Abstract
NLR (nucleotide-binding domain, leucine-rich repeat) proteins are intracellular regulators of host defense and immunity. One NLR gene, NLRP12/Monarch-1, has emerged as an important inhibitor of inflammatory gene expression in human myeloid cells. This is supported by genetic analysis linking the loss of a functional NLRP12 protein to hereditary periodic fever. NLRP12 transcription is diminished by specific TLR stimulation and myeloid cell maturation, consistent with its role as a negative regulator of inflammation. The NLRP12 promoter contains a novel Blimp-1/PRDM1 binding site, and Blimp-1 reduces NLRP12 promoter activity, expression and histone 3 acetylation. Blimp-1 associates with the endogenous NLRP12 promoter in a TLR-inducible manner and mediates the down-regulation of NLRP12 expression by TLR agonists. As expected, the expression of NLRP12 and Blimp-1 is inversely correlated. Analysis of Blimp-1-/- murine myeloid cells provides physiologic evidence that Blimp-1 reduces NLRP12 gene expression during cell differentiation. This demonstrates a novel role for Blimp-1 in the regulation of an NLR gene.
Introduction
The recognition of microbial components during host infection is a key step in activating the innate immune response followed by the induction of inflammatory gene expression. Pattern-recognition receptors exemplified by Toll-like receptors (TLRs) are key regulators in host response to bacterial, viral and fungal components (1, 2). Central to this response is the ability to upregulate genes involved in host protection, innate and adaptive immune cell recruitment and pathogen clearance. Although the TLR-mediated inflammatory response is critical for providing immune defense against pathogens, dysfunctional responses may lead to both acute and chronic inflammatory states manifested as sepsis, inflammation and autoimmunity (3). Therefore, the intensity and duration of TLR responses must be tightly controlled.
In addition to TLR, recent research has focused on the discovery and characterization of a new immune gene family, the NLR (nucleotide binding domain, leucine rich repeat containing) gene family (4), also known as CATERPILLER, NOD-LRR, NACHT-LRR or NOD-like receptor (5-8). Mutations in several NLR genes have been associated with human disease states including autoimmunity and inflammation. Members of the NLR gene family are involved in regulating cellular activation after exposure to specific or multiple pathogen-derived products (9). NOD2 mediates cellular responses to peptidoglycan derived muramyl dipeptide (MDP), leading to activation of inflammatory cytokines and the release of antimicrobial peptides from cytosolic granules (10-12). Nlrp1b/Nalp1b, regulates disease susceptibility of some murine strains to anthrax lethal toxin (13). NLRC4/IPAF mediates caspase-1 and IL-1 processing in response to the flagellin of Salmonella typhimurium and Legionella pneumophila (14-16). Naip5 mediates host susceptibility to the intracellular pathogen Legionella pneumophila (17). NLRP3/cryopyrin/NALP3 mediates caspase-1 and IL-1 processing in response to an array or stimuli (18-21). The more recently described NLRX1 gene negatively regulates the intracellular type I interferon signaling pathway in mitochondria (22). Taken together these data suggest that the NLR genes are involved in regulating a variety of host defense processes.
The relevance of NLR genes is most apparently revealed by the genetic analysis of patients suffering from immune and inflammatory disorders. Mutations in CIITA result in the immunodeficiency Type II Bare Lymphocyte Syndrome (Group A) (23). NOD2 mutations are associated with increased susceptibility to inflammatory Crohns’ disease, and to a granulomatous disorder known as Blau syndrome (24-26). Hyperactive mutants of the NLRP3 (CIAS1) gene predispose patients to a variety of autoinflammatory disorders classified as cryopyrin-associated periodic syndrome (CAPS), which falls under the general classification of hereditary periodic fever (HPF) (27-32). Mutations in NLRP1/NALP1 has been genetically linked with vitiligo-associated autoimmune disease (33) while NLRP7/NALP7 has been associated with hydatidiform mole (34). These studies underscore the contention that NLR family members are important players in both the maintenance of normal immune responses and the onset of inflammatory disorders.
We and others have identified an NLR family member Monarch-1/PYPAF7, recently renamed NLRP12 (35-37). NLRP12 is expressed primarily by cells of the myeloid-monocytic lineage, including monocytes, granulocytes and eosinophils in humans (36). Although it has been suggested that some NLR proteins are involved in “sensing” microbial components, currently there is no known ligand for NLR proteins in general, and NLRP12 in specific. In vitro studies in human cells employing both gene transfection and small heteroduplex RNA (shRNA)-mediated gene silencing suggest that NLRP12 functions as a negative regulator of TLR and tumor-necrosis factor receptor (TNFR) induced NF-κB signaling in human cells. NLRP12 blocks IRAK-1 hyperphosphorylation/activation (37) and facilitates the degradation of NF-κB inducing kinase (NIK) leading to reduced NF-κB activation (38). A recent study of patients with the clinical manifestation of HPF but without mutations in several HPF-linked genes revealed a new genetic link to NLRP12 (39). In contrast to the genetic linkage of NLRP3 and CAPS wherein a variety of hyperactive, single nucleotide missense mutants have been found in patients, the known disease-linked NLRP12 mutants are represented by a splicing variant that caused an 11 residue insertion and truncation of the LRR region in one family, and a nonsense mutation that truncated a portion of the NBD domain and all of the LRR region in a second family (39). These truncated NLRP12 proteins failed to inhibit NF-κB activation, supporting the conclusion that NLRP12 exhibits an inhibitory function.
In contrast to most innate immune genes, human NLRP12 expression is down-regulated after activation of myeloid cells with either TLR2 or TLR4 agonists, M. tuberculosis, or exposure of cells to TNFα or IFNγ (36, 37). Based on our current understanding of the function of NLRP12, we hypothesize that NLRP12 is present prior to immune stimulation to maintain a quiescent phenotype, but during infection its expression is reduced to allow for a full inflammatory response to occur. At the center of this hypothesis is the downregulation of NLRP12 by TLR and during states of myeloid cell differentiation. However, the mechanism by which TLRs and myeloid cell differentiation processes regulate NLR gene expression is an uncharted territory. In this work, we show that the transcriptional silencer, Blimp-1/PRDM1 is involved in the transcriptional repression of NLRP12 after TLR activation or after myeloid cell differentiation.
Blimp-1 is a DNA binding factor that recruits histone deacetylases, histone lysine methyltransferases, histone arginine methyltranserses and co-repressors to induce promoter silencing (40-43). The role for Blimp-1 in regulating B cell differentiation into antibody-producing plasma cells has received the most attention (44). However, the role of Blimp-1 extends significantly beyond B cell differentiation, as it also controls T cell homeostasis and activation (45, 46). Most relevant to this work, Blimp-1 expression is induced during differentiation of myeloid cell lines, whereby it reduces c-myc expression and promotes the upregulation of myeloid-monocytic markers, CD11b and CD11c (47).
In this report we identify Blimp-1 as a key regulator of TLR-mediated, and differentiation-associated reduction of NLRP12 in human myeloid-monocytic cells. We propose that Blimp-1 is important in restricting NLRP12 expression, thereby modulating the extent by which NLRP12 inhibits inflammatory signaling. These data provide a new linkage between Blimp-1 and the innate immune TLR and NLR pathways.
Materials and Methods
Mice
Prdm1F/F and prdm1F/- mice (48) were crossed with Rosa26ERCre/+ mice to generate controls (Rosa26+/+prdm1F/F ) and experimental (Rosa26ERCre/+prdm1F/F) mice. Mice were injected with 1.5ml of 10mg/ml tamoxifen in sunflower seed oil (0.5ml/day over three days) to induce gene deletion 3 weeks prior to tissue harvest. Mice were housed in the barrier facility of Columbia University (New York, New York). All experiments were approved by the institutional animal care and use committee of Columbia University.
Reagents
LPS derived from Escherichia coli 011:B5 (Sigma-Aldrich) was used at a final concentration of 100 ng/ml; phorbol myristate acetate (PMA) (Sigma-Aldrich) was used to differentiate cells at a concentration of 50 ng/ml. Antibodies included anti-Flag M2 (Sigma); anti-p65 (Rockland); anti-Blimp-1 (AbCam), anti-acetylated-H3 (Upstate); mouse IgG (Upstate).
Tissue culture cells and conditions
HEK293T, undifferentiated HL-60 cells, undifferentiated U937 cells and primary adherent blood cells were maintained as previously described (37) . All cells were grown at 37°C with 5% CO2. HL-60 cells were differentiated with 1.25% DMSO as described below. U927 cells were stimulated with 100 ng/ml LPS or differentiated with 50 ng/ml PMA.
Primary cell isolation and stimulation
Human granulocytes were isolated from buffy coats (American Red Cross) using the Lymphocyte Separation Media (ICN, Costa Mesa, CA). For adherent cell purification, cells were plated and allowed to adhere for 2 hours at 37°C. After three washes with PBS to remove non-adherent cells, adherent cells were stimulated for the indicated time points with 100 ng/ml LPS from Escherichia coli (LPS 011:B5; Sigma-Aldrich), or where indicated 100 ng/ml K12 LPS, 0.1 μg/ml Pam3Cys, 1 × 108 cells/ml HKLM or 1 μg/ml Flagellin (Invivogen).
Isolation and differentiation of murine bone marrow cells
Bone marrow cells from either wild-type C57BL/6N or treated /Rosa26//^ERCre/+ prdm1//^flox/flox/ and /Rosa26//^+/+ prdm1^flox/flox / mice were flushed from the tibiae and femurs and cultured at 1 × 106 cells/ml in MEM medium supplemented with 5% GM-CSF [culture supernatant from X63 cells transfected with murine GM-CSF cDNA (provided by Dr. Chris Nicchitta, Duke University, Durham, NC)]. Total cells (adherent and non-adherent) were isolated at the indicated time points and RNA was prepared using Trizol Reagent (Invitrogen) and analyzed for NLRP12 expression by qPCR.
Expression plasmids
The Flag-tagged Blimp-1 expression vector was previously described (49). A sequence containing 1158 bp of the NLRP12 promoter was cloned into pGL3-Basic (Promega) luciferase reporter vector and called Mon-1 luc. Site directed mutagenesis was used to mutate four base pairs of the Blimp-1 binding site in using the QuikChange Site Directed Mutagenesis Kit (Stratagene). Primers were NLRP12 5′- GGAAGAGACAGCAGACTCGAGAATCTTTTTCATC-3′ and 5′- GATGAAAAAGATTCTCCTCTCTGCTGTCTCTTCC-3′. To generate the pGL4.10 based luciferase vectors the following Forward primer sequences were used, each containing KPNI sites for cloning: pGL4-1412; 5′- GCGGTACCGAGAATTAGCCAGGCGTGTTG -3′; pGL4-852 5′- GCGGTACCGCATGGTGGCGCATGTCTGTGGT -3′; pGL4-399 5′- GCGGTACCCTGGAAGAGGCGCAGTCGCAGTT -3′; a common reverse primer was used for all PCR reactions and contains a HindIII site for cloning 5′- GCGAAGCTTCCTGGAGGCTGAGATGCTCCTATG -3′.
Identification of a potential Blimp-1 binding site in the NLRP12 promoter
The NCBI and Transcription Element Search Software (TESS) (http://agave.humgen.upenn.edu/utess/tess) databases were used to search for potential transcription factor binding sites within the NLRP12 promoter.
Luciferase Reporter Gene Assays
For transfections, 1×106 U937 cells were transfected with AMAXA nucleofector (AMAXA) with 0.2 μg phRL-luc, 0.5 μg of NF-κB luc, Mon-1 luc or mutMon-1 luc. Where indicated, cells were transfected with 1.0 μg of Flag-Blimp-1 or stimulated with LPS. Firefly luciferase values were normalized to Renilla luciferase control values when harvested 24 h post-transfection.
EMSA
HEK293T cells were transfected with pcDNA3 or a Flag-tagged Blimp-1 using Fugene 6. EMSA was performed as previously described (50). For supershifts, 1 μl of anti-Flag M2 antibody (Sigma) or anti-p65 antibody (Rockland) was added and the binding reaction was allowed to proceed for an additional 15 min. Protein-DNA complexes were resolved on a non-denaturing polyacrylamide gel. Oligo pairs were annealed in STE buffer (10 mM Tris pH 8.0, 50 mM NaCl, 1mM EDTA) and T4 kinase-labeled with [α-32p] CTP. Primers used for NLRP12 EMSA were 5′- GGAAGAGACAGCAGAAGTGAAAATCTTTTTCATC-3′ and 5′- GATGAAAAAGATTTTCACTTCTGCTGTCTCTTCC-3′. The consensus Blimp-1 binding site is underlined.
DNA affinity purification assays
DAPA experiments were performed using the μMACS Streptavidin Kit (Miltenyi Biotec). Briefly, 1 μg of 5′-biotinylated, duplexed oligo corresponding to the Blimp-1 binding site in the NLRP12 promoter (shown above for EMSA) was coupled to streptavidin beads for 30 min at 4°C followed by addition of nuclear extract (500 ng) for 30 min. DNA—protein complexes were immobilized magnetically and washed four times with 20 mM HEPES—KOH pH 7.9, 100 mM KCl and 2.5 mM MgCl2. Specifically bound proteins were eluted with 20 mM HEPES—KOH pH 7.9 and 1 M NaCl. Eluates were electrophoresed on denaturing polyacrylamide gels for subsequent immunoblot analysis using anti-Blimp-1.
Chromatin immunoprecipitation assays
ChIP analysis was performed as previously described (51) with some modifications. Briefly, protein—DNA complexes were crosslinked with for 10 min with formaldehyde and cells were harvested. Cells were either snap-frozen in dry ice/EtOH and stored at −80°C or immediately used. DNA in lysed cells was sonicated to lengths of 500–1000 bp and anti-Blimp-1 (AbCam), anti-acetylated-H3 (Upstate) or mouse IgG (Upstate) was used for immunoprecipitation overnight at 4°C. SYBR Green quantitative qPCR was performed and values were expressed as a percent of total input DNA for each time point. Primers to amplify the NLRP12 promoter were: 5′- CCTGGAAGAGGCGCAGTCGCAGTT-3′ (forward) and 5′- CCGAGGAGCCACTTGGCTGATGAA-3′ (reverse). For visualization, PCR fragments from the ChIP analysis were run on an agarose gel.
Immunoprecipitation and Western analysis
The U937 cell line was stimulated with 100 ng/ml LPS or 50 ng/ml PMA for the indicated time. Cells were lysed in lysis buffer (50mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40) supplemented with Complete protease inhibitor cocktail (Roche). Protein concentrations were determined by Bradford assay (BioRad) and equivalent amounts of cellular extract were used in subsequent immunoprecipitations using rabbit polyclonal antisera against human NLRP12 as described with polyclonal goat anti-Blimp-1 (AbCam) or mouse monoclonal anti-GAPDH (Chemicon). Immunoblots were visualized by enhanced chemiluminescence (Pierce).
Measurement of NLRP12 mRNA Stability
Nuclear versus cytoplasmic transcripts were isolated using the PARIS Kit following the manufacturers protocol (Ambion), and qPCR was performed with primers recognizing process mRNA. Measurement of DRB regulated mRNA stability was performed as previously described (52). HL-60 cells were treated with 1.25% DMSO for 4 days to induce differentiation to neutrophils and expression of NLRP12. Cells were replated and stimulated with LPS, exposed to 50 μM 5,6-dichloro-1-ß-ribofuranosyl benzamidazole (DRB; Sigma-Aldrich) dissolved in 0.1% DMSO, or exposed to 0.1% DMSO as a vehicle control. NLRP12 expression was quantified by qPCR and normalized to the housekeeping gene GusB expression levels.
RNA Preparation and QPCR
Total RNA was isolated according using Trizol reagent (Invitrogen). qPCR results are normalized to 18S r RNA or GUSB mRNA internal controls and are expressed in relative numbers. Primers were: Human NLRP12 F 5′-AGAGGACCTGGTGAGGGATAC-3′, R 5′- CTTCCAGAAGGCATGTTGAC-3′, probe 5′- CCCGTCCTCACTTGGGAACCA-3′. 18S F 5′-GCTGCTGGCACCAGACTT-3′, R 5′-CGGCTACCACATCCAAGG-3′, probe 5′-CAAATTACCCACTCCCGACCCG-3′. GUSB (ABI: Hs99999908_m1). Mouse NLRP12 F 5′-CGACCCACCAGAACCTTCAG -3′, R 5′-CAGGCAGCATGTTCCTTTCC -3′, probe 5′-TCCAGACTCAGTCCACATACTTAC-3′.
Results
NLRP12 expression is regulated by specific TLRs
NLRP12 is expressed by cells of the myeloid and monocytic lineage (35, 37). Myeloid and monocytic cells are involved in the innate immune response to pathogens, and recognize invading microorganisms via Toll-like receptors (TLRs) on their cell surface (53). We have recently shown that NLRP12 is a negative regulator of TLR1/2 and TLR4 signaling in human monocytic-macrophage cell lines, and that ablation of NLRP12 increases pro-inflammatory gene expression (37). Moreover, NLRP12 inhibits CD40-mediated NF-κB activation by promoting the protein degradation of NF-κB inducing kinase (NIK) (38). NLRP12 mRNA expression is reduced within one hour of TLR1/2, TLR4 or microbial pathogen stimulation (37) and to a lesser extent after exposure of cells to TNFα or (36). To determine if NLRP12 expression is regulated by other TLR agonists, we exposed primary human granulocytes for 1 hour to Heat Killed Listeria monocytogenes (HKLM) (TLR2), the synthetic analog of double stranded RNA (PolyI:C) (TLR3), E. Coli K12 rough LPS (TLR4), and S. thyphimurium flagellin (flagellin) (TLR5) (Figure 1A). Pam3Cys4 (TLR1/2) was used as a control as we have previously observed that NLRP12 expression is down-regulated in response to stimulation with this agonist. NLRP12 mRNA expression was dramatically reduced after exposure of granulocytes to Pam3Cys4, HKLM and K12 LPS, but not in response to stimulation with poly I:C or flagellin, although these cells were determined to express and be responsive to TLR3 and TLR5 stimulation (Figure 1A and Figure 1B). Functional TLR3 and TLR5 receptors have been previously observed in granulocytes (54, 55). The observation that NLRP12 expression was not altered to the same extent in response to each TLR agoinst suggests that down-regulation of NLRP12 is mediated by specific TLRs.
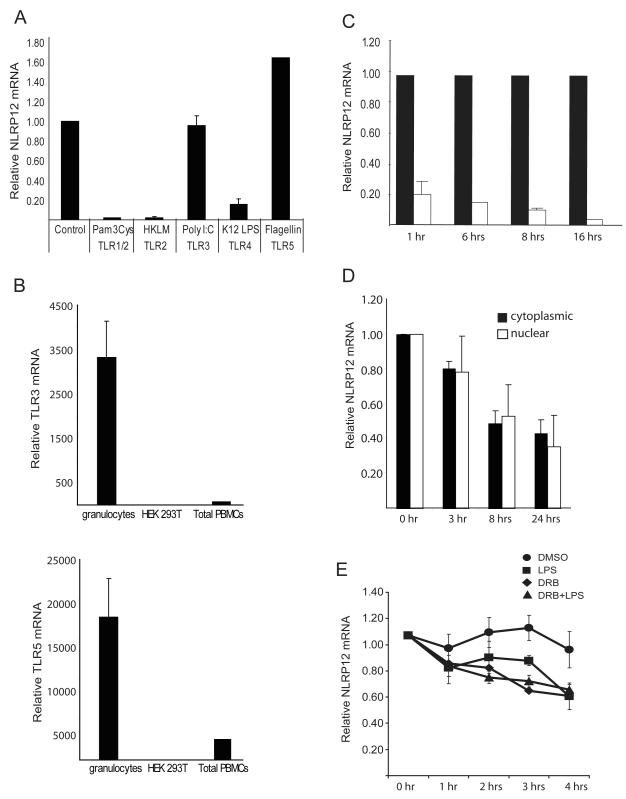
FIGURE 1.
NLRP12 expression is regulated by specific TLRs. (A) Human primary granulocytes were stimulated with the indicated TLR agonists for 1 hour. NLRP12 expression was analyzed by qPCR analysis of total RNA. NLRP12 expression was normalized to the housekeeping gene β-glucuronidase (GusB). Data are an average of three separate experiments. (B) TLR3 and TLR5 expression in total human peripheral blood cells, granulocyte isolated from human peripheral blood cells by ficoll separation, and in the HEK293T cell line were assessed by qPCR. TLR expression was normalized to the expression of GusB. (C) NLRP12 expression in human adherent peripheral blood cells stimulated for 1, 6, 8 and 16 hours with phenol-purified LPS (TLR4 agonist). NLRP12 expression was normalized to the expression of 18S rRNA and represented as fold difference compared to control. Error bars represent the SEM of three separate cell preparations and experiments. (D) The reduction of nuclear verses cytoplasmic NLRP12 expression after TLR activation. U937 cells were stimulated with 500 ng/ml LPS for 3, 5, 8, 18 and 24 hours followed by isolation of nuclear verses cytoplasmic mRNA as described in materials and methods. NLRP12 mRNA abundance was then quantified by qPCR and normalized to GusB. (E) The stability of the NLRP12 mRNA is similar in cells treated with a transcriptional inhibitor or stimulated with LPS. The human pre-monocyte cell line HL-60 was treated with 1.25% DMSO for 3 days to induce differentiation into NLRP12 expressing neutrophils. Differentiated HL-60 cells were then stimulated with LPS (squares), treated with DRB (triangles), treated with DRB plus stimulated with LPS, or exposed to 0.1% DMSO (circles) as a vehicle control for 0, 1, 2, 3, and 4 hours. NLRP12 mRNA abundance was then quantified by qPCR. The amount of NLRP12 mRNA at the time before DRB addition (0 hr) was set to 1, and the amount remaining at the indicated time points was determined. Error bars represent the SEM of five separate experiments.
To assess the kinetics of NLRP12 down-regulation by TLR stimulation, human primary adherent peripheral blood mononuclear cells (PBMC) were isolated and exposed to purified lipopolysaccaride (LPS) from E. coli which activates TLR4 (Figure 1C). NLRP12 expression is down-regulated 1 hr after TLR stimulation, and it remained low 6 hours, 8 hours and 16 hours post-stimulation. This indicates the sustained down-regulation of NLRP12 in primary human myeloid cells via TLR.
TLR stimulation affects NLRP12 mRNA transcription
The sustained decrease in NLRP12 expression following TLR stimulation led us to examine whether NLRP12 is regulated at the level of mRNA stability, transcription, or both. We first examined the transcription rate of the NLRP12 gene by comparing nuclear and cytoplasmic NLRP12 mRNA levels after exposure of U937 cells to LPS (Figure 1D). This approach has been proven as a sensitive and reliable method often used as an alternative to a nuclear run-on assay (52). U937 cells were stimulated for the indicated time points with LPS followed by separate isolation of nuclear and cytoplasmic mRNA followed by qPCR analysis of NLRP12 transcripts. The levels of NLRP12 mRNA were similar in both the nuclear and cytoplasmic compartments, suggesting that the reduction of NLRP12 mRNA caused by TLR signaling is partly or primarily attributed to an effect on transcription. As an alternative measure of mRNA stability, we used 5,6-dichlororibofuranosyl benzimidazole (DRB) to chemically block gene transcription in the HL-60 cell line. The human monocyte HL-60 cell line was treated with DMSO for three days to promote granulocyte differentiation and NLRP12 expression. It was necessary to use the HL-60 cell line as addition of DMSO to the U937 cell line promotes differentiation induced down-regulation of NLRP12 (K.L. Williams, unpublished observations). Differentiated HL-60 neutrophils were then analyzed for the level of NLRP12 transcripts after TLR stimulation, blocking transcription with DRB in DMSO, with both LPS and DRB, or with DMSO alone (Figure 1E). NLRP12 mRNA was relatively stable in the untreated control cells, however its level was reduced in cells treated with the transcriptional inhibitor DRB or with LPS alone. The reduction with these two treatments is not statistically different as assessed by a Students t test (p=0.17). The inclusion of both DRB and LPS did not cause an additive effect. Taken together, these results suggest that the decline in NLRP12 mRNA is significantly altered by the transcriptional inhibitor DRB, and that the reduction of NLRP12 mRNA caused by TLR signaling is attributed to an effect on transcription.
The NLRP12 promoter contains a potential novel Blimp-1 binding site
Our data thus far suggest that the down-regulation of NLRP12 mRNA after TLR stimulation involves transcriptional regulation. To identify potential factors involved in down-regulating NLRP12, we analyzed the first 622 bp sequence of the NLRP12 promoter for transcription factor-binding sites using the Transcription Element Search Software (TESS) (http://agave.humgen.upenn.edu/utess/tess). We identified several transcription factor binding sites including PU.1, which is involved in myeloid cell activation and differentiation (56) (Figure 2A). Sequence analysis also revealed a potential binding site for Blimp-1 encoded by the gene, PRDM1 located at nucleotide position -80 relative to the transcription start site (Figure 2A) (57). All previously identified Blimp-1 binding sites contain the consensus sequence (A/C)AG(T/C)GAAAG(T/C)(G/T) with the exception of the NLR gene CIITA which contains the sequence (A/C)AG(T/C)GAAAT(T/C)(G/T) (49, 57, 58). Analysis of the putative Blimp-1 binding site in NLRP12 suggests that this site contains the sequence (A/C)AG(T/C)GAAAA(T/C)(C), which is different from the previously identified consensus sequence (Figure 2B).
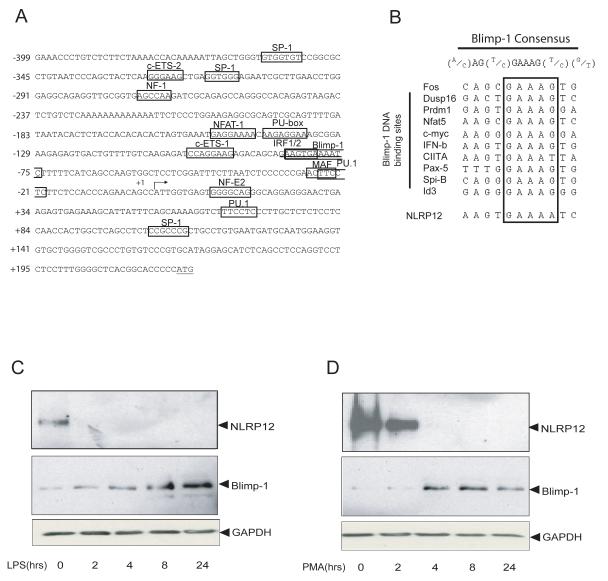
FIGURE 2.
The NLRP12 promoter contains a potential Blimp-1 binding site. (A) Sequence of the human NLRP12 promoter region from − 399 to +223 bp. Several transcription factor DNA binding sites are shown. The DNA binding site for Blimp-1 is underlined. The transcriptional start site is denoted by an arrow (+1), and the ATG of the translational start site is underlined. (B) Comparison of the putative Blimp-1 binding site in the NLRP12 promoter with the Blimp-1 DNA binding sequences of the ten other Blimp-1 targets (boxed). (C). U937 human monocyte cells were stimulated with LPS for 0, 2, 4, 8 and 24 hours. NLRP12 protein was detected by immunoprecipitation followed by immunoblot with two different NLRP12 antibodies as described. Blimp-1 protein expression was assessed by immunoblot. Immunoblot with anti-GAPDH indicates equal protein loading. (D). U937 cells were differentiated with PMA for 0, 2, 4, 8 and 24 hours and analyzed as in (C). Shown is a representative blot of three separate experiments.
NLRP12 and Blimp-1 protein expression are inversely correlated during TLR stimulation and myeloid cell differentiation
The identification of a potentially novel Blimp-1 binding site in the NLRP12 promoter led us to question whether Blimp-1 is important for the silencing of NLRP12 expression. We hypothesized that NLRP12 expression might be down-regulated by an increased level of Blimp-1 protein expression after TLR stimulation and cell differentiation. To assess this, we first examined the expression of NLRP12 and Blimp-1 proteins in the monocyte cell line U937 after stimulation with LPS for 0, 2, 4, 8 and 24 hours (Figure 2C, top panel). We observed a rapid down-regulation of NLRP12 protein expression with little NLRP12 protein detectable at 2 hours after LPS stimulation. In contrast, Blimp-1 protein expression was increased at 2-4 hrs after TLR stimulation, and this increase continued at 8 and 24 hours after TLR activation (Figure 2C, bottom panel). Immunoblots were probed with anti-GAPDH to indicate equal loading. While the data suggest that NLRP12 and Blimp-1 protein expression are inversely correlated, we initially observed a delay in Blimp-1 protein induction at the 2 hour time point. Our data suggest that in addition to Blimp-1 it is possible that other transcription factors are involved in the LPS mediated down-regulation of NLRP12. In addition to the Blimp-1 binding site in the NLRP12 promoter, we observed several binding sites for PU.1. PU.1 is a positive regulator of gene transcription and is expressed in monocyte cells (59). More importantly, PU.1 is down-regulated by LPS (60), which may also attribute to the rapid reduction in NLRP12 protein after LPS stimulation. These data suggest that the inhibitory effect of TLR4 activation on NLRP12 protein expression is, in part, inversely correlated with its effect on Blimp-1 protein expression.
Blimp-1 mediates cell differentiation in several cell types including myeloid cells, and its expression is increased during myeloid-monocytic differentiation (45-48, 61-64). Thus, we examined if NLRP12 expression might also be inversely correlated with Blimp-1 during myeloid differentiation. To test this, the U937 cell line was treated with PMA for 0, 2, 4, 8 and 24 hours to induce cell differentiation followed by analysis of NLRP12 and Blimp-1 protein expression (Figure 2D). NLRP12 protein expression was down-regulated beginning at two hours after PMA-induced differentiation and expression disappeared by 4 hours (Figure 2D, top panel). Conversely, the induction of Blimp-1 protein expression after PMA stimulation began four hours after PMA addition (Figure 2D, middle panel). Immunoblots were probed with anti-GAPDH to indicate equal loading. These results show that the decrease of NLRP12 during differentiation correlates with an increase in Blimp-1 protein expression. These data led us to hypothesize that the induction of Blimp-1 expression during TLR stimulation and cell differentiation might be one mechanism by which NLRP12 expression is down-regulated.
Blimp-1 binds to the NLRP12 promoter after TLR stimulation and differentiation
To address the potential role for Blimp-1 in regulating NLRP12 gene expression, we assessed if Blimp-1 could bind to the NLRP12 promoter. Electrophoretic Mobility Shift Assay (EMSA) was performed using a 30 bp oligonucleotide containing the putative Blimp-1 binding sequence 5′-AAGTGAAAATC-3′ in the NLRP12 promoter. Nuclear extracts were prepared from HEK293T cells transfected with a pcDNA3 control vector or Flag-tagged Blimp-1. EMSA revealed a DNA-protein complex in the lane containing Flag-Blimp-1 but not in the pcDNA3 vector control lane (Figure 3A, left panel, lanes 3 and 2 respectively). This complex was competed by increasing amounts of cold competitive probe indicating specificity (Figure 3A, left panel, lanes 4 and 5). Addition of a Flag antibody to the binding reaction resulted in a slower migrating band, indicating the presence of Flag-Blimp-1 in the DNA-protein complex (Figure 3A, lane 6). This complex was not shifted by a control anti-p65 antibody indicating specificity of antibody binding (Figure 3A, lane 7). These data show that Blimp-1 can bind to the novel Blimp-1 binding site found on the NLRP12 promoter.
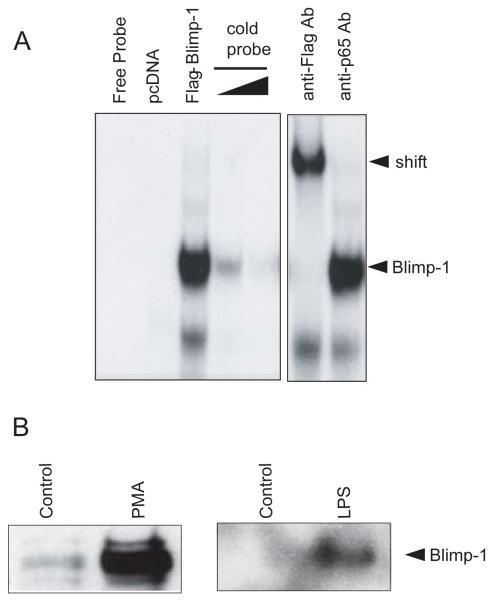
FIGURE 3.
Blimp-1 binds to the putative Blimp-1 binding site in the human NLRP12 promoter and is regulated by PMA and TLR stimulation. (A) EMSA analysis of Blimp-1 DNA binding to the novel site in the NLRP12 promoter. Nuclear extracts were obtained from HEK293T cells transfected with a pcDNA control or a Flag-Blimp-1 expression vector. EMSA was performed using the Blimp-1 binding site in the NLRP12 promoter. Binding was competed with increasing concentrations of cold probe to indicate specificity. An anti-Flag antibody was used to supershift the protein-DNA complexes to indicate specificity. A non-specific anti-p65 antibody was used as a control. A representative of three separate experiments is shown. (B). Nuclear extracts from U937 cells stimulated with PMA and LPS for 24 hours were used in a DAPA analysis of Blimp-1 binding to the NLRP12 promoter. DAPAs were performed by incubating U937 nuclear extracts with an immobilized, biotinylated double-stranded oligo corresponding to the potential Blimp-1 binding site in the NLRP12 promoter. Bound protein complexes were washed, eluted with sample buffer and subjected to SDS-PAGE and Western blot analysis with a Blimp-1-specific antibody. A representative DAPA of two separate experiments is shown.
We next used DNA affinity purification assays (DAPA) to test the binding of Blimp-1 to the NLRP12 promoter to assess if increased binding could be observed after TLR stimulation or cell differentiation. We stimulated the U937 monocyte cell line with LPS or differentiated them with PMA for 24 hours and performed DAPA analysis (Figure 3B). Nuclear extracts from control unstimulated U937 cells and LPS stimulated or PMA treated U937 cells were incubated with double-stranded biotinylated oligonucleotides containing the putative Blimp-1 binding site in the NLRP12 promoter. Bound proteins were eluted followed by immunoblotting with an antibody to Blimp-1. A dramatic increase in Blimp-1 signal was observed after cells were stimulated with PMA or LPS. These results indicate that Blimp-1 in myeloid cells can bind to the NLRP12 promoter, and that this binding is increased after TLR activation by LPS or cell differentiation with PMA.
NLRP12 expression is down-regulated by Blimp-1
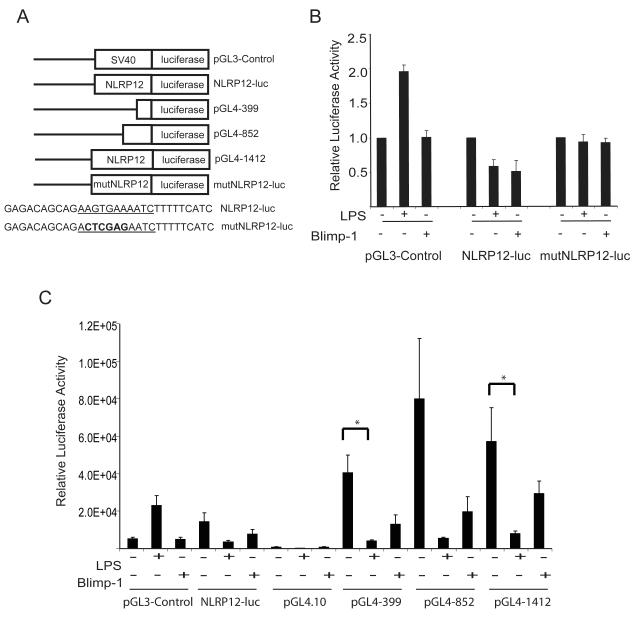
The observation that Blimp-1 binds to a novel site on the NLRP12 promoter in vitro supports the idea that Blimp-1 mediates the repression of NLRP12 expression in vivo. To test this, we generated a luciferase reporter vector driven by 1412 bp of the NLRP12 promoter (NLRP12-luc) as well as a NLRP12-luc vector in which the Blimp-1 binding site was mutated (mutNLRP12-luc) (Figure 4A). The pGL3-Control vector served as a positive control for transfection efficiency. The U937 cell line was transfected with NLRP12-luc, mutNLRP12-luc or pGL3-Control luciferase reporter vectors, and either co-transfected with Flag-Blimp-1 or stimulated with LPS (Figure 4B). We observed a similar decrease in luciferase activity in cells containing the NLRP12-luc reporter vector when stimulated with LPS or when co-transfected with Flag-Blimp-1. This is comparable to the extent that Blimp-1 reduces the pIV promoter of CIITA and to which Bach2 represses the Blimp-1/Prdm1 gene in promoter reporter assays (65, 66). Neither LPS nor Blimp-1 affected the mutNLRP12-luc vector that contained a mutated Blimp-1 site, indicating that the Blimp-1 binding site in the NLRP12 promoter is required for LPS or Blimp-1 mediated down-regulation of NLRP12. As a control, cells were transfected with a pGL3-Control vector, which contains SV40 promoter driven luciferase activity. When stimulated with LPS, pGL3-Control cells had a higher level of luciferase reporter activity, supporting previous data that SV40 promoter activity is up-regulated by LPS stimulation (67). When co-transfected with Blimp-1, we did not observe a difference in pGL3-Control reporter activity relative to control cells.
FIGURE 4.
The NLRP12 promoter is downregulated by LPS stimulation and Blimp-1 expression in human monocyte cells. (A). Schematic representation of luciferase reporter constructs generated to assess NLRP12 promoter luciferase activity. For promoter analysis, 1412 bp of the NLRP12 promoter was inserted into pGL3-luciferase to drive luciferase reporter expression (NLRP12-luc). A mutant NLRP12 promoter luciferase reporter was generated by mutating 4 bp located in the putative Blimp-1 binding site (mutNLRP12-luc). Deletions of the NLRP12 promoter were also cloned into pGL4.10-luciferase vector including a 399 bp fragment (pGL4-399), a 852 bp fragment (pGL4-852), and the full length 1412 fragment (pGL4-1412). (B). The U937 human monocyte cell line was transfected with the pGL3-Control vector, Mon-1 luc, or mutMon-1 luc. After transfection, cells were either left unstimulated, stimulated with LPS or transfected with a Flag-tagged Blimp-1 expression vector. Twenty-four hours later, cells were lysed and luciferase levels were determined as described in Materials and Methods. Error bars represent the SEM of four separate experiments. (C). The U937 human monocyte cell line was transfected with the pGL3-Control vector, NLRP12-luc, pGL4.10-luciferase vector, pGL4-399, pGL4-852 and pGL4-1412. After transfection, cells were either left unstimulated, stimulated with LPS or transfected with a Flag-tagged Blimp-1 expression vector. Twenty-four hours later, cells were lysed and luciferase levels were determined as described in Materials and Methods. Error bars represent the SEM of three separate experiments.
In order to confirm these results, we also cloned shorter regions of the NLRP12 promoter and the full length 1412 bp fragment into the pGL4.10 luciferase vector. The pGL4.10 luciferase vector was chosen due to the improved levels of signal to noise ratio as compared to the older pGL3 luciferase version (Figure 4A). The U937 cell line was transfected with NLRP12-luc, pGL3-Control, pGL4.10 vector, pGL4 containing 399 bp of the NLRP12 promoter (pGL4-399), pGL4 containing 852 bp of the NLRP12 promoter (pGL4-852) and pGL4 containing 1412 bp of the NLRP12 promoter (pGL4-1412) luciferase reporter vectors. Cells were either co-transfected with Flag-Blimp-1 or stimulated with LPS (Figure 4C). Consistent with our previous observations, we observed a decrease in luciferase activity in cells containing all of the NLRP12 reporter vectors when stimulated with LPS or when co-transfected with Flag-Blimp-1. A Students T Test was used to determine significance where indicated. Taken together, these data suggest that the NLRP12 promoter is regulated by Blimp-1, and that a functional Blimp-1 binding site is necessary for LPS or Blimp-1 mediated down-regulation of NLRP12 promoter.
TLR stimulation induces Blimp-1 binding to the NLRP12 promoter in vivo
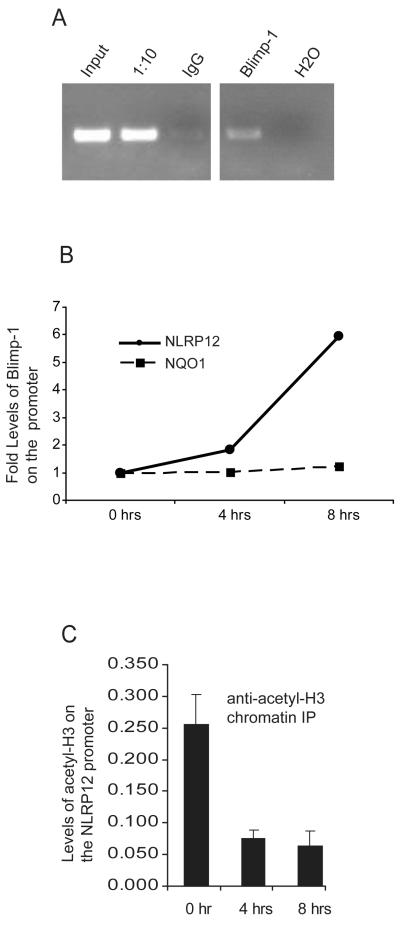
Our observations suggested that Blimp-1 binds the NLRP12 promoter in vitro as assessed by EMSA and DAPA analysis, and that Blimp-1 can repress an NLRP12 promoter luciferase reporter construct. To determine if Blimp-1 binds to the NLRP12 promoter in vivo in an LPS induced manner, we performed chromatin immunoprecipitation assays (ChIP). U937 cells were stimulated with LPS for 8 hours followed by immunoprecipitation of protein-DNA complexes with anti-IgG or with anti-Blimp-1. PCR using NLRP12 promoter specific primers which result in a 300 bp product surrounding the Blimp-1 site was performed on input DNA, a 1/10 dilution of input DNA, and ChIP performed with anti-IgG or anti-Blimp-1 antibodies followed by visualization by gel electrophoresis (Figure 5A). Samples precipitated with the anti-Blimp-1 antibody show that endogenous Blimp-1 is recruited to the NLRP12 promoter in vivo after TLR stimulation. To assess the kinetics of Blimp-1 binding we then stimulated U937 cells for 0, 4 and 8 hours followed by ChIP with anti-IgG or anti-Blimp-1 antibodies as described above (Figure 5B). Blimp-1 association with the endogenous NLRP12 promoter increased by two-fold 4 hours after LPS stimulation, and six-fold by 8 hours. Primers specific for the NQO1 promoter which is not regulated by Blimp-1 were used as a negative control (Figure 5B). These data suggest that Blimp-1 associates with the NLRP12 promoter in an LPS induced manner.
FIGURE 5.
Blimp-1 is recruited to the NLRP12 promoter after TLR stimulation in vivo. (A) Human U937 cells were stimulated with LPS for 8 hours followed by CHIP using a control IgG antibody, an antibody to acetylated Histone3, and an antibody to Blimp-1. DNA was eluted from the CHIP and subjected to PCR with primers specific for the NLRP12 promoter. PCR products were analyzed on an agarose gel. Lanes include PCR product obtained from input DNA, a 1/10 dilution of input DNA, immunoprecipitation with an IgG control antibody, immunoprecipitation with an anti-Blimp-1 antibody, and water as a PCR control. Data are representative of three separate experiments. (B) Chromatin Immunoprecipitation was performed to confirm the interaction of Blimp-1 with the NLRP12 promoter in vivo. The human monocyte cell line U937 was stimulated with LPS for 1, 4 and 8 hours. Chromatin bound Blimp-1 complexes were immunoprecipitated using an anti-Blimp-1 antibody followed by analysis of complexed chromatin by qPCR using primers specific for the NLRP12 promoter. qPCR using primers specific for the NQO1 promoter were used as a negative control. qPCR values have been normalized to input. Precipitation with an unrelated antibody was used as negative control. Data are a representative of three separate experiments. (C) Chromatin Immunoprecipitation was performed to measure acetylated-Histone 3 (H3) levels with the NLRP12 promoter in vivo. The human monocyte cell line U937 was stimulated with LPS for 1, 4 and 8 hours. Chromatin bound acetylated-H3 complexes were immunoprecipitated using an anti- acetylated-H3 antibody followed by analysis of complexed chromatin by qPCR using primers specific for the NLRP12 promoter. qPCR values have been normalized to input. Data are an average of four experiments.
Transcriptional silencing is often associated with decreased histone acetylation. Blimp-1 recruits the G9a methyltransferase, and confers transcriptional silencing by inducing histone H3 acetylation and methylation (42). To assess if the binding of Blimp-1 to the NLRP12 promoter is correlated with a decrease in H3 acetylation, we stimulated U937 cells for 0, 4 and 8 hours followed by ChIP with anti-IgG or anti-acetylated Histone 3 antibodies. We observed that acetylated-Histone 3 levels were reduced by four-fold at the four and eight hour time point, suggesting that the NLRP12 promoter is being transcriptionally silenced (Figure 5C). These data show that LPS reduced transcriptional activity of the NLRP12 promoter is characterized by decreased histone acetylation and increased association of Blimp-1.
Endogenous Nlrp12 gene expression is regulated by Blimp-1 in primary cells
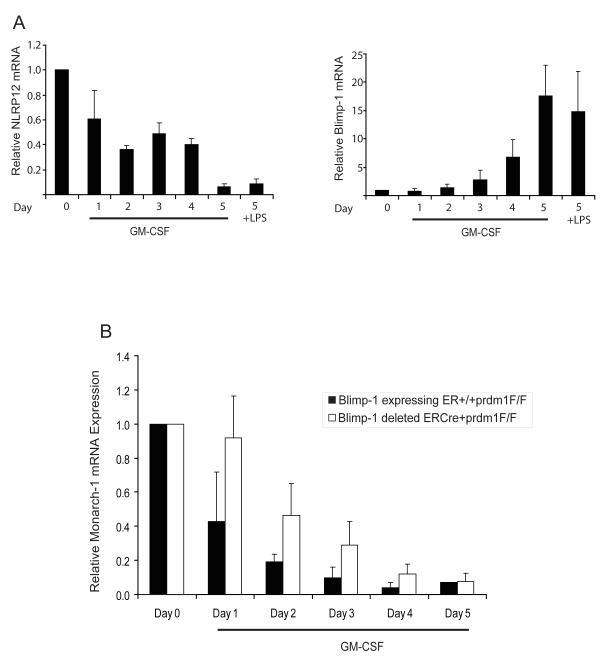
The observation that NLRP12 and Blimp-1 protein expression is inversely correlated in human myeloid cells during LPS stimulation and differentiation led us to examine the expression of these two genes in primary myeloid cells from mice. However, we have not observed decreased Nlrp12 expression upon LPS stimulation of mouse bone marrow cells, likely reflecting species-variation in the regulation of this gene. Thus we concentrated on Nlrp12 expression during myeloid differentiation. In vitro differentiation of murine bone marrow with GM-CSF induces the differentiation of hematopoietic progenitors into more mature dendritic cells and macrophages. To determine if NLRP12 and Blimp-1 expression is altered during differentiation of mouse bone marrow cells, we isolated bone marrow from C57BL/6 mice and differentiated these cells in vitro for a period of five days with GM-CSF. We observed a decrease in NLRP12 expression over the GM-CSF differentiation period as assessed by qPCR but not by LPS stimulation of murine cells as expected (Figure 6A). Conversely, Blimp-1 expression was increased during the differentiation period. Thus, NLRP12 and Blimp-1 expression are inversely correlated during differentiation of murine bone marrow cells with GM-CSF in vitro.
FIGURE 6.
Reduced expression of NLRP12 during murine bone marrow differentiation is regulated by Blimp-1. (A) Murine bone marrow cells were isolated from C57BL/6 mice and differentiated in vitro with GM-CSF or stimulated with LPS for 3 hrs. Blimp-1 and NLRP12 expression were assessed by qPCR analysis of RNA isolated at the indicated time points. Data are a representative of three separate experiments. (B) Murine bone marrow cells were isolated from control ER+/+prdm1F/F mice or from ERCre/+prdm1F/F (48) mice in which Blimp-1 expression is knocked out under the control of the ER promoter . Bone marrow cells were differentiated in vitro with GM-CSF. NLRP12 expression was assessed by qPCR analysis of RNA isolated at the indicated time points. Data are an average of at least three separate experiments and error bars are calculated using SEM values.
Next, we determined whether Blimp-1 mediates the transcriptional repression of NLRP12 during murine bone marrow differentiation. We obtained cells from Rosa26ERCre/+prdm1F/F Cre-lox mice made from the Rosa26 ES cell line which ubiquitously expresses lacZ in adult mouse tissues as described previously (68). For our studies, Blimp-1 null bone marrow cells were obtained from Rosa26ERCre/+prdm1F/F mice following treatment of the mice with tamoxifen to activate the Cre recombinase resulting in deletion of the Blimp-1 gene (68). Isolated Blimp-1-deficient bone marrow cells were differentiated in vitro with GM-CSF for the indicated time points (Figure 6B). GM-CSF caused a steady decrease in NLRP12 expression over a period of 5 days as shown in the control Blimp-1 expressing cells from Rosa26+/+prdm1F/F mice, although the data did not reach statistical significance. From days 1-4 of GM-CSF treatment, NLRP12 was reduced in the Blimp-1-deficient Rosa26ERCre/+prdm1F/F cells but to a much lesser extent relative to the control Blimp-1 expressing Rosa26+/+prdm1F/F cells. By day 5, expression was reduced in both cell types regardless of the presence of Blimp-1 (Figure 6B). It may by possible that additional transcription factors including PU.1 are also involved. The data indicate that Blimp-1 reduces NLRP12 during the early points of GM-CSF stimulation but its expression is reduced by additional mechanism(s) after prolonged GM-CSF stimulation. Thus, inhibition of NLRP12 expression observed during differentiation of murine bone marrow cells is mediated in part by Blimp-1.
Discussion
Based on in vitro analysis as well as the analysis of patients with NLRP12 splicing or nonsense mutations, this gene has emerged as a negative regulator of inflammatory gene induction. NLRP12 modulates TLR inflammatory gene induction by its inhibition of IRAK-1 hyperphosphorylation (37) and NIK activation (38). The negative regulatory functions of NLRP12 have been shown to require ATP binding (69). In human myeloid cells, NLRP12 expression is down-regulated both by cellular activation through TLR stimulation as well as developmentally after exposure of myeloid cells to TNFα and IFNγ (36). While NLRP12 is expressed in monocytes, it is not expressed in differentiated macrophages suggesting that the regulation of NLRP12 is under developmental control (35-37). Thus, the differentiation of monocytes into macrophages, and the inflammatory response after TLR stimulation involves the repression of NLRP12.
The observation that NLRP12 expression is down-regulated after TLR mediated activation of human myeloid cells and by myeloid differentiation of monocytes to macrophages is unusual for most innate immune genes. More often, proteins which function as negative regulators of TLR signaling are induced after activation and are thought to inhibit the signaling pathways. An example of TLR induced expression of negative regulators includes but the induction of IRAK-M (70)as well as the induction of A20 (71) which are both induced 2-3 hours after LPS exposure and “turn off” downstream activation by associating with TLR pathway signaling proteins to once expressed. Based on our current understanding of the function of NLRP12 we hypothesize that NLRP12 is present prior to immune stimulation to maintain a quiescent phenotype by inhibiting inappropriate TLR activation in the absence of appropriate signals. We further hypothesize that during states of infection NLRP12 initially functions to modulate over-zealous immune responses by associating with and regulating the activity of TLR signaling proteins including IRAK-1 and NIK (37, 38). Since we have observed that NLRP12 regulates both TLR and TNFR pathways, we hypothesize that at later time points after TLR activation or during states of myeloid differentiation NLRP12 expression is decreased in order to allow a more complete immune response to occur. The hypothesis that NLRP12 initially functions to modulate over-zealous immune responses is supported by the recent observation that humans with mutations in NLRP12 are predisposed to the autoimmune syndrome known as hereditary periodic fever (39). Future characterization of the function of NLRP12 will likely lead to a greater understanding of the role of NLRP12 down-regulating after TLR activation and myeloid differentiation.
This report shows that Blimp-1 represses the NLRP12 promoter in transformed and primary myeloid-monocytic cells. This is mediated by the binding of Blimp-1 to the NLRP12 promoter in intact cells resulting in a correlative loss of histone acetylation. The regulation of NLRP12 gene expression by Blimp-1 is most convincingly shown using a Blimp-1 Cre-knockout mouse where the down-regulation of NLRP12 during GM-CSF-dependent differentiation of bone marrow progenitors is lessened in Blimp-1-/- cells. These data indicate that Blimp-1 reduces NLRP12 expression during cell differentiation and TLR signaling.
Blimp-1 has long been implicated as a master regulator of B cell differentiation into plasma cells (72, 73). It is expressed in antibody-secreting plasma cells and plasmablasts, is required for the formation of immunoglobulin secreting plasma cells (48, 61) and is induced after TLR activation in B cells (74). Most relevant to this work, Blimp-1 is induced during differentiation of myeloid cell lines, whereby it promotes the induction of macrophage cell morphology (47). Here we confirm that increased Blimp-1 causes the reduction of NLRP12 expression during TLR-mediated activation and PMA-mediated differentiation.
During myeloid cell differentiation and activation, key transcription factors including PU.1, GATA-1, Ikaros, C/EBPα, C/EBPβ and Blimp-1 are shown to play important roles in mediating cell development and function (75). Blimp-1 has more recently been shown to regulate differentiation of myeloid cells to macrophages (76). The alteration of Blimp-1 expression during myeloid differentiation was demonstrated by the observation that Blimp-1 is induced during M-CSF-driven differentiation of murine myeloid progenitors, and is rapidly expressed following PMA stimulation of human myeloid cell lines (47, 62, 77). Blimp-1 regulates key differentiation genes including c-myc (78), CIITA (49), Spi-B, Id3 (64); and Pax5 (63). These results support a model that places Blimp-1 at the center of the transcriptional network that controls cellular terminal differentiation and activation. Consistent with findings that Blimp-1 is involved in recruiting factors that directly modify histones to cause gene silencing (42), we find an inverse correlation between enhanced Blimp-1 binding to the NLRP12 promoter and a decrease in H3 acetylation that is associated with gene transcription.
In addition to Blimp-1, it is necessary to evoke another unknown molecule(s) or mechanism(s) for a more complete inhibition of NLRP12 expression during myeloid treatment. While PMA-induced myeloid differentiation resulted in a near-complete depletion of NLPR12 expression, Blimp-1 only partially inhibited NLRP12 promoter activity in a cell line and Blimp-1 gene deletion only partially rescued Nlrp12 expression in differentiating murine mononuclear cells. Future identification of this second inhibitor will be important to fully understand how NLRP12 is regulated.
In summary, we propose that the transcriptional silencing of the NLRP12 gene, a negative regulator of inflammatory gene induction, is essential for the development of a proper immune response. NLRP12 gene silencing is mediated by Blimp-1 during both innate immune activation and cellular differentiation. This work is the first to link the transcriptional repressor, Blimp-1, to NLR gene silencing during TLR activation and myeloid differentiation.
Acknowledgements
We thank Erna Magnúsdóttir for her assistance with the Rosa26ERCre/+prdm1F/F mice. We thank Zhengmao Ye for his assistance with site-directed mutagenesis.
This work was supported by NIH grants HL-30923 and HL-084917 support to JRW; AI057175, AI063031 to JPT; NRSA, UNC CFAR, Amgen/FOCIS Fellowship Award, SERCEB and support from The American Cancer Society to KLW.
Abbreviations
- NLR
nucleotide-binding domain, leucine-rich repeat
- MDP
muramyl dipeptide
- CAPS
cryopyrin-associated periodic syndrome
- HPF
hereditary periodic fever
- shRNA
small heteroduplex RNA
- NIK
NF-κB inducing kinase
- PBMC
peripheral blood mononuclear cells
- DRB
5,6-dichlororibofuranosyl benzimidazole
- DAPA
DNA affinity purification assays
References
- 1.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr Opin Immunol. 2003;15:396–401. doi: 10.1016/s0952-7915(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 3.Ehlers M, Ravetch JV. Opposing effects of Toll-like receptor stimulation induce autoimmunity or tolerance. Trends Immunol. 2007;28:74–79. doi: 10.1016/j.it.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 6.Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 9.Lamkanfi M, Kanneganti TD, Franchi L, Nunez G. Caspase-1 inflammasomes in infection and inflammation. J Leukoc Biol. 2007;82:220–225. doi: 10.1189/jlb.1206756. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 11.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 13.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 14.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 15.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Nunez G. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 16.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, Brady G, Meshinchi S, Jagirdar R, Gewirtz A, Akira S, Nunez G. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 17.Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, Sadigh K, Abney AL, Bernstein-Hanley I, Dietrich WF. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 19.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 20.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 21.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, Flavell RA. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–1850. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 24.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 25.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 26.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 27.Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow JE, Jr., Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O’Shea JJ, Kastner DL, Goldbach-Mansky R. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 2002;46:3340–3348. doi: 10.1002/art.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, Rubin BI, Kim HJ, Brewer C, Zalewski C, Wiggs E, Hill S, Turner ML, Karp BI, Aksentijevich I, Pucino F, Penzak SR, Haverkamp MH, Stein L, Adams BS, Moore TL, Fuhlbrigge RC, Shaham B, Jarvis JN, O’Neil K, Vehe RK, Beitz LO, Gardner G, Hannan WP, Warren RW, Horn W, Cole JL, Paul SM, Hawkins PN, Pham TH, Snyder C, Wesley RA, Hoffmann SC, Holland SM, Butman JA, Kastner DL. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1beta inhibition. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins PN, Lachmann HJ, Aganna E, McDermott MF. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 2004;50:607–612. doi: 10.1002/art.20033. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman HM, Rosengren S, Boyle DL, Cho JY, Nayar J, Mueller JL, Anderson JP, Wanderer AA, Firestein GS. Prevention of cold-associated acute inflammation in familial cold autoinflammatory syndrome by interleukin-1 receptor antagonist. Lancet. 2004;364:1779–1785. doi: 10.1016/S0140-6736(04)17401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, Fain PR, Spritz RA. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 34.Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, Slim R. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–302. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 36.Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. Cutting edge: Monarch-1: A pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol. 2003;170:5354–5358. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 37.Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, Kurtz S, Coffield VM, Accavitti-Loper MA, Su L, Vogel SN, Braunstein M, Ting JP. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, Ting JP. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 39.Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, Grimprel E, Landman-Parker J, Hentgen V, Marlin S, McElreavey K, Sarkisian T, Grateau G, Alnemri ES, Amselem S. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A. 2008;105:1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 42.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciammas R, Davis MM. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J Immunol. 2004;172:5427–5440. doi: 10.4049/jimmunol.172.9.5427. [DOI] [PubMed] [Google Scholar]
- 45.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 46.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 47.Chang DH, Angelin-Duclos C, Calame K. BLIMP-1: trigger for differentiation of myeloid lineage. Nat Immunol. 2000;1:169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 49.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 50.Mayo MW, Norris JL, Baldwin AS. Ras regulation of NF-kappa B and apoptosis. Methods Enzymol. 2001;333:73–87. doi: 10.1016/s0076-6879(01)33046-x. [DOI] [PubMed] [Google Scholar]
- 51.Sitcheran R, Gupta P, Fisher PB, Baldwin AS. Positive and negative regulation of EAAT2 by NF-kappaB: a role for N-myc in TNFalpha-controlled repression. Embo J. 2005;24:510–520. doi: 10.1038/sj.emboj.7600555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landmann S, Muhlethaler-Mottet A, Bernasconi L, Suter T, Waldburger JM, Masternak K, Arrighi JF, Hauser C, Fontana A, Reith W. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J Exp Med. 2001;194:379–391. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 54.Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 55.Muzio M, Bosisio D, Polentarutti N, D’Amico G, Stoppacciaro A, Mancinelli R, van’t Veer C, Penton-Rol G, Ruco LP, Allavena P, Mantovani A. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 56.Dakic A, Wu L, Nutt SL. Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis? Trends Immunol. 2007;28:108–114. doi: 10.1016/j.it.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 58.Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci U S A. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 60.Ishii J, Kitazawa R, Mori K, McHugh KP, Morii E, Kondo T, Kitazawa S. Lipopolysaccharide suppresses RANK gene expression in macrophages by down-regulating PU.1 and MITF. J Cell Biochem. 2008;105:896–904. doi: 10.1002/jcb.21886. [DOI] [PubMed] [Google Scholar]
- 61.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 62.Chang DH, Cattoretti G, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 63.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 65.Tooze RM, Stephenson S, Doody GM. Repression of IFN-gamma induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling. J Immunol. 2006;177:4584–4593. doi: 10.4049/jimmunol.177.7.4584. [DOI] [PubMed] [Google Scholar]
- 66.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 67.Macchi M, Bornert JM, Davidson I, Kanno M, Rosales R, Vigneron M, Xiao JH, Fromental C, Chambon P. The SV40 TC-II(kappa B) enhanson binds ubiquitous and cell type specifically inducible nuclear proteins from lymphoid and non-lymphoid cell lines. Embo J. 1989;8:4215–4227. doi: 10.1002/j.1460-2075.1989.tb08607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–1476. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by Monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2007 doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pathak SK, Basu S, Bhattacharyya A, Pathak S, Kundu M, Basu J. Mycobacterium tuberculosis lipoarabinomannan-mediated IRAK-M induction negatively regulates Toll-like receptor-dependent interleukin-12 p40 production in macrophages. J Biol Chem. 2005;280:42794–42800. doi: 10.1074/jbc.M506471200. [DOI] [PubMed] [Google Scholar]
- 71.Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 72.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 73.Turner CA, Jr., Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 74.Genestier L, Taillardet M, Mondiere P, Gheit H, Bella C, Defrance T. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J Immunol. 2007;178:7779–7786. doi: 10.4049/jimmunol.178.12.7779. [DOI] [PubMed] [Google Scholar]
- 75.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Kallies A, Nutt SL. Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol. 2007;19:156–162. doi: 10.1016/j.coi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 77.Schliephake DE, Schimpl A. Blimp-1 overcomes the block in IgM secretion in lipopolysaccharide/anti-mu F(ab’)2-co-stimulated B lymphocytes. Eur J Immunol. 1996;26:268–271. doi: 10.1002/eji.1830260142. [DOI] [PubMed] [Google Scholar]
- 78.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]