted by the black triangle.
Mechanisms of Nuclease-Induced Instability in Mammalian Cells.
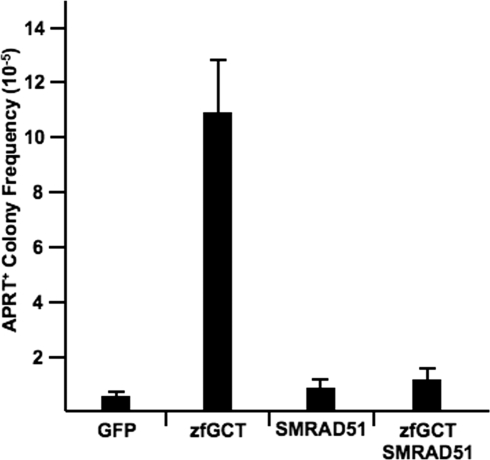
It is likely that the zfGCT nuclease destabilizes CAG repeats via a DNA repair pathway that is initiated in response to DSBs. Two major pathways—NHEJ and HR—can repair DSBs. The deletions and insertions identified among the characterized mutations constitute strong evidence that a portion of the zfGCT-induced breaks must be repaired by NHEJ. The repeat contractions, however, could arise by NHEJ, using individual CAG repeats as microhomology for end joining, or through HR, which could use the sister chromatid as the repair template or anneal complementary single-stranded CAG sequences on either side of the break. To determine the role of HR in zfGCT-induced repeat contractions, we used a mutant form of RAD51, SMRAD51, which blocks templated repair and partially inhibits single-strand annealing in CHO cells (38). Expression of SMRAD51 in APRT(CAG)95 cells did not affect cell viability or the background frequency of APRT+ colony formation, whereas coexpression of SMRAD51 with the zfGCT nuclease substantially inhibited the production of APRT+ colonies (Fig. 6). The extent of inhibition is somewhat surprising, given that 44% of characterized events were either deletions or insertions, consistent with NHEJ (Table 1). Nevertheless, these results suggest that repair of zfGCT-induced DSBs in CAG repeat tracts may involve both the NHEJ and HR pathways.
Fig. 6.
Suppression of zfGCT-induced APRT+ colony formation by SMRAD51. A total of 50 μg of DNA was transfected for each condition. The frequencies represent the average of 6 independent experiments. Standard deviations are indicated as the vertical bars. The average values (×10 − 6) for each condition are: GFP (6.0 ± 2.0), zfGCT (107 ± 21), SMRAD51+GFP (8.6 ± 4.9), and SMRAD51+zfGCT (11 ± 6.3).
Discussion
In this study, we show that ZFNs can cleave CAG repeats in mammalian cells. We assembled 2 ZFNs—zfGCT and zfGCA—that were designed to bind to the complementary strands of CAG repeats, allowing them to heterodimerize through their FokI nuclease domains and cleave the DNA. We found, however, that the individual ZFNs were capable of cleaving plasmid DNA, presumably via homodimerization and misrecognition of GCA triplets by GCT fingers, and vice versa. This result may not be surprising given that the GCT and GCA fingers we used differed by a single amino acid in the recognition segment: QSGDLTR for GCA fingers vs. QSSDLTR for GCT fingers. The underlined residues, which make direct contact with DNA bases, are identical in the 2 ZFNs.
Using selection assays in human and CHO cells, we showed that expression of zfGCT, but not of zfGCA, increased the frequency of HPRT+ and APRT+ cells in treated populations. This stimulation by zfGCT was slightly enhanced by coexpression of zfGCA. As far as we are aware, cleavage of a target sequence composed of nonidentical left and right half-sites by the same ZFN is previously undescribed. This dual recognition means that only a single nuclease need be delivered to induce breaks in CAG repeats, a feature that may prove advantageous for some applications.
Three lines of evidence indicate that zfGCT introduces DSBs into CAG repeat tracts in cells. First, analysis of the altered CAG repeats in the surviving colonies showed that several carried insertions of extrachromosomal DNA within the repeat. In 9 of 10 of these insertions, the lengths of CAG repeat on either side of the inserted DNA sum to about the length of the CAG tract in the parental cells, consistent with insertion at a DSB within the repeat tract. Second, the microhomologies found at insertion and deletion junctions in the altered repeat tracts are consistent with repair by NHEJ, one mechanism for repair of DSBs. Third, expression of a mutant form of Rad51 that interferes with repair of DSBs was found to suppress the ability zfGCT to stimulate HPRT+ and APRT+ colony formation. Collectively, these observations argue that zfGCT cleaves genomic DNA in cells the same way it cuts plasmid DNA in vitro.
These experiments provide a critical proof-of-principle demonstration that ZFNs can be designed to cleave repeat sequences in cells. Long repeat tracts present a special challenge for ZFN design because they offer many potential opportunities for nonproductive binding, with the nuclease domains improperly juxtaposed. For this reason, it was unclear, a priori, whether cleavage of longer tracts would be more efficient, or less efficient, than cleavage of shorter tracts. As we show here, a 30% decrease in CAG tract length reduced cleavage efficiency in cells by 2- to 3-fold. This steep length-dependence suggests, for example, that the extremely long repeats typical of many disease loci would be highly preferred targets for ZFN cleavage, relative to the hundreds of short CAG repeat tracts in the human genome.
The 2 ZFNs that we tested in this study were built by modular assembly of 3 identical zinc fingers, directed either at GCT or at GCA triplets. Modular assembly of identical fingers is unlikely to yield the most active nuclease, given that the binding of individual fingers is known to depend in part on their neighboring fingers in the nuclease (5, 29). Cleavage efficiency of ZFNs can be improved by iterative methods based on phage display or the bacterial 2-hybrid system (39, 40). Recently, ZFNs that had been optimized by the bacterial 2-hybrid method were shown to perform better than those assembled from modules (41). Other, proprietary methods for designing zinc-finger proteins, such as those used by Sangamo Biosciences, might also be used to build improved repeat-specific nucleases. In addition, it is likely that manipulation of the length of the amino acid spacer between the zinc fingers and the FokI nuclease domain, which defines the optimum spacing for dimerization, may also enhance cleavage efficiency (15, 30). These considerations suggest that cleavage efficiency of CAG-specific ZFNs may be amenable to significant improvement.
It is difficult to estimate the absolute cleavage efficiency of zfGCT from the frequencies of APRT+ and HPRT+ cells that arise in our selection assay because the break must be processed in a way that eliminates the ability of the repeat to interfere with splicing of the reporter gene. In the majority of cases (56%) the CAG tract in surviving cells had contracted to a shorter length, one that allows adequate production of normal mRNA. We do not know what fraction of all breaks is healed by the large contractions, deletions, and insertions detectable by our assay, versus the short contractions, expansions, and other repair events to which our assay is blind.
The repeat instability generated by zfGCT-induced cleavage raises the question of what role, if any, DSBs might play in human repeat diseases. In yeast, CAG repeats ranging from 130 to 250 units were shown to induce DSBs at a high enough frequency to be directly detectable by Southern blot analysis (42). In mammalian cells carrying CAG repeats of 98 or 183 units, breaks were detected indirectly via stimulation of gene rearrangements, albeit with a lower frequency than in yeast (28). A low frequency of spontaneous breakage is also supported by the absence of fragile sites associated with the myotonic dystrophy locus from patient cells that carried several thousand CAG repeats (43). In mice with CAG repeat tracts of ≈370 units, the absence of DNA-PKcs (which functions in NHEJ) or of Rad54 (which functions in HR) each had subtle effects on the distribution of repeat lengths found in the quadriceps, an affected muscle with elevated repeat mosaicism in both myotonic dystrophy patients and this mouse model (44). These observations leave open the possibility that DSBs may play a role in CAG repeat instability. zfGCT should prove a useful reagent for probing the mechanism of repair of DSBs in CAG repeat tracts.
A ZFN directed at CAG repeat tracts might also prove useful as a therapeutic reagent for treatment of patients. If DSBs in CAG tracts are repaired predominantly by contraction and deletion, which is consistent with our results, then delivery of a CAG-specific ZFN to affected cells could promote reduction in the size of repeat tracts and alleviate repeat-associated symptoms. Even insertions within a repeat tract may interfere with the pathology of many repeats. Such a reagent should have 2 properties. First, it should cleave CAG repeats with high efficiency and specificity. We do not know the absolute efficiency of cleavage by zfGCT, but it is not toxic to cells, suggesting a reasonable specificity (45). Second, it should preferentially target the expanded CAG repeats that characterize a disease locus, rather than the shorter repeats at the normal locus and the other short CAG repeat tracts scattered throughout the genome. As we have shown here, cleavage by zfGCT exhibits a significant length dependence that should help focus its activity on expanded repeats. Treatments that can promote the reduction of large CAG repeat tracts, or interrupt them, might form the basis for a future therapeutic approach designed to prevent or delay the onset of late-stage neurological disorders caused by expanded CAG repeat tracts (46).
Materials and Methods
Plasmid Construction.
The zinc finger DNA-binding domains were assembled from overlapping primers by PCR (47). Each zinc finger DNA binding domain is based on the previously described CP-1 backbone (48). Our primers incorporated the key recognition amino acids at positions -1 to 6 relative to the start site of the helix portion of the zinc fingers, using QSGDLTR to recognize GCA and QSSDLTR for GCT (5). To produce plasmids with the complete ZFN, the PCR products, ≈300 bases in length, were cloned in-frame into a modified pET15B vector using the NdeI and SpeI restriction sites, to create a fusion of the DNA binding domain to the L6 glycine-serine linker and the FokI endonuclease domain (15). Plasmid pET15B-GCA encodes zfGCA and plasmid pET15B-GCT encodes zfGCT. The pET15B vector drives expression of the nuclease using a T7 promoter and includes an N-terminal 6x-His tag for protein purification. To create expression plasmids for use in mammalian cell culture, the coding sequence for each ZFN was amplified with an extended primer that appended an NLS (PKKKRKV) and an HA tag (YPYDVPDYA) to the N terminus of the sequence. The final PCR product was cloned into the pcDNA 3.1 vector using the Directional TOPO Cloning kit from Invitrogen. Nucleotide sequences of zfGCA and zfGCT are available on request.
Two plasmids were used in the in vitro cleavage assay. The pCR2.1-CAG38 plasmid contains a 370-nt sequence that includes 38 CAG repeats from (31). This sequence was PCR amplified and cloned into the pCR2.1 vector using the TOPO cloning kit from Invitrogen. We confirmed the size and orientation of the CAG repeat sequence by restriction digests and DNA sequencing. The pCR2.1-EMPTY plasmid is the pCR2.1 vector without an insert. The vectors were linearized with NcoI before treatment with the nucleases.
Protein Purification and in Vitro Assay.
Plasmids pET15B-GCA and pET15B-GCT were transformed into BLR21(DE3)pLysS host cells from Novagen to express protein for purification. Cells were grown in the presence of tetracycline, ampicillin, chloramphenicol, and 0.1 mM ZnCl2. The proteins were purified according to procedures described elsewhere (49, 50). The best protein fractions, as determined by a Bradford assay and gel electrophoresis, were pooled and further purified using a Heparin-Sepharose column. After the second purification step, the best final protein fractions were stored for future use in the in vitro assays. Purified ZFNs were quantified from a protein standard curve using a BSA standard and the Protein Assay Reagent from Pierce. To confirm the quantification, protein samples were also analyzed by gel electrophoresis. The in vitro cleavage assays were conducted using equal amounts of the ZFNs, ≈440, 220, and 44 nanograms for each enzyme in a reaction volume of 20 μL. The digestion reactions were carried out as described (9).
Cell Lines and Growth Conditions.
Construction of the APRT(CAG)61 and APRT(CAG)95 CHO cell lines is described elsewhere (31), as is the FLAH25 HT1080 cell line, referred to in this article as HPRT(CAG)95 (24). The HPRT(CAG)68 HT1080 cell line was derived from the same transfection that produced the FLAH25 cell line, but it was found to contain only 68 copies of the CAG repeat. CHO cells were grown in DMEM with high glucose and 10% FCS, nonessential amino acids, pyruvate, penicillin and streptomycin. HT1080 cells were grown in DMEM-F-12 medium (DMEM-F12) supplemented with 10% FCS, nonessential amino acids, pyruvate, penicillin and streptomycin. Cell culture reagents were purchased from Invitrogen. All cells were incubated at 37 °C with 5% CO2. For the HPRT(CAG)95 cell line, the pTRE-CMVmini promoter controls transcription of the HPRT reporter gene, so doxycycline (2 μg/mL) was added to the medium to induce maximal transcription during treatment with the nucleases and during selections (24).
APRT+ CHO cells were selected by plating 500,000 cells on 10-cm plates supplemented with ALASA (25 μM alanosine, 50 μM azaserine, 100 μM adenine) for 2 weeks. For the HPRT(CAG)95 and HPRT(CAG)68 cell lines, HPRT+ cells were selected by plating 500,000 cells on 10-cm plates supplemented with HAT (0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymine) plus doxycycline for 2 weeks, with the addition of fresh doxycycline after the first week of selection. The resulting colonies were then picked for analysis or stained with 1% Coomassie blue for counting. The frequencies of HPRT+ or APRT+ colonies were calculated as the number of colonies surviving selection, divided by the number of viable cells.
Cell Transfections.
Human and CHO cells were transfected at 70–90% confluence in 10-cm culture dishes using 37.5 μL of Lipofectamine-LTX with 15 μL of Plus Reagent and 60 μL of Lipofectamine 2000 (Invitrogen), respectively, with plasmids expressing the zfGCA nuclease, the zfGCT nuclease, or SMRAD51. For each transfection in CHO cells, 25 μg of each plasmid was used along with 25 μg of another plasmid or 25 μg of a control plasmid expressing GFP for a total of 50 μg. Each experiment also included a set of control cells transfected with 50 μg of the GFP plasmid only. In human cells, the total amount of transfected material was 15 μg, using 7.5 μg of each individual plasmid. Opti-MEM (Invitrogen) and a reduced serum medium were used to dilute the plasmids and the Lipofectamine reagent for the transfections. HPRT+ or APRT+ selections were initiated 72 h after transfections. At the same time cells were replated for selection, a sample was plated for cell survival in the absence of selection. Colony formation for human cells averaged 28% for GFP transfections, 32% for zfGCT, and 37% for zfGCA; for CHO cells, they averaged 63% for GFP, 65% for zfGCT, and 72% for zfGCA.
Supplementary Material
Acknowledgments.
We thank Bernard S. Lopez (Institut de Radiobiologie Cellulaire et Moléculaire, Fontenay-aux-Roses Cedex, France) for generously contributing the SMRAD51 expression plasmid. We are grateful to Jon Trautman (University of Utah School of Medicine, Salt Lake City) for performing some of the in vitro assays and to W. Justin Cordill (Baylor College of Medicine, Houston) for his RT-PCR analyses of nuclease expression. We also thank the members of the Wilson and Carroll laboratories for helpful comments and discussion. This work was supported by a T-32 National Eye Institute training grant (EY07001, to D.M.), a National Institute of Diabetes and Digestive and Kidney Diseases training grant (DK007696, to K.S.), and grants from the National Institutes of Health (GM38219 and EY11731, to J.H.W., and GM58504, to D.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902420106/DCSupplemental.
References
- 1.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 2.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 3.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: Selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 6.Dreier B, et al. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 7.Segal DJ, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 8.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 9.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 10.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 11.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 17.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 18.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 19.Albrecht A, Mundlos S. The other trinucleotide repeat: Polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 21.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: Mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 22.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG. CAG trinucleotide repeats in mammalian cells. Hum Mol Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dion V, Lin Y, Hubert L, Jr, Waterland RA, Wilson JH. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum Mol Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankowski C, Nasar F, Nag DK. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc Natl Acad Sci USA. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meservy JL, et al. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol Cell Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez CL, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: The inter-domain linker as a major determinant of target site selectivity. Mol Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorbunova V, et al. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol Cell Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burman RW, Popovich BW, Jacky PB, Turker MS. Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet. 1999;8:2293–2302. doi: 10.1093/hmg/8.12.2293. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier R, Farrell BT, Miret JJ, Lahue RS. Mechanistic features of CAG*CTG repeat contractions in cultured cells revealed by a novel genetic assay. Nucleic Acids Res. 2005;33:5667–5676. doi: 10.1093/nar/gki880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert S, Lopez BS. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 2000;19:3090–3099. doi: 10.1093/emboj/19.12.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebar EJ, Pabo CO. Zinc finger phage: Affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 40.Thibodeau-Beganny S, Joung JK. Engineering Cys2His2 zinc finger domains using a bacterial cell-based two-hybrid selection system. Methods Mol Biol. 2007;408:317–334. doi: 10.1007/978-1-59745-547-3_17. [DOI] [PubMed] [Google Scholar]
- 41.Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- 42.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 43.Wenger SL, Giangreco CA, Tarleton J, Wessel HB. Inability to induce fragile sites at CTG repeats in congenital myotonic dystrophy. Am J Med Genet. 1996;66:60–63. doi: 10.1002/(SICI)1096-8628(19961202)66:1<60::AID-AJMG13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 44.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornu TI, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 46.Gomes-Pereira M, Monckton DG. Chemical modifiers of unstable expanded simple sequence repeats: What goes up, could come down. Mutat Res. 2006;598:15–34. doi: 10.1016/j.mrfmmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Segal DJ. The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods. 2002;26:76–83. doi: 10.1016/S1046-2023(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 48.Krizek BA, Amann BT, Kilfoil VJ, Merkle DL, Berg JM. A consensus zinc finger peptide: Design, high-affinity metal binding, a pH-dependent structure, and a His to Cys sequence variant. J Am Chem Soc. 1991;113:4518–4523. [Google Scholar]
- 49.Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.