Abstract
Genetic studies are refining our understanding of neurodevelopmental mechanisms in autism. Some autism-related mutations appear to disrupt genes regulated by neuronal activity, which are especially important in development of the postnatal nervous system. Gene replacement studies in mice indicate that the developmental window to ameliorate symptoms may be wider than previously anticipated.
Autism is classified as a “pervasive developmental disorder”—pervasive because it affects many aspects of cognition and behavior and developmental because autistic symptoms emerge during development from infancy or perhaps from birth. Yet, a key question about autism is at what stage during brain development does the primary lesion occur? Several recent findings in genetic models of autism suggest that substantial improvement in some behavioral or neurobiological defects can result from gene replacement or pharmacological treatment after “development” is largely complete. Does this mean that autism is less a fixed defect in development, and more a disorder of late postnatal development, plasticity, or even adult function? Here, we consider the role of genes and early development in autism, providing a context for consideration of treatment strategies.
Genes and Autism
Autism is regarded as a highly genetic disorder, although the genes involved have proved difficult to identify (see Essay by D.H. Geschwind, page 391 of this issue). A focus on the genetic nature of autism provides a unique opportunity to consider its pathophysiological mechanisms. Perhaps surprisingly, indistinguishable autistic disorders can clearly be caused by many different genetic changes, a phenomenon generally referred to as genetic heterogeneity. The first genes implicated in autism were associated with broader syndromes that included autistic symptoms rather than with pure or nonsyndromic autism (for which the child psychiatrist Leo Kanner is generally credited with providing the original description). For instance, prominent autistic symptoms accompany a genetic metabolic disorder called phenylketonuria (PKU) (Fombonne, 1999). In addition, children with mutations in the genes associated with tuberous sclerosis (TSC1, TSC2) or in the PTEN tumor suppressor gene show prominent autistic symptoms, although these patients would not generally be diagnosed with autism because they show broader symptoms (such as tumors or benign malformations, called hamartomata, of many structures including brain, skin, and kidney). Deletions of chromosome 22q11 (the region normally associated with velo-cardiofacial syndrome) are seen in about 1% of autistic children but again are typically accompanied by other features as well (cleft palate, congenital heart disease). Inverted duplications of proximal chromosome 15q are also associated with autistic symptoms but often cause prominent mental retardation as well.
Important clues about the mechanisms underlying autism come from the monogenic disorders Rett’s syndrome and Fragile × syndrome (see Essay by Kelleher and Bear, page 401 of this issue). Rett’s syndrome, caused by mutations in the human MECP2 gene, which encodes the methyl-cytosine binding protein, occurs almost exclusively in girls (mutations in this X-linked gene are lethal to males). Affected girls show normal development for 1–2 years, followed by the appearance of stereotyped repetitive hand movements and a regression of neurological and social skills. Similarly, one of the best-known disorders associated with autistic symptoms is the Fragile × syndrome. Males with Fragile × syndrome typically have developmental delays (for example, delays in walking or language) and somatic dysmorphology (for example, large ears, a large head), and so Fragile × is generally not referred to as autism per se. Although there are many syndromic forms of autism, it is also important to stress that 40% to 60% of autistic children show some degree of mental retardation (Giacometti et al., 2007), although measuring IQ in autism may pose challenges. Also, some pediatric neurology clinics report that ~30% of autistic patients suffer from seizures (Fombonne, 1999).
As clinical genetic testing becomes more frequent, more autistic children will receive specific genetic diagnoses. The same genetic defects associated with broader syndromes are now recognized to cause a spectra of abnormalities, including those found in some higher-functioning children with autism. For instance, deletion of 22q11 (Vorstman et al., 2006), Fragile × gene abnormalities, or milder MECP2 mutant alleles (Moretti and Zoghbi, 2006) are occasionally seen in children without the typical broader syndromes associated with these conditions. Why similar genetic abnormalities cause a range of phenotypes is an enduring mystery and may reflect the involvement of other genetic or nongenetic factors.
One of the areas that has seen the most rapid recent progress in autism genetics has come with the realization that up to 7%–10% of children with autism have a variety of de novo chromosomal deletions and duplications (Sebat et al., 2007;Marshall et al., 2008; Kim et al., 2008). These deletion syndromes typically cause a spectrum of phenotypes that includes autism. Some chromosome aberrations have been helpful in identifying specific genes mutated in autism, and some of these genes appear to have brain-specific functions. For instance, × chromosome deletions implicated the NLGN3 and NLGN4 genes (Jamain et al., 2003) encoding neuroligins 3 and 4, which are synaptic adhesion molecules. Subsequent studies identified an NLGN4 mutation inherited in a large family with many affected males showing mental retardation and/or autism (Laumonnier et al., 2004). These findings suggest that NLGN3 and NLGN4 gene mutations are causes, albeit rare, of autism, mental retardation, and potentially other neuropsychiatric syndromes. The SHANK3 gene, which encodes a cytoplasmic binding partner of the neuroligins, was also identified based on a chromosomal deletion (Durand et al., 2007). Chromosome deletions (Kirov et al., 2007; Moessner et al., 2007) and translocations (Kim et al., 2008) involving the NRXN1 neurexin gene, which encodes an extracellular binding ligand for neuroligins, have implicated this gene in autism as well. Rare changes in CNTNAP2, encoding contactin associated protein-like 2 (a neurexin superfamily member), are associated with autism, and common alleles of CNTNAP2 may increase the risk of developing autism. Even these mutations, which can affect the neuronal synapse, tend not to be highly specific for “pure autism” but rather cause a broader mental retardation phenotype in some patients and pure autism in others.
Why do many mutations associated with autism occur de novo, meaning they are present in the child but not in the parents or earlier ancestors? This is actually not surprising. Autistic children rarely marry and have children, so that the condition is subject to “negative evolutionary selection,” with affected patients having decreased fertility. This negative selection means that for mutations to be observed in the population they must frequently occur as de novo mutations. Such negative evolutionary selection is common to mutations associated with other severe diseases, especially those of childhood, for example mental retardation, epilepsy, or congenital heart disease (Lupski, 2007). A recent study focused on identifying autosomal recessive loci in pedigrees with autism and recent shared ancestry (Morrow et al., 2008). Autosomal recessive mutations, like sporadic mutations, also frequently reduce reproductive fitness. In this study, consistent with the finding of copy number variations (CNVs) in autism, multiple heterogeneous loci were implicated, but these loci represent potential inherited causes. Hence, by tracing shared ancestry this approach may provide an important way to identify inherited loci in heterogeneous neurodevelopmental conditions that reduce reproductive fitness.
Is it unusual that mutant forms of many different genes cause an overlapping or indistinguishable autistic phenotype? On the contrary, it appears to be the rule, at least for neurological disorders of the cerebral cortex, such as dementia, mental retardation, or epilepsy. Disorders that affect the cerebral cortex and its associated forebrain neural systems may uniquely define this genetic heterogeneity, perhaps because the forebrain requires many genes (likely >10,000) for normal development and function, yet the plasticity of forebrain structures appears tailor-made to redistribute and remap critical functions in response to perturbation. Hence, the cerebral cortex may provide essentially a “plastic reserve” to ameliorate genetic defects, which in turn results in there being a small number of stable, recognizable, abnormal states that multiple mutant genes converge upon.
Gleaning Cellular Mechanisms from Genetics
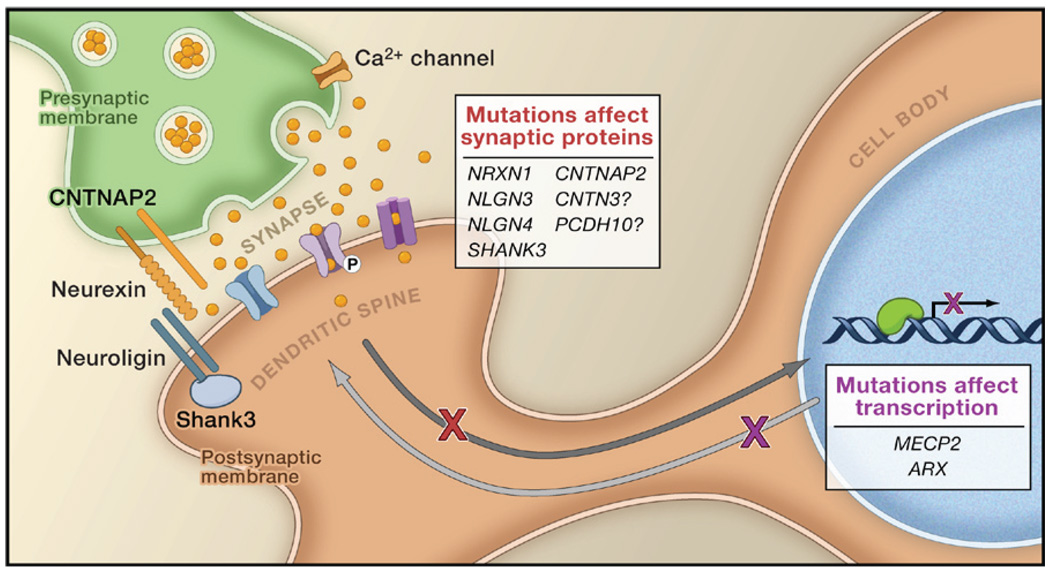
When surveying the cellular mechanisms that flow from the genetic defects associated with autism, one of the most striking revelations is the important role of genes that encode proteins of the neuronal synapse (Zoghbi, 2003) (Figure 1). NLGN3/NLGN4 and NRXN1 encode a ligand-receptor pair that mediates synaptic development, and SHANK3 encodes a cytoplasmic adaptor protein that binds to neuroligin (Durand et al., 2007) . On the other hand, synaptic activity-related signals are transduced into alterations in protein synthesis within the synaptic spine, and several other genes associated with autism mediate these changes in protein synthesis, including FMR1, PTEN, and TSC1/TSC2 (see Essay by Kelleher and Bear, page 401 of this issue). Hence, the importance of this synaptic activity-related response is very clear. Interestingly, a recent study suggested that the synapse-promoting function of neuroligin 1 in mice is dependent on synaptic activity (Chubykin et al., 2007). Autism genes identified in another genome-wide study showed significant association with those identified in an independent screen for genes regulated by neuronal activity or by transcription factors regulated by neuronal activity (Morrow et al., 2008). In this study, the transcription factor MEF2 was found to regulate two genes implicated in the whole-genome study of autism pedigrees, namely PCDH10 (protocadherin10, another neuronal cell adhesion protein) and DIA1 (deleted-in-autism1, also known as c3orf58, a new protein found in the Golgi) (Figure 1). Finally, consistent with the association of activity-regulated genes with autism, a conditional MEF2c knockout mouse has abnormalities in synaptic maturation and behavior related to Rett’s syndrome, an autism-related disorder (Li et al., 2008).
Figure 1. The Potential Relationship of Autism Genes to Synaptic Function.
Synaptic activity stimulates signaling pathways that ultimately result in the binding in the nucleus of trancsciption factors to specific DNA sequences necessary to activate target genes, Which in turn modify synaptic efficacy. Gene mutations associated with autism affect synaptic proteins such as neuroligins 3 and 4 (NLGN4, NLGN4), neurexin 1 (NRXN1), contactin associated protein-like 2 (CNTNAP2), and SHANK3. These proteins include adhesion molecules of the synapse or cytoplasmic molecules that associate with synaptic receptors. In addition, some genes, such as MECP2 and ARX, which encode proteins that regulate DNA transcription, are associated with autistic features when mutated. Moreover, some of the targets of these activity-dependent transcription factors are mutated in recessive autism. Some autism mutations appear to involve not the coding sequence of genes but rather noncoding sequences that may regulate patterns of gene transcription. For example, PCDH10 and CNTN3 encode proteins that are potentially associated with the synapse yet autism mutations do not alter the coding part of these genes. Together these data suggest that heterogeneous causes of autism may be associated with alterations in activity-dependent synaptic plasticity.
Other genes associated with autism seem to affect neuronal excitability, or the ratio of excitation to inhibition (Rubenstein and Merzenich, 2003). Indeed, mutations in neuroligins appear to affect the balance between excitatory and inhibitory synapses (Tabuchi et al., 2007). MeCP2 mutations in mice also appear to regulate the balance of excitatory and inhibitory synapses (Chao et al., 2007; Dani et al., 2005). Furthermore, evidence is accumulating that several genes associated with autism play roles in the development of cortical inhibitory interneurons including c-Met (Campbell et al., 2006, 2007), DLX (Hamilton et al., 2005), and ARX, where rare, partially inactivating mutations cause mental retardation or autism (Kato et al., 2004; Turner et al., 2002). A common theme of these studies is that autism may relate less to generic synaptic function and more to levels of excitation or inhibition, and the balance between the two.
One unifying hypothesis, consistent with current findings, is that genes associated with autism may encode proteins that mediate activity-dependent changes in neuronal function (Hong et al., 2005; Zoghbi, 2003). These activity-dependent changes require gene transcription and ultimately result in synaptic plasticity, learning, and memory formation. A recent analysis of recessive autism mutations shows that 5% of these represent homozygous deletions, completely removing both copies of specific gene segments (Morrow et al., 2008). Homozygous deletions often occur near activity-regulated genes, either removing the gene entirely or sometimes removing only noncoding sequences outside the region of the gene that codes for the protein. In addition, two other studies have implicated noncoding variants (potentially with regulatory activity) with autism (Campbell et al., 2006; Sutcliffe et al., 2005). These presumptive regulatory mutations may ultimately complete an “autism pathway” by showing autismassociated disruptions at each step from the synapse to activity-dependent gene activation. Hence, autism may reflect a disorder of regulation of gene expression level under specific circumstances, such as activity-dependent learning. This may explain the sometimes milder syndrome of autism compared to mental retardation, where specific genes are typically inactivated completely by mutations that disrupt those parts of the gene that encode the protein.
The Specificity of Autism
Whereas synaptic changes presumably underlie all forms of learning, the synaptic view of autism does not provide an explanation for the preferential loss of linguistic and social functions in autism. Although there is little data on this point, several hypotheses seek to explain this specificity. In one view, specific neural circuits essential for social behavior (Belmonte and Bourgeron, 2006; Levitt et al., 2004; Rubenstein and Merzenich, 2003) are preferentially disrupted by mutations in either synaptic or other proteins. For example, vasopressin and oxytocin regulate social behavior in mammals (Caldwell et al., 2008; Campbell, 2008), and hence synaptic defects in oxytocin and vasopressin pathways could cause defects in human social behavior (Insel and Young, 2001). Such a social learning model may also relate to the observed male-female differences in autism prevalence (boys are typically affected at least four times more commonly than girls). The basis for this male-female difference in autism prevalence is not understood, but one hypothesis is that the male brain may be built in such a way as to be particularly susceptible to abnormalities in pathways that control social behavior (Baron-Cohen and Belmonte, 2005; Gomot et al., 2006). In addition, some of the cognitive features of higher-functioning autistic patients (with splinter skills or relatively spared abilities) or even the outstanding gifts of so-called autistic savants remain to be explained.
Another hypothesis to explain the specificity of autism for affecting language and social behavior holds that these are simply the most difficult tasks ever learned by the human brain, so that mild global defects in synaptic function may be sufficient to impair them. Language acquisition, at ~2 years of age is one of the last developmental milestones, whereas learning about social relationships continues into adulthood. The late appearance of these landmarks may reflect the complex circuitry they require, which may be susceptible to subtle defects that spare simpler tasks. Alternatively, genes implicated in autism may include those that have been under recent evolutionary selection (Baron Cohen and Belmonte, 2005). After all, the ability of humans to live in large social groupings—that is, civilization—was a recent evolutionary development, occurring only after the appearance of anatomically modern humans, and probably as recently as 50,000 years ago. There is evidence that some genes (Williamson et al., 2007) and genomic regions (16p11.2) in which mutations cause autism show signatures of recent evolutionary selection.
A consideration of autism as an evolutionarily recent disorder also suggests that disruption of evolutionarily new parts of the brain may relate to autism. Among these more newly evolved regions of the brain is the frontal cortex (Hill and Walsh, 2005), and recent evidence shows that disorders of frontal cortex development produce social defects. For example, the secreted signaling protein Fgf17 is essential for normal development of the frontal cortex of mice (Cholfin and Rubenstein, 2007). Fgf17 mutations in mice result in defects in social behavior, which may be a model for autism (Scearce Levie et al., 2007). Fgf17 signaling uses receptor tyrosine kinases and therefore is likely to be modulated by proteins encoded by TSC1, TSC2, and PTEN; perhaps mutations in this signaling pathway have early effects on frontal cortex patterning, as well as later effects on synaptic signaling, each contributing susceptibility to autism.
Apparent Reversal of Neurobiological Defects in the Adult
A remarkable finding in some recently studied animal models of autistic symptoms has been the surprising degree to which autistic symptoms have been apparently reversed or ameliorated by replacing or modulating gene function after birth or even in the adult. The first example of this came with a Drosophila model of Fragile × syndrome (McBride et al., 2005). Normally, the mutant flies have a defect in social/courtship learning and also have defects in the axons of neurons clustered in a brain region known as the mushroom body. The Fragile × disease pathway was modulated by administration of several drugs including metabotropic glutamate antagonists that can decrease the excessive protein translation caused by mutations in FMR (the protein mutated in Fragile × syndrome; see the Essay by Kelleher and Bear, page 401 of this issue) or even using lithium carbonate, which modulates downstream signaling of the Fragile × pathway through the kinase GSK-3β. Remarkably, in flies born with fixed defects in social behavior the phenotype could be rescued almost completely in adulthood by administration of metabotropic glutamate antagonists or even lithium (McBride et al., 2005).
An even more remarkable example of the reversibility of autism-related genetic and neurobiological defects comes from the study of mouse models of Rett’s syndrome. The Bird and Jaenisch labs created mice that lacked MeCP2, and when the mice were born they developed the typical symptoms of Rett’s syndrome (Giacometti et al., 2007; Guy et al., 2007). Remarkably, reinstating the MeCP2 gene after birth caused partial (Giacometti et al., 2007) or seemingly complete (Guy et al., 2007) resolution of the abnormalities. These findings suggest that MECP2 is not essential for the earliest wiring of the nervous system but instead is required later for activity-dependent processes. This is consistent with the normal development of Rett’s syndrome in girls until 18–24 months of age, and it highlights the potential for reversing the symptoms of Rett’s syndrome in humans, if effective therapies can be found.
The apparent phenotypic reversibility of some autism-associated genetic defects suggests a refinement of autism as a paradigmatic “developmental disorder,” as it may be partially distinct from mental retardation. Development of the nervous system is characterized by several phases that occur in sequence. Early stages of neuronal production, axonal outgrowth, and initial connectivity are driven by intrinsic genetic programs, which are largely independent of synaptic activity (Sur and Rubenstein, 2005). At later stages, refinement of synapses relies on neuronal activity, which in turn induces activitydependent transcriptional changes that underlie synaptic plasticity and ultimately learning (Hong et al., 2005). Mental retardation often reflects mutations in genes that sculpt early development of brain architecture, leading to fixed defects in connectivity. On the other hand, defects in autism appear more closely tied to later developmental steps that depend on synaptic activity and activity-dependent changes. This may explain the later age of diagnosis of autism, the preferential effects on skills that appear later in childhood like language and social behavior, and perhaps the greater likelihood of improvement in some fortunate children. The later onset of autism-specific defects in synaptic activity is an optimistic sign that, if we can develop medications to modulate these synaptic changes, we may be able to provide better therapies for this devastating disorder.
ACKNOWLEDGMENTS
C.A.W. is supported by Autism Speaks (NAAR), the NIMH (1R01 MH083565), the Nancy Lurie Marks (NLM) Family Foundation, the Simons Foundation, and the Anne and Paul Marcus Foundation and is an Investigator of the Howard Hughes Medical Institute. E.M.M. is supported by the NIMH (1K23MH080954) and the Rappaport Research Scholarship in Neuroscience at MGH and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. J.L.R.R. is supported by Nina Ireland, Autism Speaks (NAAR), Cure Autism Now, the Simons Foundation, the Althea Foundation, Larry L. Hillblom Foundation, NINDS (R01 NS34661), and NIMH (RO1 MH49428).
REFERENCES
- Baron-Cohen S, Belmonte MK. Annu. Rev. Neurosci. 2005;28:109–126. doi: 10.1146/annurev.neuro.27.070203.144137. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Bourgeron T. Nat. Neurosci. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- Caldwell HK, Lee HJ, Macbeth AH, Young WS., 3rd Prog. Neurobiol. 2008;84:1–24. doi: 10.1016/j.pneurobio.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Biol. Psychol. 2008;77:1–10. doi: 10.1016/j.biopsycho.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, et al. Proc. Natl. Acad. Sci. USA. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, D’Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, Persico AM. Ann. Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Proc. Natl. Acad. Sci. USA. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Südhof TC. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Proc. Natl. Acad. Sci. USA. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, et al. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fombonne E. Psychol. Med. 1999;29:769–786. doi: 10.1017/s0033291799008508. [DOI] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Proc. Natl. Acad. Sci. USA. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomot M, Bernard FA, Davis MH, Belmonte MK, Ashwin C, Bullmore ET, Baron-Cohen S. Neuroimage. 2006;29:475–484. doi: 10.1016/j.neuroimage.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SP, Woo JM, Carlson EJ, Ghanem N, Ekker M, Rubenstein JL. BMC Genet. 2005;6:52. doi: 10.1186/1471-2156-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RS, Walsh CA. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Hong EJ, West AE, Greenberg ME. Curr. Opin. Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Insel TR, Young LJ. Nat. Rev. Neurosci. 2001;2:129–136. doi: 10.1038/35053579. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Paris Autism Research International Sibpair Study. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Das S, Petras K, Kitamura K, Morohashi K, Abuelo DN, Barr M, Bonneau D, Brady AF, Carpenter NJ. Hum. Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Am. J. Hum. Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O’Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. Hum. Mol. Genet. 2007;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Trends Neurosci. 2004;27:400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Proc. Natl. Acad. Sci. USA. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Nat. Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, et al. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. Curr. Opin. Genet. Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scearce-Levie K, Roberson ED, Gerstein H, Cholfin JA, Mandiyan VS, Shah NM, Rubenstein JL, Mucke L. Genes Brain Behav. 2007;7:344–354. doi: 10.1111/j.1601-183X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L, Li C, Folstein SE, Blakely RD. Am. J. Hum. Genet. 2005;77:265–279. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Partington M, Kerr B, Mangelsdorf M, Gecz J. Am. J. Med. Genet. 2002;112:405–411. doi: 10.1002/ajmg.10714. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, Beemer FA, Swaab H, Kahn RS, van Engeland H. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]