Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia in adults requiring hospitalization and is characterized by rapid and irregular activation of the atria. Loss of effective atrial contraction leads to decreased ventricular filling and cardiac output, while stasis of blood in the atria increases the risk for thromboembolic stroke1. Although most AF develops as a consequence of other systemic processes such as hypertensive, valvular or coronary artery disease, 10–20% occurs in individuals without underlying risk factors. Genetic factors are implicated in the development of isolated AF, with nearly 40% of cases having a positive family history.2,3 Disease-causing mutations have been found in cardiac potassium channel genes, although mutations in sodium channels and connexins have also been implicated.4 An infrequent genetic etiology is mutation in the gene encoding for lamin A/C, LMNA, but the AF is typically associated with a combined cardiac and skeletal myopathy.5
The LMNA gene resides on chromosome 1q21 and it encodes for two isoforms, lamin A and C, which are generated by alternative splicing (Figure 1D). Lamins A and C are intermediate filament proteins that form the nuclear lamina, function as a nuclear scaffold to maintain the structure and size of the nucleus and are also implicated in transcriptional regulation, nuclear pore positioning and heterochromatin organization.6 Mutations in nuclear lamins and lamin-associated proteins cause more than 16 distinct human diseases, termed “laminopathies”. Lamin A/C is expressed in multiple cell types and mutations in LMNA result in a range of phenotypes including premature aging syndromes, types of muscular dystrophy, a subset of lipodystrophies, bone dysplasias and cardiovascular diseases.7 The cardiac involvement in laminopathies usually presents with progressive atrioventricular block (AVB), dilated cardiomyopathy (DCM), sudden cardiac death (SCD) and infrequently, atrial arrhythmias. It remains unknown how mutations in these ubiquitously expressed proteins cause such heterogeneous phenotypes with variable penetrance.
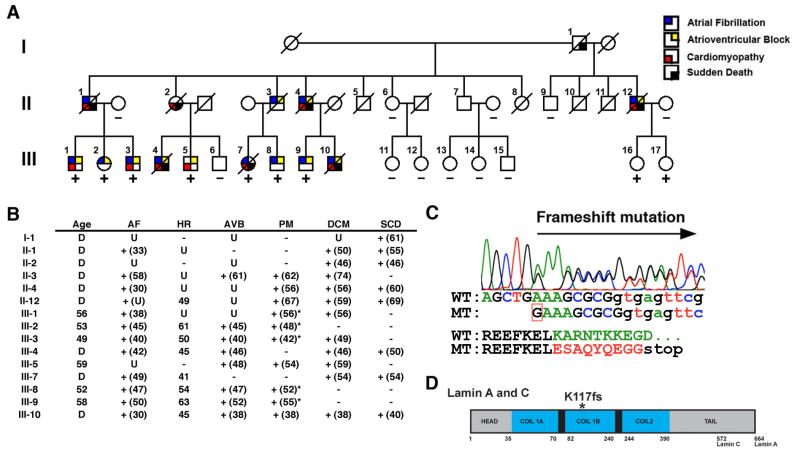
Figure 1. Novel LMNA mutation (K117fs) segregates with atrial fibrillation, atrioventricular block, and dilated cardiomyopathy.
(A) Three generation family had 15 affected members exhibiting autosomal dominant cardiac disease. Square, males; circles, females; slash, deceased. (B) Phenotype of affected family members. Age at presentation or implantation is noted in parentheses. AF, atrial fibrillation; HR, heart rate; AVB, atrioventricular block; PM, pacemaker implantation; *, implantation of cardiac defibrillator; DCM, dilated cardiomyopathy; SCD, sudden cardiac death; D, deceased; U, unknown. (C) Sequence chromatogram demonstrating single nucleotide insertion (red box) on mutant (MT) allele compared to wildtype (WT). The insertion alters amino acid sequence (in red) after codon 117 and predicts a premature stop codon. (D) Schematic of lamin A/C protein. Alternative splicing results in 572 and 664 amino acid lamin C and lamin A proteins. *, K117fs mutation in the α-helical rod domain (in blue).
Here, we present a large family in which a novel mutation of LMNA segregates with affected members with cardiac disease. The phenotype consists of early-onset AF and progressive AVB that is followed by DCM and SCD. In this report, we discuss the clinical and genetic findings and emphasize that LMNA mutations are a potential cause of early-onset AF and progressive conduction system disease.
Case Report
We identified a family of Han Chinese ancestry spanning 3 generations with autosomal dominant cardiac disease that had 15 affected members (Figure 1A). The disease was 100% penetrant in family members over 40 years of age and no members exhibited muscular dystrophy or other organ system involvement as commonly described in other laminopathies.7 Clinical histories are summarized in Figure 1B and were obtained by review of available medical records, detailed family histories and a previous case report.8 The predominant disease presentation consisted of AF with a slow ventricular response (heart rate < 65 beats/minute) at a median age of 42 years (Figure 1B). For individuals in which detailed information is available, the AF did not require anti-arrhythmic medications except for III-2 and III-9, who were treated with disopyramide (Figure 1B). Milder degrees of AVB were diagnosed at 46.5 years (median age) and progressed to 3rd degree AVB by 54.5 years (median age) with subsequent pacemaker implantation in 10 subjects. Additionally, 8 family members experienced SCD between 40–69 years of age (median=54.5 years). The only exception is II-3, who died at 81 years of age from non-cardiac causes. SCD occurred between 5–30 years after initial presentation, including 3 subjects (II-4, II-12 and III-10) who had previously received pacemakers. Five members have received an implantable cardiac defibrillator (ICD) (asterisks, Figure 1B). A high incidence of DCM was found as 12 members were diagnosed at a median age of 54 years (range 38–74 years). Of 12 subjects with AF, 10 went on to develop DCM with 6 members experiencing a delay of at least 8 years after initial presentation. The rapid progression of the disease is exemplified in subject III-7, who had AF at 49 years of age and at 53 years of age had normal ventricular function with a shortening fraction of 46% (normal=25–53%) and a normal left ventricular end diastolic diameter of 48 mm (normal=35–57mm) by echocardiogram. At 54 years of age, III-7 had SCD and had evidence of DCM on autopsy. Interestingly, III-7 was also noted to have QT interval prolongation prior to SCD but no other family members possess this electrocardiographic abnormality.
Mutation Detection
Genetic studies were performed after obtaining informed consent in accordance with a University of Texas Southwestern Medical Center Institutional Review Board approved protocol. DNA was extracted from peripheral blood lymphocytes (PBL) or buccal cells of 20 available family members (Puregene, Gentra Systems). Autosomal genome linkage analysis performed with microsatellite DNA markers (ABI Mapping Set v2.5) surrounding previously described AF loci implicated the chromosome 1q21 locus in which LMNA resides.5 The exons of LMNA were amplified by polymerase chain reaction (PCR) with the BD Biosciences Advantage GC Genomic PCR kit, under conditions previously described.9 The amplified products were sequenced (ABI 3730 DNA sequencer, Applied Biosystems) and analyzed.
Genetic Analysis
Direct sequencing of LMNA revealed a heterozygous insertion of a guanine between nucleotides 348 and 349 causing a frameshift mutation at codon 117 (Figure 1C). This mutation (K117fs) near the end of exon 1 predicts a severely altered protein that contains 9 incorrect amino acids in the highly conserved region of the α-helical rod domain followed by a premature stop codon and results in a truncated protein (Figure 1C, 1D). The mutation was present in all affected family members tested along with two currently asymptomatic carriers, III-16 and III-17, who are currently under 40 years of age (Figure 1A). The mutation was absent in unaffected family members tested and in 378 chromosomes of normal control individuals of mixed ancestry.
Lamin A/C Expression
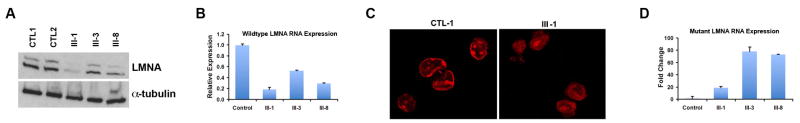
The expression of lamin A/C was studied in affected family members using cultured PBLs. Protein and mRNA were isolated using standard methods and demonstrated variable reduction in lamin A/C protein and mRNA levels in affected family members when compared to controls (Figure 2A, 2B). III-1 had the lowest expression levels of lamin A/C protein and similarly a very low level of mRNA expression. Consistent with this, III-1 also had almost no detectable staining of lamin A/C in the nuclear envelopes of cultured lymphocytes (Figure 2C). The mutant RNA was detected at variable levels in affected members presumably due to variable extent of nonsense mediated mRNA decay (Figure 2D).10 However, we were not able to detect the truncated K117fs mutant protein by Western blot (data not shown).
Figure 2. Expression of Lamin A/C in cultured lymphocytes.
Variable expression levels of lamin A/C protein (A) and mRNA (B) are found in affected members (III-1, III-3, and III-8) compared to controls (CTL1, CTL2) by Western blotting with lamin A/C antibody and real time quantitative PCR (RT-qPCR). Normalization to GAPDH for RT-qPCR and α-tubulin for Western blotting was performed. (C) Immunofluorescence staining of LMNA in lymphocytes from controls showed staining at nuclear envelope, whereas in affected member (III-1) there was almost no detectable nuclear envelope staining at the same level of exposure. (D) Variable quantities of lamin A/C mutant RNA were found in lymphocytes of III-1, III-3 and III-8 by RT-qPCR. Expression of mutant RNA was normalized to 1 in control sample.
Discussion
This family exemplifies the progressive nature of conduction system disease in subjects who harbor mutations in LMNA. Conduction abnormalities have long been recognized as a component of cardiac laminopathies since the first description of a large family with autosomal dominant Emery-Dreifuss muscular dystrophy, which is characterized by a skeletal and cardiac myopathy along with progressive heart block.11 Subsequently, mutations in LMNA were identified in individuals with isolated cardiac involvement, specifically DCM, AVB and infrequently atrial arrhythmias, including AF.9 In previous reports, the clinical presentation of AF in relation to the diagnosis AVB or DCM is not well-defined and our kindred uniquely demonstrates the early-onset nature of AF prior to the development of DCM and SCD in individuals with a mutation in LMNA.
Sudden cardiac death is increasingly being recognized as a manifestation of LMNA mutations.12 The etiology for SCD remains unknown but ventricular arrhythmias are suspected, not AVB, as SCD is not preventable by pacemaker implantation. We identified a similar phenotype in our pedigree, although, one member, but none of the others, was also diagnosed with long QT syndrome. This individual unfortunately suffered from SCD, and the contribution of the long QT syndrome is unknown. Other family members who suffered SCD did not have long QT syndrome noted on available electrocardiograms. The relationship between LMNA mutations and long QT syndrome is not clear, but prolongation of the QT interval was recently reported to occur in a subset of patients with Hutchinson-Gilford progeria.13 The role for lamin A/C in QT interval prolongation requires further investigation.
Lamin A/C mutations identified in individuals with isolated cardiac disease are predominantly found in the rod domain although no clear genotype-phenotype relationship exists.9,14 The majority of the currently identified predominantly cardiac disease-causing mutations are missense mutations and surprisingly only a few are nonsense or frameshift mutations that result in premature lamin termination.15,16 It is not clear whether the truncated lamins cause disease primarily by haploinsufficiency or by exerting dominant-negative effects. In support of the dominant-negative hypothesis, investigators described a novel nonsense mutation (R321X) in LMNA in a family with cardiac disease. Low quantities of the mutant protein were found in human tissues and overexpression of the mutant protein resulted in mis-localization of lamin A in HeLa cells.16 In our family, we did identify a much larger than expected decrease in wildtype mRNA and protein levels than would be predicted by haploinsufficiency. We also demonstrated the expression of the mutant mRNA but were unable to identify the K117fs truncated protein in PBLs. Further study is required to determine if this LMNA mutation causes cardiac disease via dominant-negative effects or haploinsufficiency.
Our report describes a family in which members with isolated AF in young adults was the first manifestation of a potentially lethal mutation in LMNA. In our kindred, three family members presented for medical care with AF, then went on to develop AVB while two members presented with both AF and AVB. The identification of a novel mutation in LMNA in this pedigree with isolated and highly penetrant cardiac disease highlights the need for further studies to determine the prevalence of LMNA mutations in patients with isolated AF, especially those with a positive family history for DCM or SCD. Similarly, mutations in the sodium channel SC5NA have also been reported in individuals with early-onset AF who develop late-onset AVB and DCM.17 These studies highlight a subset of patients who present with early-onset isolated AF that require close monitoring for the development of DCM and genetic testing for inherited arrhythmia syndromes. The phenotype of SCD appears to be unique to mutations in LMNA and placement of an ICD to prevent SCD has been suggested in these patients who will likely succumb to ventricular arrhythmias.18 The diagnosis of the underlying genetic etiology may lead to a diminution of SCD by intervening at an early stage of the disease process, when only AF or mild conduction system disease is present. Ultimately, an increased mechanistic understanding of cardiac conduction disease in the setting of LMNA mutations is needed to result in the development of novel therapeutics and prevention strategies that will increase survival of affected individuals.
Acknowledgments
Financial Support: H.L.Y. is supported by grants from NIH/GM and The Welch Foundation and V.G. is supported by grants from NIH/NHLBI and March of Dimes Birth Defects Association.
The authors thank family members for their participation; S. Tsai and T. Hyatt in the McDermott Center for Human Growth and Development for technical support; J. Chen for translation of medical records; M. Maitra and H. Hsia for helpful discussions; and P. J. Kannankeril for review of the manuscript.
Footnotes
Disclosures: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke. 1991;22:983–8. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 2.Ellinor PT, Yi BA, MacRae CA. Genetics of atrial fibrillation. Med Clin North Am. 2008;92:41–51. doi: 10.1016/j.mcna.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gollob MH. Begetting atrial fibrillation: Connexins and arrhythmogenesis. Heart Rhythm. 2008;5:888–91. doi: 10.1016/j.hrthm.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Darbar D. Genetics of atrial fibrillation: rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5:483–6. doi: 10.1016/j.hrthm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatkin D, Otway R, Vandenberg JI. Genes and atrial fibrillation: a new look at an old problem. Circulation. 2007;116:782–92. doi: 10.1161/CIRCULATIONAHA.106.688889. [DOI] [PubMed] [Google Scholar]
- 6.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–52. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 7.Rankin J, Ellard S. The laminopathies: a clinical review. Clin Genet. 2006;70(4):261–74. doi: 10.1111/j.1399-0004.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Gao B, Zhao X. A case report of family dilated cardiomyopathy. China Ji Shui Tan Department of Medicine Hospital Report. 1995;10:34. [Google Scholar]
- 9.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–24. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 10.Frischmeyer PA, van Hoof A, O’Donnell K, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 11.Bonne G, Di Barletta MR, Varnous S, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–8. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 12.van Berlo JH, de Voogt WG, van der Kooi AJ, et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J Mol Med. 2005;83:79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- 13.Merideth MA, Gordon LB, Clauss S, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebillon P, Bouchier C, Bidot LD, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–7. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLeod HM, Culley MR, Huber JM, et al. Lamin A/C truncation in dilated cardiomyopathy with conduction disease. BMC Med Genet. 2003;4:4. doi: 10.1186/1471-2350-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiger SK, Bar H, Ehlermann P, et al. Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. J Mol Med. 2008;86:281–9. doi: 10.1007/s00109-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 17.Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meune C, Van Berlo JH, Anselme F, et al. Primary prevention of sudden death in patients with lamin A/C gene mutations. N Engl J Med. 2006;354:209–10. doi: 10.1056/NEJMc052632. [DOI] [PubMed] [Google Scholar]