Abstract
Neurodegeneration and gliosis are the main neuropathological features of prion diseases. However, the molecular mechanisms involved in these processes remain unclear. Several studies have demonstrated changes in the expression of apoptotic factors and inflammatory cytokines in animals with experimental infection. Here we present the expression profiles of 15 genes implicated in the intrinsic and extrinsic apoptotic pathways in the central nervous systems of sheep naturally infected with scrapie. Expression changes obtained by real-time RT-PCR were also compared with the extent of classical scrapie lesions, such as prion deposition, neuronal vacuolisation, spongiosis, and astrogliosis as well as with the activation of caspase-3, using a stepwise regression. The results suggest that the factors assessed participate in apoptotic or inflammatory functions, depending on the affected area. The mitochondrial apoptosis pathway was associated with prion deposition in the prefrontal cortex (the less affected area), and with activation of caspase-3-mediated cell death via over-expression of BAK. In addition to its known association with astroglial activation, the extrinsic apoptosis pathway was also related to cell death and neuronal vacuolisation.
Keywords: apoptosis, prion, scrapie, mitochondrial pathway, extrinsic pathway
1. INTRODUCTION
Prion diseases or Transmissible Spongiform Encephalopathies (TSE), which include Creutzfeldt-Jakob Disease (CJD) in humans, Bovine Spongiform Encephalopathy (BSE) in cattle, and scrapie in sheep and goats, are fatal disorders that are produced by prions, which are a type of unconventional agent that targets the central nervous system (CNS) in mammals. All forms of prion disease are associated with an accumulation of abnormal isoforms of the prion protein (PrPSc) in nervous tissues. The neurological lesions of these diseases include spongiform changes, gliosis, and neuronal degeneration [4, 19]. Astrocytes also appear to be targets for the scrapie agent in the CNS [8, 22], and the disease condition results in widespread astrogliosis.
Neurodegeneration is the main neuropathological feature of prion diseases; however, the molecular mechanisms involved in prion-induced cell death remain unclear. Several studies of scrapie-infected sheep, mice, and hamsters have shown that apoptosis is a common cause of neuronal cell death in animal prion disease [34, 42]. Similarly, apoptosis has been considered as a common form of neuronal death in CJD and fatal familial insomnia [12].
Initiation of apoptosis has been broadly separated into two main pathways: (1) the extrinsic or death receptor pathway and (2) the intrinsic or mitochondrial pathway [9, 16]. Mitochondria play a central role in the regulation of apoptosis [3, 21]. Numerous signalling molecules, as well as pathological stimuli, converge in this organelle, inducing the release of pro-apoptotic molecules into the cytoplasm. Mitochondrial membrane permeability is directly controlled by the Bcl-2 family of proteins [13, 33], and the sensitivity of cells to apoptotic stimuli can depend on the balance of pro and anti-apoptotic Bcl-2 proteins. The role of Bcl-2 family proteins in prion disease has not been extensively investigated.
Furthermore, few studies have been devoted to investigating the implications of the extrinsic apoptosis pathway in the neuropathology induced by prions [17, 26, 34]. This regulatory pathway is also related to the inflammation induced in different neurodegenerative diseases [28, 44]. In addition, the neuronal apoptosis described in some TSE has been associated with the presence of inflammatory cells and cytokines [32, 38].
Analysis of factors involved in the regulation of these apoptotic pathways will be useful in clarifying the pathogenic mechanisms responsible for prion diseases. Here we report the gene expression analysis of fifteen genes that encode regulatory factors involved in the mitochondrial and extrinsic pathways of apoptosis in different regions of the CNS of sheep naturally infected with scrapie. Finally, we investigated the relationship between the regulation of these pathways and the presence of neuropathological features of scrapie.
2. MATERIALS AND METHODS
2.1. Sample selection
Thirteen female Rasa aragonesa sheep were included in the present study. Scrapie was diagnosed in vivo in 8 sheep by means of neurological examination and/or third eyelid biopsy [40]. When these animals were sacrificed, all exhibited clinical signs of scrapie in a terminal state. The diagnosis was confirmed using rapid tests and immunohistochemical techniques to detect PrPSc [2]. The animals were from a closed regulatory monitored flock and from the Spanish Scrapie Surveillance Programme. Five control animals of the same breed were selected from a different flock in which no scrapie cases have been reported to date. All animals analysed were 3 to 6 years old and displayed the ARQ/ARQ genotype for PRNP, which is the most susceptible genotype in this breed of animals [1]. Care and handling of the animals were performed according to the rules established by the National Research Council’s guide.
Animals were sacrificed by intravenous injection of sodium pentobarbital and exsanguination. Necropsy did not reveal any other pathological signs upon physical examination of the scrapie-infected or control animals. Immediately after sacrifice, samples from the medulla oblongata, diencephalon, cerebellum, and prefrontal cortex were harvested from each sheep. Because the histopathological lesions of scrapie are bilateral, half of these samples were placed in RNAlater® (Ambion, Austin, TX, USA) for 24 h at 4 °C and then frozen at –80 °C until RNA extraction. The other half were formalin-fixed and paraffin-embedded for further histopathological analysis.
2.2. Total RNA isolation and cDNA synthesis
Total RNA was isolated from tissue samples using the RNeasy Lipid Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In order to avoid contamination with genomic DNA, the samples were treated with two units of TURBO DNase (Ambion) for 25 min at 37 °C. The RNA concentration was assessed with a NanoDrop ND-100 Spectrophotometer (Wilmington, DE, USA), and its quality was determined by denaturing formaldehyde agarose gel electrophoresis, which revealed sharp and intense 18S and 28S rRNA bands. Complementary DNA for each animal and CNS region was synthesised from 2 μg of total RNA using random hexamers and the Superscript First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA). Parallel reactions without reverse transcriptase enzyme were conducted to confirm the complete elimination of genomic DNA.
2.3. Real-time quantitative PCR
The expression of fifteen genes involved in apoptosis regulation was evaluated using quantitative real-time RT-PCR. Primer express 2.0 software (Applied Biosystems, Foster City, CA, USA) was used to design primers and probes based on known ovine and bovine sequences. Primer design accounted for gene structure, and combinations of forward and reverse primers were designed in different exons. When using heterologous primers, the resulting PCR fragments were purified with either the NucleoSpin Extract kit (Macherey-Nagel, Düren, Germany) or reactive EXOSAP-IT (USB, Cleveland, OH, USA) and bidirectionally sequenced with the PCR primers using the Big Dye Terminator Cycle Sequencing kit and ABI PRISM 3100 system (Applied Biosystems). The identity of the PCR products was confirmed by BLAST comparison with the GenBank database. The accession numbers for the new ovine cDNA sequences and the length of the RT-PCR products are shown in Table I.
Table I.
Genes, GenBank accession numbers of the sequences used for primer design and origin species. Primers (F: forward and R: reverse) and TaqMan probes (P) used for gene amplification. Length of the amplicon and GenBank accession numbers for the new ovine cDNA sequences. Real-time RT-PCR conditions: annealing temperature, primer and probe concentrations, correlation coefficient (r 2) and slope of the standard curve.
| Genes | GenBank | Species | Primers and Probes | bp | New ovine sequences | PCR conditions |
|||
|---|---|---|---|---|---|---|---|---|---|
| Ta | (nM) | r2 | Slope | ||||||
| AIF | AF529274 | Ovine | F: TGGAGGCACAGCTGCTTTT | 94 | 59 | 900 | 0.991 | −3.44 | |
| R: CGGCAGCTCAGGATCTTCA | 900 | ||||||||
| APAF1 | XM_617966 | Bovine | F: GCCAAGCAGGAGGTCGATAA | 117 | EF447257 | 60 | 900 | 0.995 | −3.24 |
| R: GCAAGCATGGTAAACAGCATCT | 900 | ||||||||
| BAD | NM_001035459 | Bovine | F: CAGAGTTTGAGCCGAGTGAG | 110 | AY547260 | 60 | 300 | 0.982 | −3.58 |
| R: GGCTGTTAGCCAGTGCTTG | 300 | ||||||||
| BAK | AF164518 | Ovine | F: CCCAGAGCCTACCAGCACC | 164 | 60 | 300 | 0.997 | −3.3 | |
| R: ACAGGCTGGACGCGATCTT | 300 | ||||||||
| BCL2 | AY423861 | Ovine | F: TGGTGGAGGAGCTCTTCAGG | 65 | 60 | 300 | 0.995 | −3.39 | |
| R: TCCGAACTCAAAGAAGGCCA | 300 | ||||||||
| P: ATGCGCCCCCAGTTCACCCC | 150 | ||||||||
| BCL-XL | AF164517 | Ovine | F: CAGGCGATGAGTTTGAACTG | 121 | 60 | 300 | 0.996 | −3.32 | |
| R: TCCCGGAAGAGTTCATTCAC | 300 | ||||||||
| P: CGACCTGACGTCCCAGCTCCACA | 150 | ||||||||
| BCL-XS | AY568555 | Ovine | F: TCAGAGCTTTGAACAGGACACG | 98 | 60 | 300 | 0.993 | −3.42 | |
| R: TCAGGAACCAGCGGTTGAAG | 300 | ||||||||
| FAS | AB011671 | Ovine | F: CTGGAGCAAGTTCCTGCCAA | 105 | 60 | 300 | 0.996 | −3.14 | |
| R: CTCCGTCGCGTTTGC | 900 | ||||||||
| FASL | XM_584322 | Bovine | F: GAAGAGGAGGGACCACAACACA | 147 | EF458007 | 61 | 900 | 0.990 | −3.32 |
| R: CTTTGGCTGGTGGACTCTCTGA | 900 | ||||||||
| P: TGGTGGCCCTGGTTGGATTGGG | 200 | ||||||||
| MCL1 | AF144097 | Ovine | F: GGCTTTCCAAGGCATGCTT | 112 | 60 | 900 | 0.997 | −3.32 | |
| R: GAGTCACAATCCTGCCCCAG | 900 | ||||||||
| P: CGAGTGATGGTTCATGTTTTCAGTGACG | 150 | ||||||||
| PARP1 | D90073 | Bovine | F: AGTGAAGGCCACGATTGAGAA | 147 | EF458008 | 59 | 900 | 0.991 | −3.32 |
| R: TCAGACACGACACGGATGTTG | 900 | ||||||||
| TGFB1 | X76916 | Ovine | F: GCACGTGGAGCTGTACCAGAA | 125 | 60 | 900 | 0.998 | −3.32 | |
| R: GCCACTGCCGCACAACTC | 900 | ||||||||
| TNFA | X56756 | Ovine | F: CCCTTCCACCCCCTTGTTC | 277 | 60 | 900 | 0.979 | −3.32 | |
| R: GGCTCTTGATGGCAGAGAGGAT | 900 | ||||||||
| TNFR1 | U90937 | Bovine | F: CTCAGGACCCAGGCACTACAG | 134 | EF447258 | 59 | 900 | 0.998 | −3.25 |
| R: CCCGCAAATGATGGAGTAGAG | 900 | ||||||||
| TNFR2 | NM_001040490 | Bovine | F: GATGGGTCCTGTCTTGGTGTGT | 144 | EU542424 | 60 | 300 | 0.993 | −3.32 |
| R: CAAAACAACAAGGGCTCCAGAA | 300 | ||||||||
Gene expression was analysed using SYBR® Green (PE Applied Biosystems) assays. A dissociation curve protocol was run after every real-time RT-PCR reaction in order to identify the presence of spurious PCR bands or high levels of primer dimers. These artefacts were observed for the BCL2, BCL-X L, MCL1, and FASL genes. In these cases, expression was analysed using TaqMan probes designed to recognise the exon-exon boundaries. The appropriate primers and probes used for amplification of all genes, as well as their concentrations, are shown in Table I.
Real-time RT-PCR amplifications were performed in an ABI-Prism 7000 Sequence Detection System (PE Applied Biosystems). All real-time RT-PCR reactions were run in triplicate in a total reaction volume of 10 μL using 10-20 ng of cDNA as the template. Amplification of cDNA by PCR was achieved using universal cycling conditions, with an initial 10 min activation and denaturation step at 95 °C, followed by 40 cycles of 15 s at 95 °C and 30 s at the suitable annealing temperature (Tab. I). The levels of gene expression were determined by the comparative Ct method.
A normalisation factor (NF) was used to determine the expression level of each gene in each sample, according to the recommendation of Vandesompele et al. [39]. The NF was calculated as the geometric mean of the quantity (Q) of the 3 or 4 most stable housekeeping genes (from a total of eight) in each tissue (GAPDH, SDHA, and HPRT for the prefrontal cortex; YWHAZ, SDHA, GAPDH, and G6PDH for the diencephalon; G6PDH, SDHA, and ACTB for the cerebellum; and G6PDH, GAPDH, and RPL32 for the medulla oblongata). Primers and probes used for housekeeping gene amplification as well as the procedure for arranging these genes based on the order of their stability will be described elsewhere (Lyahyai et al., unpublished results1).
2.4. Histopathology and immunohistochemistry
The histopathological study of samples obtained from the medulla oblongata, diencephalon, prefrontal cortex, and cerebellum was performed using haematoxylin-eosin stained slices (one from each animal). The extent of neuronal vacuolation and neuropil spongiosis was evaluated in these stained sections and scored on a scale ranging from 0–5 (0 = no neuron/neuropil vacuolation or gliosis and 5 = substantial lesions throughout the region).
PrPSc detection was performed for adjacent sections, as previously described [15]. Briefly, after pretreatment with 98% formic acid, hydrated autoclaving, and digestion with proteinase K (4 μg/mL), the sections were incubated with endogenous peroxidase blocking reagent (DAKO, Glostrup, Denmark) for 10 min. The sections were then incubated with monoclonal primary antibody L42 (R-Biopharm, Germany; dilution 1:500) at RT for 30 min. The enzyme-conjugated polymer Envision (DAKO, Glostrup, Denmark; 30 min) was used as the visualisation system, and DAB (DAKO; 10 min) was used as the chromogen. The sections were counterstained by treatment with haematoxylin.
Astrogliosis was evaluated on the basis of glial fibrillary acidic protein (GFAP) immunostaining, and induction of caspase-dependent apoptosis was evaluated by immunohistochemical detection of the active form of caspase-3. In both cases, after heat-induced epitope retrieval pretreatment with citrate buffer (pH 6.0), the sections were incubated for 1 h at RT with the appropriate primary antibody. The antibodies employed recognise GFAP (rabbit polyclonal, 1:400, DAKO) and caspase-3 (mouse monoclonal Ab-4, 1:40, Calbiochem, Darmstadt, Germany), and the caspase-3 antibody preferentially reacts with the 11 kDa caspase-3 molecule, which is the activated form of caspase-3. In routine immunoreactions, sheep lymph node tissue sections were included as positive controls, whereas omission of primary antibodies in control and scrapie slides served as negative controls.
Global quantification of PrPSc, GFAP, and caspase-3 was also determined using the same scoring system ranging from 0 to 5 (0 = weak immunoreactivity and 5 = strong immunoreactivity). The subjective scores were carried out by two observers.
2.5. Statistics
Quantitative results obtained from real-time RT-PCR assays are expressed as mean ± standard error of the mean values. Gene expression means for the control group were arbitrarily standardised to 1.0, and data for the scrapie-infected group were compared based on this calibrator. The student t-test analysis was applied to determine whether the differences observed between groups were statistically significant (P < 0.05).
To score the neuropathological and immunohistochemical features of scrapie, the differences between controls and scrapie-infected sheep were assessed using the non-parametric Mann-Whitney test. The effect of area on the intensity of anatomopathological lesions and immunostaining was analysed using the Kruskal-Wallis test. The scores were plotted as a function of the anatomical area, and data are expressed as mean ± standard deviation.
Principal component analysis was performed using the covariance matrix to study the magnitude of co-expression for apoptotic pathways. Genes were grouped according to their participation in the extrinsic and mitochondrial apoptosis pathways and their known functions in these pathways (see Tab. II). The previously reported expression data for BAX, which is a pro-apoptotic BCL2 member, were included in these analyses [24].
Table II.
Association between gene expression and prion-related lesions in 4 CNS areas. Genes are classified in 7 groups according to their role in apoptotic pathways. P1: BAX, BAD, BAK, BCLXS, BCL2, BCLXL, MCL1, AIF, APAF1, PARP1 (mitochondrial pathway); P1a: BAX, BAD, BAK, BCLXS (pro-apoptotic Bcl-2 family factors); P1b: BCL2, BCLXL, MCL1 (anti-apoptotic Bcl-2 family factors); P1c: AIF, APAF1, PARP1 (other apoptosis factors); P2: FAS, FASL, TNFA, TNFR1, TNFR2, TGFB (extrinsic pathway); P2a: FAS, TNFR1, TNFR2 (receptors); P2b: FASL, TNFA, TGFB (ligands). Lesions: Pr (prion deposition); Sp (neuropil spongiosis); Vac (neuronal vacuolisation) and Ag (astrogliosis). PC: percentage of the variability explained by the first principal component of each group. Significant P-values (P < 0.05) obtained using a stepwise regression between the first principal component of each group and the lesions are shown. (ns: not statistically significant).
We also studied the relationship between the gene expression profiles of each gene or the first principal component of the apoptotic pathways and histopathological lesions by a stepwise regression with forward selection and backward elimination set at P < 0.1. The model used for analysis of each combination of gene or principal component and tissue was the following:
where y i is the gene expression profile of the ith individual; pr i, nv i, sp i, as i, and cp i represent numerical evaluations of the ith individual for PrPSc, neuronal vacuolisation, spongiosis, astrogliosis, and activation of caspase-3, respectively; b 1, b 2, b 3, b 4 , and b 5 are the slopes of the multiple regression associated with each variable; and e i is the residual.
Finally, a Bonferroni correction was performed within tissue to correct for multiple testing. The statistical analyses were performed using either SPSS (Chicago, IL, USA) or STATISTIX (Analitical Software, Tallahassee, FL, USA).
3. RESULTS
3.1. Gene expression profiles
The expression of 15 genes that encode proteins involved in the regulation of the mitochondrial and extrinsic pathways of apoptosis was analysed in 4 different areas of the CNS of scrapie-infected and control sheep.
Samples that were treated and untreated with reverse transcriptase showed differences of more than 6 cycles for every gene, indicating that genomic DNA was successfully removed (data not shown). Standard curves for all analysed factors displayed appropriate slopes and correlation values (Tab. I).
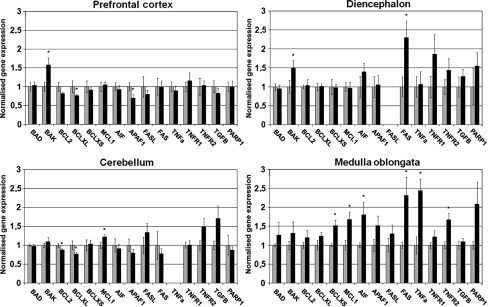
The expression profiles varied according to the area analysed (Fig. 1). In scrapie-infected medulla oblongata, 3 genes involved in the regulation of the mitochondrial pathway of apoptosis (BCL-X S , MCL1, AIF) and 3 genes involved in the extrinsic pathway (TNFA, TNFR2, FAS) were significantly over-expressed.
Figure 1.
Expression profiles of 15 genes implicated in the intrinsic and extrinsic apoptotic pathways in the medulla oblongata, diencephalon, cerebellum, and prefrontal cortex of control (grey bar) and naturally-infected scrapie sheep (black bar). Significant differences were detected using the Student t-test. * P < 0.05.
In the diencephalon, only the mRNA levels of the pro-apoptotic genes BAK and FAS were significantly increased. The level of FASL was very low in both scrapie and control diencephalons, which rendered quantification difficult.
In the cerebellum, changes in expression were only significant for anti-apoptotic genes. BCL2 and BCL-X L were down-regulated, and an increase in MCL1 was observed. Similarly, slight but significant decreases in BCL-X L and APAF1 were detected in scrapie-infected prefrontal cortex. In this tissue, BAK was up-regulated. The differences in the expression of genes involved in the extrinsic pathway of apoptosis were not significant in either the prefrontal cortex or cerebellum.
3.2. Scrapie-related lesions
The neuropathological features of scrapie were evaluated in the 4 CNS areas of 5 control and 9 scrapie-infected sheep.
Neuronal vacuolisation was only detected in affected animals, and the highest lesion levels were observed in the medulla oblongata and diencephalon (Fig. 2). The Kruskal-Wallis test revealed a significant effect of area with regards to this feature. Besides the high variability observed in the scrapie group, the differences compared to controls were statistically significant for the diencephalon (P < 0.05).
Figure 2.
Scoring of histopathological lesions in each analysed area. Scores for neuronal vacuolation, neuropil spongiosis, prion deposition and reactive gliosis (GFAP) are shown as means ± standard error. Black bars: control sheep, grey bars: scrapie-infected sheep. Areas: medulla oblongata (MO), diencephalon (D), prefrontal cortex (PFC), and cerebellum (Cb). Student t-test * P < 0.05, ** P < 0.01.
Neuropil spongiosis was observed in both groups of animals (Fig. 2). In the infected sheep, spongiosis scoring was stable in all analysed regions. In contrast, the control group displayed significant differences between areas (Kruskal-Wallis P < 0.05) due to the low levels of spongiosis observed in the prefrontal cortex. The Mann-Whitney test showed that the increase in lesions in scrapie animals was statistically significant (P < 0.05) in all areas except the cerebellum.
The effect of the CNS area in prion deposition was also statistically significant (P < 0.01). The origin of this difference was the low level of PrPSc immunostaining observed in the prefrontal cortex (Fig. 2).
Astrogliosis was evaluated by GFAP detection. A generalised increase in the astroglial marker GFAP was noticed in the scrapie-affected cases (Fig. 2). Apparent hyperplasia and hypertrophy of stellate-shaped astrogial cells was observed in both the grey and white matter when compared to control brains, indicative of reactive astrogliosis. Differences between areas were not observed in either control or infected sheep.
Finally, cell death was also analysed by immunodetection of the activated form of caspase-3 using an antibody that preferentially reacts with the activated form of this protein. In general, very weak staining was observed in the different CNS sections, and it was not significantly different between the scrapie and control groups (data not shown).
3.3. Relationship between expression profiles and histopathological lesions
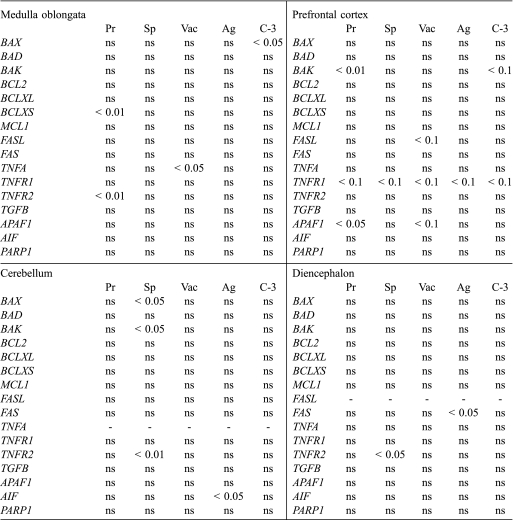
The analysed genes were grouped according to their known functions in the apoptotic pathways. In most cases, the first principal component represented more than 60% of the total variability, indicating a common regulation of the genes involved in each specific pathway (Tab. II).
Prion deposition was positively associated with the first principal component of the mitochondrial apoptosis pathway in the prefrontal cortex, as demonstrated by the statistical significance of its covariate in the stepwise regression analysis. This association is apparent in the up-regulation of BAK (Tab. III). The expression change contributes to the positive association observed between the pro-apoptotic group of Bcl-2 factors and the activation of caspase-3. The extrinsic pathway was also associated with prion immunostaining in the same region.
Table III.
Association between individual gene expression levels and histopathological scrapie lesions in four analysed CNS areas. The table shows significant P-values (P < 0.1) obtained by Stepwise regression analysis. (ns: not statistically significant). Lesions: Pr (prion deposition); Sp (neuropil spongiosis); Vac (neuronal vacuolisation) and Ag (astrogliosis).
Astrogliosis was positively associated with the extrinsic pathway in the medulla oblongata and prefrontal cortex.
Neuropil spongiosis exhibited a significant association with both the first principal component of the death receptor group and that of the ligand group in the cerebellum. The covariate of the ligand sub-group was also related to neuronal vacuolisation in the medulla oblongata.
Finally, after Bonferroni correction for multiple testing, the stepwise regression between BAK and prion deposition in the prefrontal cortex remained significant.
4. DISCUSSION
Apoptosis has been widely considered as one of the main types of Programmed Cell Death that causes neurodegeneration in prion diseases [10, 23, 37]. A comprehensive understanding of the molecular mechanisms that regulate apoptosis and their relationship to the presence of PrPSc or other prion-related lesions may provide new targets for future treatments of human diseases.
In the present work, the expression profiles of fifteen genes involved in the mitochondrial and extrinsic pathways of apoptosis were analysed in the CNS of sheep that were naturally infected with scrapie. Distinct expression profiles were found in each CNS region, indicating that different mechanisms could be involved in TSE-related cell death, depending on the area (Fig. 1).
On the one hand, expression changes compatible with apoptosis induction were detected via over-expression of pro-apoptotic genes (BAK, BCL-X S, AIF, FAS, TNFA, TNFR2) in different areas or down-regulation of the anti-apoptotic genes BCL2 and BCL-X L in the prefrontal cortex. The reduced mRNA levels of BCL2 and BCL-X L play a primary role in PrP-/- neuronal cell death [30], as well as in other neurodegenerative diseases exhibiting anomalous protein accumulation, such as spinocerebellar ataxia types 3 [7] and 7 [41] or Alzheimer disease [46].
On the other hand, the decrease in APAF1 expression in scrapie prefrontal cortex and the up-regulation of MCL1 observed in the medulla oblongata and cerebellum of scrapie animals suggest the presence of anti-apoptotic mechanisms. In this context, the Apaf-1 deficiency confers resistance to cyt-c-dependent caspase activation and apoptosis in vitro [18, 43] and in vivo [36, 45]. In addition, the interaction of Mcl-1 with different pro-apoptotic Bcl-2 members, such as Bax and Bak, inhibits the opening of the mitochondrial permeability transition pore [5, 47] and subsequent activation of caspase-3. These observations indicate that both pro and anti-apoptotic mechanisms are activated in prion-infected tissues.
In order to determine any possible association between the scrapie lesions and the induction of a specific apoptotic pathway, stepwise regressions were performed between the gene expression data and the prion-related lesions. Although we are aware that the histopathological valuation does not correspond to a real scale, our procedure can reliably detect the dependence between the different parameters. However, these results are not meant to serve in a predictive capacity.
Using this statistical methodology, prion deposition was positively associated with the first principal component of the mitochondrial apoptosis pathway in the prefrontal cortex, which was one of the less affected areas. This was mainly evident for BAK expression. The reliability of this BAK/prion association was confirmed after Bonferroni correction. Over-expression of BAK has been reported in other neurodegenerative diseases, such as Alzheimer disease [20, 29], but never before in prion diseases. In our study, up-regulation of BAK was associated with the presence of the active form of caspase-3 in the prefrontal cortex. This result indicates that this factor can play an important role in neurodegeneration during the early stages of prion disease, when the levels of PrPSc are still relatively low. In addition, the neuroprotective role of PrPC seems to be specific for BAX-mediated apoptosis, and it is unable to arrest BAK-mediated cellular death [27].
Different factors comprising the death receptor pathway of apoptosis have been implicated in the neuronal death that occurs in different prion disease models [35]. For instance, cerebella from CJD patients exhibit an increase in Fas and Fas ligand proteins [26]. Although the increase in FASL, TNFR2, and TGFB observed herein was not significant, the stepwise regression analysis revealed a positive association between the sub-groups of this pathway and spongiosis. This specific apoptosis pathway could then participate in scrapie-induced cellular loss in the cerebellum.
Our statistical analysis also demonstrated a relationship between the ligand sub-group, and neuronal vacuolisation in the medulla oblongata. To our knowledge, this is the first time that this relationship has been reported, although a known association exists between Fas and TNF-alpha with glial and neuronal vacuolisation in other brainstem neurodegenerative models [6].
Our research shows that the extrinsic pathway of apoptosis had a positive association with astrogliosis in the medulla oblongata and the prefrontal cortex. This association has been previously reported. TNFR2 can participate in cellular apoptosis [14], as well as in inflammation [31], and the increase in Fas has also been associated with reactive gliosis and neuroinflammation [11, 25].
In conclusion, our findings indicate that the mitochondrial apoptosis pathway is activated during early stages of prion deposition via up-regulation of BAK. In addition, induction of the extrinsic pathway is related to the reactive gliosis and neuronal loss that occur in sheep naturally infected with scrapie. These data provide new clues to further understand the molecular pathways that regulate cell death in prion diseases.
Acknowledgments
J. Lyahyai and C. Serrano were supported by doctoral grants from the University of Zaragoza and DGA (B171/2004), respectively. This work was partially financed by the research projects EET20-03/09890 and UZ2007-Bio-10.
Footnotes
Lyahyai J., Serrano C., Ranera B., et al., Effect of scrapie on the stability of housekeeping genes, Anim. Biotechnol. (submitted).
References
- 1.Acin C., Martin-Burriel I., Monleon E., Rodellar C., Badiola J.J., Zaragoza P.. PrP polymorphisms in Spanish sheep affected with natural scrapie. Vet. Rec. 2004;155:370–372. doi: 10.1136/vr.155.12.370. [DOI] [PubMed] [Google Scholar]
- 2.Bolea R., Monleon E., Schiller I., Raeber A.J., Acin C., Monzon M.. et al. Comparison of immunohistochemistry and two rapid tests for detection of abnormal prion protein in different brain regions of sheep with typical scrapie. J. Vet. Diagn. Invest. 2005;17:467–469. doi: 10.1177/104063870501700511. [DOI] [PubMed] [Google Scholar]
- 3.Brenner C., Cadiou H., Vieira H.L., Zamzami N., Marzo I., Xie Z.. et al. Bcl-2 and Bax regulate the channel activity of the mitochondrial adenine nucleotide translocator. Oncogene. 2000;19:329–336. doi: 10.1038/sj.onc.1203298. [DOI] [PubMed] [Google Scholar]
- 4.Budka H., Aguzzi A., Brown P., Brucher J.M., Bugiani O., Gullotta F.. et al. Neuropathological diagnostic criteria for Creutzfeldt-Jakob disease (CJD) and other human spongiform encephalopathies (prion diseases) Brain Pathol. 1995;5:459–466. doi: 10.1111/j.1750-3639.1995.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 5.Cuconati A., Mukherjee C., Perez D., White E.. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe W., Stoica G., Lynn W., Wong P.K.. Neurodegeneration induced by MoMuLV-ts1 and increased expression of Fas and TNF-alpha in the central nervous system. Brain Res. 1998;779:1–8. doi: 10.1016/s0006-8993(97)00929-3. [DOI] [PubMed] [Google Scholar]
- 7.Chou A.H., Yeh T.H., Kuo Y.L., Kao Y.C., Jou M.J., Hsu C.Y.. et al. Polyglutamine-expanded ataxin-3 activates mitochondrial apoptotic pathway by upregulating Bax and downregulating Bcl-xL. Neurobiol. Dis. 2006;21:333–345. doi: 10.1016/j.nbd.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Diedrich J.F., Bendheim P.E., Kim Y.S., Carp R.I., Haase A.T.. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc. Natl. Acad. Sci. USA. 1991;88:375–379. doi: 10.1073/pnas.88.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elmore S.. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ettaiche M., Pichot R., Vincent J.P., Chabry J.. In vivo cytotoxicity of the prion protein fragment 106–126. J. Biol. Chem. 2000;275:36487–36490. doi: 10.1074/jbc.C000579200. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer I., Puig B., Krupinsk J., Carmona M., Blanco R.. Fas and Fas ligand expression in Alzheimer’s disease. Acta Neuropathol. 2001;102:121–131. doi: 10.1007/s004010000325. [DOI] [PubMed] [Google Scholar]
- 12.Gray F., Chretien F., Adle-Biassette H., Dorandeu A., Ereau T., Delisle M.B.. et al. Neuronal apoptosis in Creutzfeldt-Jakob disease. J. Neuropathol. Exp. Neurol. 1999;58:321–328. doi: 10.1097/00005072-199904000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Green D.R., Kroemer G.. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 14.Grell M., Zimmermann G., Hulser D., Pfizenmaier K., Scheurich P.. TNF receptors TR60 and TR80 can mediate apoptosis via induction of distinct signal pathways. J. Immunol. 1994;153:1963–1972. [PubMed] [Google Scholar]
- 15.Hardt M., Baron T., Groschup M.H.. A comparative study of immunohistochemical methods for detecting abnormal prion protein with monoclonal and polyclonal antibodies. J. Comp. Pathol. 2000;122:43–53. doi: 10.1053/jcpa.1999.0343. [DOI] [PubMed] [Google Scholar]
- 16.Igney F.H., Krammer P.H.. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 17.Jamieson E., Jeffrey M., Ironside J.W., Fraser J.R.. Apoptosis and dendritic dysfunction precede prion protein accumulation in 87V scrapie. Neuroreport. 2001;12:2147–2153. doi: 10.1097/00001756-200107200-00021. [DOI] [PubMed] [Google Scholar]
- 18.Jia L., Srinivasula S.M., Liu F.T., Newland A.C., Fernandes-Alnemri T., Alnemri E.S., Kelsey S.M.. Apaf-1 protein deficiency confers resistance to cytochrome c-dependent apoptosis in human leukemic cells. Blood. 2001;98:414–421. doi: 10.1182/blood.v98.2.414. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y.S., Carp R.I., Callahan S., Wisniewski H.M.. Incubation periods and histopathological changes in mice injected stereotaxically in different brain areas with the 87V scrapie strain. Acta Neuropathol. (Berl.) 1990;80:388–392. doi: 10.1007/BF00307692. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura Y., Shimohama S., Kamoshima W., Ota T., Matsuoka Y., Nomura Y.. et al. Alteration of proteins regulating apoptosis, Bcl-2, Bcl-x, Bax, Bak, Bad, ICH-1 and CPP32, in Alzheimer’s disease. Brain Res. 1998;780:260–269. doi: 10.1016/s0006-8993(97)01202-x. [DOI] [PubMed] [Google Scholar]
- 21.Kroemer G., Zamzami N., Susin S.A.. Mitochondrial control of apoptosis. Immunol. Today. 1997;18:44–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- 22.Lazarini F., Boussin F., Deslys J.P., Tardy M., Dormont D.. Astrocyte gene expression in experimental mouse scrapie. J. Comp. Pathol. 1994;111:87–98. doi: 10.1016/s0021-9975(05)80114-8. [DOI] [PubMed] [Google Scholar]
- 23.Liberski P.P., Sikorska B., Bratosiewicz-Wasik J., Gajdusek D.C., Brown P.. Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: from apoptosis to autophagy. Int. J. Biochem. Cell Biol. 2004;36:2473–2490. doi: 10.1016/j.biocel.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Lyahyai J., Bolea R., Serrano C., Vidal E., Pumarola M., Badiola J.J.. et al. Differential expression and protein distribution of Bax in natural scrapie. Brain Res. 2007;1180:111–120. doi: 10.1016/j.brainres.2007.08.085. [DOI] [PubMed] [Google Scholar]
- 25.Martinez M., Fernandez-Vivancos E., Frank A., De la Fuente M., Hernanz A.. Increased cerebrospinal fluid fas (Apo-1) levels in Alzheimer’s disease. Relationship with IL-6 concentrations. Brain Res. 2000;869:216–219. doi: 10.1016/s0006-8993(00)02363-5. [DOI] [PubMed] [Google Scholar]
- 26.Puig B., Ferrer I.. Cell death signaling in the cerebellum in Creutzfeldt-Jakob disease. Acta Neuropathol. 2001;102:207–215. [PubMed] [Google Scholar]
- 27.Roucou X., Giannopoulos P.N., Zhang Y., Jodoin J., Goodyer C.G., LeBlanc A.. Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ. 2005;12:783–795. doi: 10.1038/sj.cdd.4401629. [DOI] [PubMed] [Google Scholar]
- 28.Saas P., Boucraut J., Quiquerez A.L., Schnuriger V., Perrin G., Desplat-Jego S.. et al. CD95 (Fas/Apo-1) as a receptor governing astrocyte apoptotic or inflammatory responses: a key role in brain inflammation? J. Immunol. 1999;162:2326–2333. [PubMed] [Google Scholar]
- 29.Sajan F.D., Martiniuk F., Marcus D.L., Frey W.H. 2nd, Hite R., Bordayo E.Z., Freedman M.L.. Apoptotic gene expression in Alzheimer’s disease hippocampal tissue. Am. J. Alzheimers Dis. Other Demen. 2007;22:319–328. doi: 10.1177/1533317507302447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakudo A., Lee D.C., Saeki K., Matsumoto Y., Itohara S., Onodera T.. Tumor necrosis factor attenuates prion protein-deficient neuronal cell death by increases in anti-apoptotic Bcl-2 family proteins. Biochem. Biophys. Res. Commun. 2003;310:725–729. doi: 10.1016/j.bbrc.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 31.Santee S.M., Owen-Schaub L.B.. Human tumor necrosis factor receptor p75/80 (CD120b) gene structure and promoter characterization. J. Biol. Chem. 1996;271:21151–21159. doi: 10.1074/jbc.271.35.21151. [DOI] [PubMed] [Google Scholar]
- 32.Sawada M.. Brain cytokine network and novel characteristics of microglia. Nihon Shinkei Seishin Yakurigaku Zasshi. 1999;19:151–154. (in Japanese) [PubMed] [Google Scholar]
- 33.Shimizu S., Narita M., Tsujimoto Y.. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 34.Siso S., Puig B., Varea R., Vidal E., Acin C., Prinz M.. et al. Abnormal synaptic protein expression and cell death in murine scrapie. Acta Neuropathol. (Berl.) 2002;103:615–626. doi: 10.1007/s00401-001-0512-6. [DOI] [PubMed] [Google Scholar]
- 35.Stobart M.J., Parchaliuk D., Simon S.L., Lemaistre J., Lazar J., Rubenstein R., Knox J.D.. Differential expression of interferon responsive genes in rodent models of transmissible spongiform encephalopathy disease. Mol. Neurodegener. 2007;2:5. doi: 10.1186/1750-1326-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoka V., Turk V., Bredesen D.E.. Differential regulation of the intrinsic pathway of apoptosis in brain and liver during ageing. FEBS Lett. 2006;580:3739–3745. doi: 10.1016/j.febslet.2006.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theil D., Fatzer R., Meyer R., Schobesberger M., Zurbriggen A., Vandevelde M.. Nuclear DNA fragmentation and immune reactivity in bovine spongiform encephalopathy. J. Comp. Pathol. 1999;121:357–367. doi: 10.1053/jcpa.1999.0333. [DOI] [PubMed] [Google Scholar]
- 38.Van Everbroeck B., Dewulf E., Pals P., Lubke U., Martin J.J., Cras P.. The role of cytokines, astrocytes, microglia and apoptosis in Creutzfeldt-Jakob disease. Neurobiol. Aging. 2002;23:59–64. doi: 10.1016/s0197-4580(01)00236-6. [DOI] [PubMed] [Google Scholar]
- 39.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F.. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vargas F., Lujan L., Bolea R., Monleon E., Martin-Burriel I., Fernandez A.. et al. Detection and clinical evolution of scrapie in sheep by 3rd eyelid biopsy. J. Vet. Intern. Med. 2006;20:187–193. doi: 10.1892/0891-6640(2006)20[187:daceos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 41.Wang H.L., Yeh T.H., Chou A.H., Kuo Y.L., Luo L.J., He C.Y.. et al. Polyglutamine-expanded ataxin-7 activates mitochondrial apoptotic pathway of cerebellar neurons by upregulating Bax and downregulating Bcl-x(L) Cell. Signal. 2006;18:541–552. doi: 10.1016/j.cellsig.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 42.Williams A., Lucassen P.J., Ritchie D., Bruce M.. PrP deposition, microglial activation, and neuronal apoptosis in murine scrapie. Exp. Neurol. 1997;144:433–438. doi: 10.1006/exnr.1997.6424. [DOI] [PubMed] [Google Scholar]
- 43.Wolf B.B., Schuler M., Li W., Eggers-Sedlet B., Lee W., Tailor P.. et al. Defective cytochrome c-dependent caspase activation in ovarian cancer cell lines due to diminished or absent apoptotic protease activating factor-1 activity. J. Biol. Chem. 2001;276:34244–34251. doi: 10.1074/jbc.M011778200. [DOI] [PubMed] [Google Scholar]
- 44.Wyss-Coray T., Mucke L.. Inflammation in neurodegenerative disease – a double-edged sword. Neuron. 2002;35:419–432. doi: 10.1016/s0896-6273(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 45.Yakovlev A.G., Ota K., Wang G., Movsesyan V., Bao W.L., Yoshihara K., Faden A.I.. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J. Neurosci. 2001;21:7439–7446. doi: 10.1523/JNEUROSCI.21-19-07439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao M., Nguyen T.V., Pike C.J.. Beta-amyloid-induced neuronal apoptosis involves c-Jun N-terminal kinase-dependent downregulation of Bcl-w. J. Neurosci. 2005;25:1149–1158. doi: 10.1523/JNEUROSCI.4736-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou P., Qian L., Bieszczad C.K., Noelle R., Binder M., Levy N.B., Craig R.W.. Mcl-1 in transgenic mice promotes survival in a spectrum of hematopoietic cell types and immortalization in the myeloid lineage. Blood. 1998;92:3226–3239. [PubMed] [Google Scholar]