Abstract
BACKGROUND & AIMS
Eosinophilic esophagitis (EE) occurs in families, but neither familial EE clustering nor phenotype/genotype of familial compared with sporadic EE have not been reported.
METHODS
Record review confirmed patient kinship and provided clinical information. Slide review confirmed the diagnosis (threshold peak number •24 eosinophils/hpf).
RESULTS
Fifty-nine members (41 males, 18 females) of 26 families were 3 months to 47 years of age (mean age 10.3 years) at diagnosis. The only recorded race was Caucasian. In four families, a parent of an affected male had EE. The most common complaint at diagnosis was dysphagia (68% of patients). Endoscopy showed esophageal mucosal furrows (93% of patients), and exudates (44%). Fifty-one percent had asthma. Skin prick tests to food and aeroallergens were positive in 76% and 71%, respectively. Familial EE characteristics (clinical, endoscopic, pathological, and global esophageal transcript expression profile analysis) were similar to sporadic EE, except among patients with mucosal furrows: familial patients had lower peak eosinophil counts in the distal esophagus (P = 0.03) compared with sporadic patients. The basic characteristics of EE (e.g., eosinophil levels, rate of atopy) did not vary with patient age. Using genome wide microarray analysis, no significant differences (P<0.05, FDR) were observed between familial and sporadic EE. Among all patients, chest pain was more common in females (P = 0.02), and thickened mucosa in males (P = 0.006).
CONCLUSIONS
These data support a familial pattern of inheritance of EE and a pathogenesis shared with sporadic EE. EE should be considered in symptomatic family members of patients who have EE.
INTRODUCTION
Eosinophilic gastrointestinal diseases (EGID) exhibit increased numbers of eosinophils in one or more parts of the gastrointestinal (GI) tract1. Primary EGID is diagnosed if a known etiology (parasitic infections, allergies, drug reactions, hypereosinophilic syndrome, etc) is not identified. Eosinophilic esophagitis (EE), a form of EGID, is a chronic, relapsing inflammatory disease manifesting large numbers of intraepithelial eosinophils in esophageal mucosal biopsies2 and characteristic endoscopic findings3,4. EE must be distinguished from gastroesophageal reflux disease (GERD)5 because EE requires treatment other than or in addition to therapy for GERD6–13.
EE patients are frequently atopic8,10,11,14. Atopic disease has a strong familial association, predominantly due to complex interactions between environmental and genetic factors, the latter accounting for ~50% of atopic disease15.
EE has stronger genetic components than other atopic diseases. Families with more than one member with EE have been reported16–20. Based on the estimated prevalence of EE as 4 per 10,000 in the pediatric population16, the sibling risk recurrence ratio for EE is very high, approximately 8021, compared to approximately 2 for asthma15. EE is linked to a single nucleotide polymorphism (SNP) in the eotaxin-3 gene based on case control and transmission disequilibrium test analyses22. Despite evidence that EE has a strong genetic component, the characteristics of the familial subset and its mode of inheritance have not been reported. We report the clinical and histopathological characteristics and genetic transcriptome of familial patients compared with EE patients who do not have affected family members (sporadic cases).
MATERIALS AND METHODS
This study was performed with the approval of the Institutional Review Board at Cincinnati Children’s Hospital Medical Center. The database in the Division of Pathology and Laboratory Medicine, which contains pathology cases from 1970 to the present, was searched (by MHC) for biopsies with the diagnosis of EE. Inclusion criteria were a threshold peak count of •24 eosinophils/hpf in an esophageal biopsy that was reviewed at the Cincinnati Center for Eosinophilic Disorders (CCED). Biopsies from other sites in the GI tract obtained at the same endoscopy as the diagnostic esophageal biopsy were not required for inclusion, and, if those biopsies were obtained, absence of pathology in those biopsies was not required for inclusion.
The hospital database was searched to confirm that patients having the same surname resided at the same address. In addition, for all families except one, review of the medical records or query of a gastroenterologist who cared for one or more members of the family confirmed the familial relationships.
A relational database of patients treated at the CCED who have EGID was created in 2001 using Microsoft Access with approval from the Institutional Review Board. Only data from patients who consented had their information entered into the database. Data included patient demographics, signs and symptoms, endoscopic findings, pathology diagnosis, results of skin prick tests, pertinent laboratory tests, treatment, and final clinical diagnosis. Information for the database was obtained mainly from hospital records retrospectively. All clinical information used in this study was gathered from the CCED database (by CK). One family member of each of the 26 familial patients registered in the database was matched as closely as possible by date of birth and gender to 26 sporadic patients who do not have affected family members. There were insufficient numbers of patients to permit random matching of familial and sporadic patients.
Signs and symptoms, endoscopic findings, and results of allergy tests for all patients were obtained from the database. Thick esophageal mucosa diagnosed at endoscopy indicates opaque mucosa in which vascular markings are not visible and appears thicker than normal when biopsy is performed, furrows indicate thick mucosa with evenly-spaced vertical lines, exudate indicates patches or plaques of white material on the mucosal surface, stricture indicates small esophageal lumen and a rigid wall, and rings indicate mucosal elevations. For patients whose diagnostic endoscopy was performed at CCED, esophageal cultures were performed if clinically indicated. None of those patients was receiving antibiotics or topical steroid therapy at the time of the diagnostic endoscopy. Evenly-spaced white patches or plaques on the vertical lines of abnormal mucosa were considered eosinophilic exudates, but randomly dispersed white patches or plaques were considered suspicious for fungal infection and were cultured. Atopy was defined as reaction to at least one antigen as demonstrated by a skin prick test at the time of EE diagnosis.23
All biopsies were fixed in formalin, and five-micron thick slides that were cut from paraffin blocks were stained with hematoxylin and eosin. The available slides of esophageal biopsies on which the diagnosis was initially made were re-reviewed to confirm the diagnosis and to more extensively quantitate eosinophilic inflammation for this report (by MHC). The number of intraepithelial eosinophils was counted in all high power fields (400X, 0.30 mm2) in one complete section of a biopsy, and the mean number and standard deviation was calculated. The peak eosinophil number was defined as the greatest number of eosinophils in a high power field. The threshold peak number used to identify EE biopsies was set at ≥24/hpf.7,10–12,22,23 Quantitative evaluation of eosinophil number was performed for most patients, but since our clinical practice is largely composed of patients referred from other locales, several initial diagnostic biopsies that had been reviewed at the CCED, confirmed to have the threshold peak eosinophil number and returned to the hospital of origin, could not be obtained for re-review for this study (Table 1).
Table 1.
Characteristics of Familial EE Patients
| Mean ± SD (peak) eosinophil no. |
||||||
|---|---|---|---|---|---|---|
| Family no. | Sex | Patients | Age at diagnosis, y | Distal esophagus | Proximal esophagus | Other biopsies |
| 1 | Female | Proband | 18 | 5.76 ± 6.73 (24) | ND | ND |
| a Male | Brother | 18 | 3.17 ± 4.13 (17) | 4.84 ± 7.15 (24) | D = eos agg; S = NDA | |
| Male | Sporadic | 15 | 13.3 ± 8.62 (32) | 8.63 ± 7.19 (21) | D, S = NDA | |
| 2 | Female | Proband | 7 | 10.7 ± 15.03 (42) | ND | D = eos cryptitis; S = mild chronic |
| a Female | Sister | 11 | 16 ± 13.11 (38) | 2.2 ± 4.41 (12) | D = NDA; S = MME + MA | |
| Female | Sporadic | 11 | 23.92 ± 14.53 (64) | ND | D, S = NDA | |
| 3 | Male | Proband | 17 | 21.5 ± 14.45 (67) | 26.6 ± 14.83 (58) | D, S = NDA |
| a Female | Sister | 20 | 16.5 ± 9.36 (32) | 0.3 ± 0.57 (21) | D, S = NDA | |
| Female | Sporadic | 20 | 31.8 ± 17.63 (84) | 41.83 ± 24.8 (101) | D, S = NDA | |
| 4 | Male | Proband | 17 | 109.2 ± 51.81 (223) | 5.6 ± 7.12 (21) | D = NDA; S = ME |
| a Male | Brother | 11 | 75 ± 36.13 (167) | 53 ± 34.58 (151) | D = ME + MA; S = NDA | |
| Male | Father | 47 | 80.3 ± 48.44 (184) | 41.5 ± 45.62 (148) | D = eos cryptitis; S = NDA | |
| Male | Sporadic | 9 | 58.06 ± 28.36 (108) | ND | D = NDA | |
| 5 | a Male | Proband | 0.08 | 12.7 ± 9 (28) | 12.7 ± 7.6 (25) | D, S, S/R = NDA |
| Female | Mother | 36 | 22.7 ± 23.2 (111) | ND | ND | |
| Male | Sporadic | 1.5 | 22.6 ± 19.3 (69) | 5.11 ± 5.33 (22) | D, S = NDA | |
| 6 | Male | Proband | 0.25 | 37 ± 20.5 (74) | 41.2 ± 20.92 (76) | D = ND; S = ME |
| a Male | Brother | 5 | 23.9 ± 14.09 (61) | 4.5 ± 7.31 (26) | D = NDA; S = NDA | |
| Male | Sporadic | 5.5 | 23.25 ± 15.17 (50) | 15.5 ± 15.77 (54) | D, S, TI = NDA C/A, T/D, S/R = ME | |
| 7 | Female | Proband | 0.5 | 87 ± 35.11 (176) | ND | D = CH; S = NDA |
| a Male | Brother | 1 | 13.8 ± 8.7 (33) | 45.8 ± 27.2 (110) | D, S, TI = NDA C/A, T/D, S/R = ME | |
| Male | Sporadic | 1 | 36.5 ± 26.7 (86) | 0.17 ± 0.41 (1) | D = NDA | |
| 8 | a Male | Proband | NA | >24 | ND | ND |
| Male | Brother | NA | >24 | ND | ND | |
| Male | Brother | NA | >24 | ND | ND | |
| Male | Brother | NA | >24 | ND | ND | |
| Male | Brother | NA | >24 | ND | ND | |
| Female | Mother | 34 | 27.3 ± 21.9 (57) | ND | ND | |
| Male | Sporadic | 5 | 1.56 ± 1.26 (4) | 38.86 ± 28.56 (105) (mid) | D, S = NDA | |
| 9 | Male | Proband | 2 | 1.5 ± 0.5 (2) | 12 ± 10.37 (28) (mid) | ND |
| a Male | Brother | 5 | 43.3 ± 17.66 (82) | 1.2 ± 1.63 (5) | D, S = NDA | |
| Male | Sporadic | 3 | 17.92 ± 7.42 (34) | 0.33 ± 0.62 (2) | D, S = NDA | |
| 10 | Female | Proband | 8 | 9.33 ± 5.91 (22) | 86.8 ± 53.32 (151) | S = NDA |
| a Female | Sister (twin) | 15 | 32.5 ± 15.57 (64) | 5.11 ± 9.5 (31) | D, S = NDA | |
| Female | Sporadic | 17 | 50 ± 26.29 (106) | 1.09 ± 1.38 (5) (mid) | D = NDA; S = CG | |
| 11 | Female | Proband | 4 | 36.9 ± 25.17 (88) | 35.1 ± 28.14 (88) | D = eos cryptitis; S = NDA |
| a Male | Brother | 3 | 39.5 ± 13.41 (62) | 36.7 ± 25.54 (86) | D, S = NDA | |
| Male | Sporadic | 2 | 33.75 ± 15.26 (54) | ND | D, S = NDA | |
| 12 | Male | Proband | 5 | 31.4 ± 22.03 (92) | ND | S = NDA |
| a Male | Brother | 1 | 13.5 ± 8.9 (39) | 12.2 ± 14.47 (67) | D, S, TI, A/C, T/D, S/R = NDA | |
| Male | Sporadic | 2 | 66.25 ± 34.38 (160) | 77.05 ± 48.55 (245) | D = eos duodenitis; S = NDA | |
| 13 | Male | Proband | 9 | 38.5 ± 26.83 (106) | 7.9 ± 7.74 (26) | D, S = NDA |
| a Female | Sister | 14 | 9.4 ± 8.98 (26) | 27.5 ± 5.36 (36) | S = ME | |
| Female | Sporadic | 11 | 8.82 ± 8.71 (33) | ND | ND | |
| 14 | a Female | Proband | 1 | 21 ± 13.89 (48) | 0.47 ± 0.7 (26) | D, S = NDA |
| Male | Brother | 7 | 17.6 ± 14.88 (61) | 1.29 ± 1.1 (3) | D, S, TI, C/A, T/D, S/R = NDA | |
| Male | Brother | 8 | 7.2 ± 9.32 (33) | 1.3 ± 2.24 (9) | D, S, TI, C/A, T/D, S/R = NDA | |
| Female | Sporadic | 1 | 5.05 ± 8.81 (33) | 6.41 ± 8.89 (27) | D, S = NDA | |
| 15 | Female | Proband | 6 | 32.1 ± 15.77 (78) | 36.1 ± 14.57 (65) | D = NDA; S = MCG |
| a Female | Sister | 11 | 51.5 ± 27.94 (121) | 39.1 ± 29.2 (104) | D, S = NDA | |
| Female | Sporadic | 7 | 18.7 ± 18.53 (62) | 13.5 ± 10.97 (42) | D = eos agg; S = NDA | |
| 16 | Male | Proband | 16 | 6.5 ± 7.89 (31) | 13.8 ± 10.26 (31) | D = NDA; S = MCG |
| a Female | Sister | 18 | 14.2 ± 22.4 (100) | ND | ND | |
| Female | Sporadic | 20 | 28.44 ± 19.46 (70) | ND | D, S, TI, C/A, T/D, S/R = NDA | |
| 17 | Male | Proband | 11 | 36.4 ± 19.62 (85) | 26.8 ± 10.4 (47) | D, S = NDA |
| a Male | Brother (twin) | 12 | 11.7 ± 11.63 (51) | 7.6 ± 7.39 (24) | D, S, TI, C/A, T/D, S/R = NDA | |
| Female | Sister | 14 | 25.5 ± 24.3 (99) | ND | D, S = NDA | |
| Male | Sporadic | 11 | 34.4 ± 24.41 (79) | 22.1 ± 20.23 (73) | D, S = NDA | |
| 18 | Female | Proband | 17 | 5.8 ± 5.2 (18) | 6.9 ± 8.66 (28) | D, S = NDA |
| a Male | Brother | 20 | 14.6 ± 11.91 (46) | 6.1 ± 7.93 (38) | D, S = NDA | |
| Male | Sporadic | 22 | 42.82 ± 35.3 (126) | ND | D, S = NDA | |
| 19 | a Male | Proband | 13 | 7.9 ± 7 (27) | 1.8 ± 3.47 (16) | D, S, TI, C/A, T/D, S/R = NDA |
| Male | Brother | 12 | 5.8 ± 4.47 (14) | 10 ± 10.5 (37) | D, S = NDA | |
| Male | Sporadic | 10 | 19.45 ± 16.28 (60) | 33.65 ± 20.56 (69) | D, S = NDA | |
| 20 | a Male | Proband | 8 | 59.8 ± 23.3 (107) | 28.7 ± 17.15 (86) | ND |
| Male | Father | 21 | 6 ± 2.16 (7) | 39.3 ± 8.99 (52) | D, S = NDA | |
| Male | Sporadic | 11 | 185 ± 76.8 (349) | 45.2 ± 43.6 (146) | D, S = NDA | |
| 21 | Male | Proband | 4 | 65.3 ± 28.52 (113) | 45.7 ± 19.26 (77) | D, S = NDA |
| a Male | Brother | 4 | 39.8 ± 20.06 (84) | 16.8 ± 16.76 (56) | D, S = NDA | |
| Male | Sporadic | 1 | 21.5 ± 16.9 (83) | 7.88 ± 9.27 (36) | D, S = NDA | |
| 22 | a Male | Proband | 5 | 16.3 ± 11.43 (33) | ND | D = NDA; S = NA |
| Female | Sister | 3 | 10.5 ± 9.52 (30) | 8.1 ± 6.57 (26) | D = NDA; S = GC | |
| Male | Sporadic | 5 | 14.7 ± 9.43 (30) | ND | D, S = NDA | |
| 23 | Male | Proband | 7 | 52.6 ± 41.99 (140) | 12.9 ± 21.58 (62) | D, S = NDA |
| a Male | Brother | 13 | 10.7 ± 9.85 (41) | 8.5 ± 8.45 (27) | D = erosive duodenitis; S = NDA | |
| Male | Sporadic | 9 | 25.39 ± 23.5 (76) | 20.15 ± 33.51 (132) | D = NDA; S = Chem g | |
| 24 | a Male | Proband | 4 | 12.7 ± 7.49 (34) | 11.4 ± 6 (24) | D, S, TI, C/A, S/R = NDA, T/D = eos agg |
| Male | Brother | 2 | 21.9 ± 8.03 (41) | 41.6 ± 16.3 (71) | D, S = NDA | |
| Male | Sporadic | 7 | 20.4 ± 27.8 (91) | 13.4 ± 11.1 (36) | D, S = NDA | |
| 25 | a Male | Proband | 9 | 17.3 ± 19.48 (70) | ND | D, S = NDA |
| Male | Brother | 2 | 34.6 ± 17.65 (60) | ND | ND | |
| Male | Sporadic | 11 | 39.1 ± 12.9 (55) | 78.4 ± 33.3 (168) | D = reac epi; S = ME | |
| 26 | a Male | Proband | 5 | 16.4 ± 7.31 (28) | 11.3 ± 9.92 (33) | D = NDA |
| Male | Brother | 5 | 42.8 ± 35.38 (129) | 21 ± 11.3 (42) | D, S = NDA | |
| Male | Sporadic | 1.3 | 33.5 ± 20.7 (61) | 12.9 ± 10.2 (35) | D, S = NDA | |
Sporadic, patient with sporadic disease; ND, not done; D, duodenum; Eos agg, eosinophil aggregate in superficial epithelium; S, stomach; NDA, no diagnostic abnormality; Eos, eosinophilic; MME, mild mucosal eosinophilia; MA, mild acute inflammation; ME, mucosal eosinophilia; S/R, sigmoid/rectum; TI, terminal ileum; C/A, cecum/ascending colon; CH, crypt hyperplasia; T/D, transverse/descending colon; NA, not available; mid, midesophagus; CG, chronic gastritis; MCG, mild chronic gastritis; Chem g, chemical gastropathy; Reac epi, reactive epithelial changes. GC, lamina propria giant cell.
Member of family matched to a patient with sporadic disease based on gender and age.
Reports of biopsies from other sites in the gastrointestinal tract that were obtained during the endoscopy that yielded the initially diagnostic esophageal biopsy were reviewed. Slides or photos of biopsies that were not reported as normal were re-reviewed. The peak number of eosinophils in normal pediatric gastrointestinal tract biopsies in Cincinnati was used as a guide to distinguish normal from abnormal biopsies.24 For purposes of this study, mucosal eosinophilia designates increased numbers of eosinophils in the lamina propria without eosinophilic cryptitis or other alterations. Eosinophilic cryptitis is the presence of multiple eosinophils in the epithelium of one or more crypts. Eosinophilic enteritis is mucosal eosinophilia plus significant architectural alterations, such as cryptitis, elongated crypts, and reactive epithelial changes.
Data were considered absent if neither positive nor negative responses were found in the database and hospital records. Data were analyzed using SAS v 9.1 (SAS Institute, Cary, NC). Simple descriptive statistics were run on each group to obtain N and percents. McNemar tests were used to test agreement between the familial and non-familial groups. Paired t-test was used to determine differences between groups. Biserial correlations were used to test correlations between eosinophil counts and endoscopic and atopic findings. Simple t-test was used to determine differences among all patients in the study. P value ≤0.05 was considered significant.
Several patients reported in this study had participated in a prior study that utilized esophageal biopsies in a genome-wide DNA microarray analysis.22 The available results of the microarray analysis for the familial and sporadic patients were extracted and compared using Welch t test with and without false discovery rate (FDR) correction.
RESULTS
EE family member relationships
Twenty-six families with more than one affected member were identified. A total of 59 patients in these families had EE (Table 1), 41 males and 18 females, who were mean age 10.3 years (range 0.25 to 47 years) at diagnosis. Affected members were siblings in 85% of families, and children and their parents in 15% of families. Four families had affected members in more than one generation. In two families, the father of a male proband had EE, in one family the mother of a male proband had EE, and in one family the mother of a male proband and all 4 of his male siblings had EE.
Clinical features of familial EE
Among all familial patients for whom the information was available, the most common complaint at diagnosis was dysphagia, reported by 68% of patients (Table 2). Emesis, abdominal pain, and heartburn were other common complaints, reported by 62%, 54% and 45% of patients, respectively. These symptoms varied with patient age. Patients who reported dysphagia were mean age 12.9 years at diagnosis, and were significantly older than patients who reported emesis (mean age 7.99 years, P<0.05), or abdominal pain (mean age 7.96 years, P<0.02), but not significantly older than patients who reported heartburn (mean age 9.76 years).
TABLE 2.
CLINICAL CHARACTERISTICS
| All familial patients N=59 | Familial patients matched to sporadic patients N=26 | Sporadic patients N=26 | All patients N=85 | |
|---|---|---|---|---|
| Signs and Symptoms | ||||
| Dysphagia | 36 (68%) N = 53 |
13 (54%) N = 24 |
10 (40%) N = 25 |
46 (59%) N = 78 |
| Emesis | 32 (62%) N = 52 |
16 (64%) N = 25 |
15 (63%) N = 24 |
47 (62%) N = 76 |
| Abdominal Pain | 27 (57%) N = 47 |
15 (65%) N = 23 |
11 (46%) N = 24 |
38 (54%) N = 71 |
| Heartburn | 23 (45%) N = 51 |
10 (43%) N = 23 |
14 (58%) N = 24 |
37 (49%) N = 75 |
| Food Impaction | 9 (17%) N= 52 |
3 (13%) N = 24 |
3 (13%) N = 24 |
12 (16%) 76 |
| Chest Pain | 12 (24%) N = 49 |
6 (26%) N = 23 |
6 (25%) N = 24 |
18 (25%) N = 73 |
| Cough | 11 (23%) N = 47 |
4 (18%) N = 22 |
2 (8%) N = 24 |
13 (18%) N = 71 |
| Nausea | 10 (22%) N = 46 |
6 (26%) N = 23 |
4 (17%) N = 24 |
14 (20%) N = 70 |
| Diarrhea | 5 (10%) N = 48 |
3 (13%) N = 24 |
4 (17%) N = 24 |
9 (13%) N = 72 |
| Choking | 7 (32%) N = 22 |
1 (11%) N = 9 |
8 (33%) N = 24 |
15 (33%) N = 46 |
| Allergic rhinitis | 36 (73%) N = 49 |
15 (65%) N = 23 |
13 (57%) N = 23 |
49 (68%) N = 72 |
| Asthma | 25 (51%) N =49 |
12 (52%) N = 23 |
13 (54%) N = 24 |
38 (52%) N = 73 |
| Allergic Conjunctivitis | 29 (59%) N = 49 |
11 (48%) N = 23 |
11 (48%) N = 23 |
41 (56%) N = 72 |
| Eczema | 22 (47%) N = 47 |
13 (57%) N = 23 |
8 (36%) N = 22 |
30 (44%) N = 69 |
| Endoscopic Findings | ||||
| Furrows | 37 (93%) N = 40 |
19 (95%) N = 20 |
16 (89%) N = 18 |
53 (91%) N = 58 |
| Thickened | 27 (68%) N = 40 |
13 (68%) N = 19 |
9 (69%) N – 13 |
36 (68%) N = 53 |
| Exudate | 19 (44%) N = 43 |
9 (45%) N = 20 |
3 (27%) N = 11 |
22 (41%) N = 54 |
| Stricture | 4 (10%) N = 42 |
0 (0%) N = 19 |
2 (12%) N = 17 |
6 (10%) N = 59 |
| Rings | 3 (7%) N = 45 |
1 (5%) N = 21 |
2 (13%) N = 15 |
5 (8%) N = 60 |
| Skin Prick Tests | ||||
| Environment | 27 (71%) N = 38 |
13 (65%) N = 20 |
13 (59%) N = 22 |
40 (67%) N = 60 |
| Food | 31 (76%) N = 41 |
14 (70%) N = 20 |
16 (80%) N = 20 |
47 (77%) N = 61 |
Sporadic patients, EE patients without affected family members, and matched for gender and date of birth to one member of each family.
Furrows, thick mucosa with vertical lines; thick mucosa, mucosa without visible vascular markings; exudate, white plaques/patches; stricture, narrow caliber; rings, mucosal ridges.
Endoscopic features of familial EE
Diagnostic endoscopy showed esophageal mucosal furrows in 93% of patients, thick mucosa in 68%, and exudate in 44%.
Atopic features of familial EE
Fifty-one percent of familial patients reported asthma, and 73% had allergic rhinitis. Over 70% of patients had skin prick tests that were reactive to either food (76%) or aeroallergens (71%) or both (63%).
Clinical, endoscopic, and pathological comparison between familial and sporadic EE
Information concerning race was available for at least one member of 17 families, and Caucasian was the only recorded race. If all members of those 17 families were the same race, then 38 familial patients were Caucasian. Information concerning race was recorded for 14 sporadic EE patients, and all were also Caucasian.
Familial patients did not have statistically significant differences in signs and symptoms, endoscopic esophageal characteristics, or skin prick test results compared to matched sporadic patients (Table 2). Among the paired patients who had furrowed mucosa at endoscopy, however, the peak number of intraepithelial eosinophils in the distal esophagus was greater in the sporadic (n=12) compared to familial (n=11, 107/hpf vs 51.5/hpf, p=0.03) patients. Peak esophageal eosinophil counts were not different between the matched pairs for patients who had dysphagia, or skin prick tests reactive to food only. There were insufficient patients with information recorded concerning mucosal exudate among the pairs for statistical analysis. Peak esophageal eosinophil number in distal esophageal biopsies in the matched pairs did not correlate with endoscopic abnormalities, or positive skin prick tests to food.
Biopsies from the distal esophagus were performed in all patients, and were frequently diagnostic, in 53/59 (90%) familial EE patients, and in 25/26 (96%) paired familial and sporadic patients. Peak eosinophil number in distal esophagus biopsies ranged from 2–223/hpf among familial patients, and from 4–349/hpf among sporadic patients. Statistically significant differences for peak and mean eosinophil counts in distal esophageal biopsies were not found in familial patients compared to sporadic patients.
Biopsies from other esophageal sites were performed less frequently than biopsies from the distal esophagus. Among patients whose biopsies were available for review, there were biopsies from sites in addition to the distal esophagus, mostly proximal, in 43/54 (71%) of familial patients, in 21/25 (84%) paired familial patients, as well as 19/26 (74%) sporadic patients. Biopsies from sites other than the distal esophagus were less frequently diagnostic than biopsies from the distal esophagus, but established the diagnosis in all seven patients (6 familial and 1 sporadic) whose distal biopsies were not diagnostic. Biopsies from esophageal sites other than the distal esophagus were diagnostic in 35/43 (81%) familial patients, in 17/21 (81%) paired familial patients, and in 15/19 (79%) sporadic patients. Peak number of eosinophils in biopsies from sites other than the distal esophagus was lower than in biopsies from the distal esophagus: the peak number ranged from 5–151/hpf among familial patients, and from 1–245/hpf among sporadic patients. Statistically significant differences for peak and mean eosinophil counts in biopsies from sites other than the distal esophagus were not found in familial patients compared to sporadic patients.
Among all 85 patients who had EE, there were not statistically significant differences in peak eosinophil count at any esophageal site for patients who had dysphagia as a presenting symptom compared to those who did not, for those who had furrows or exudates at endoscopy compared to those who did not, or for those who had reactive skin tests to food compared to those who did not.
Information concerning anaphylaxis was available in 48 patients; 25% reported anaphylactic reaction to food, 13% to nonfood, and 6% to both. Seven familial patients and 7 sporadic patients reported anaphylaxis to either food or aeroallergens; 2 familial patients and 1 sporadic patient reported anaphylaxis to both. The prevalence among familial EE patients was not significantly different from the prevalence among their matched sporadic pairs.
Biopsies from other sites
Forty-one members of 19 families had biopsies from sites in addition to the esophagus, mainly from duodenum and stomach (Table 1). Ten of 41 (24%) familial patients from 7 families had eosinophilic inflammation in extra esophageal sites. In 2 families, both members who had biopsies from extra esophageal sites had eosinophilic inflammation in one of those sites, and in another family all 3 members who had extra esophageal biopsies had eosinophilic inflammation in one of those sites. The pathology was mild focal eosinophilic duodenal cryptitis [n=3], mild mucosal eosinophilia in stomach [n=4], duodenum [n=1], or colon [n=1], and an aggregate of eosinophils in superficial duodenal epithelium [n=1]. Other pathology in extra esophageal sites in familial EE patients was mild acute inflammation (with mild eosinophilic inflammation) in stomach [n=1] or duodenum [n=1], chronic gastritis [n=2], focal duodenal crypt hyperplasia [n=1], gastric lamina propria giant cell without other inflammation [n=1] and erosive duodenitis [n=1].
Among sporadic patients, 3 of 25 (12%) who had biopsies of extra esophageal sites at the time of diagnosis had eosinophilic inflammation in 1 of those sites: eosinophilic duodenitis (1), an aggregate of eosinophils in superficial duodenal epithelium (1), and mild gastric mucosal eosinophilia (1). Other pathology in extra esophageal sites in sporadic EE patients included juvenile retention polyp in cecum/ascending colon (1), chemical gastropathy (1), chronic gastritis (1), and reactive surface duodenal epithelial changes (1). Although the prevalence of eosinophilic inflammation in extra esophageal sites was greater in familial EE patients, the difference was not statistically significant (P>0.05).
Age-related analyses
The inclusion of patients with a wide age range (including children and adults) provided a valuable opportunity to determine if there were significant differences in EE characteristics as a function of age. The greatest number of patients was in the 0–5 year age group, but 5 patients were over 20 years of age (Table 1). In this study, neither peak eosinophil number in distal esophageal biopsies, nor mean of peak eosinophil number in distal esophageal biopsies, showed a progressive decline with increasing patient age (Figure 1). Similarly, the percent of patients in each age group who were skin prick tested and had positive tests did not decline appreciably with increasing age (Figure 2).
Figure 1. Esophageal eosinophil levels as a function of patient age.
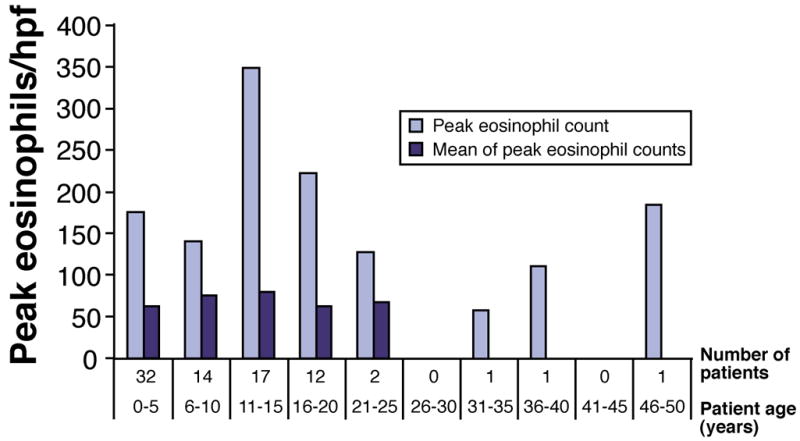
Bar graph shows peak eosinophil count in distal esophageal biopsies with increasing age. The mean of the peak eosinophil counts in biopsies from the distal esophagus is also shown; there were insufficient numbers of patients in age groups over 25 years to calculate a mean.
Figure 2. Prevalence of positive skin prick tests as a function of age.
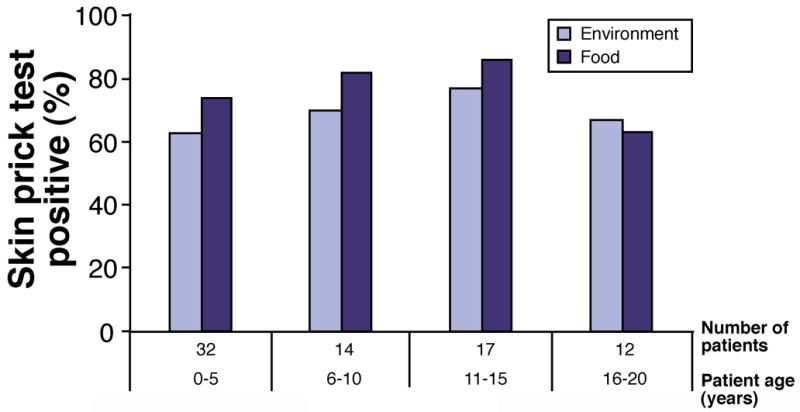
Results of skin prick tests in older age groups were not available.
Gender-related analyses
Males (N= 60, 41 familial, 19 sporadic) were compared to females (N=25, 18 familial, 7 sporadic) for the characteristics listed in Table 1. Among familial patients, there were not any gender-related differences. Among sporadic patients, chest pain (4/6 females vs 2/18 males, P=0.006), choking (4/6 females vs 4/18 males, P=0.04), and strictures (2/5 females vs 0/12 males, P=0.01) were more common in females, and thickened mucosa was more common among males (1/4 females vs 8/9 males, P=0.02). Among all patients, similar to sporadic patients, chest pain was more common in females (9/21) than males (9/52) (P=0.02), and thickened mucosa was more common in males (30/38) than females (6/15) (P=0.006). There was not a significant difference in peak eosinophil number in distal esophageal biopsies in males compared to females in any of the groups.
Intrafamilial analyses
There was insufficient information recorded for all family members to permit intrafamilial comparisons, except for family #4 (Table 1). In that family, the affected parent and each child had furrows and thick mucosa at endoscopy, the parent and one child had exudate and rings, and none had strictures. The parent was not skin prick tested, but both children reacted to both food and aeroallergens. All had allergic rhinitis, one child had asthma, and none had eczema or allergic conjunctivitis. All had dysphagia, the parent had emesis, and none had chest pain, heartburn, nausea, diarrhea or abdominal pain. In another family (#8), information concerning dysphagia was recorded for 3 family members and was reported by the parent and both children.
Gene expression in familial EE vs. non-familial EE patients
We queried if there was a unique gene expression profile associated with familial EE. In order to identify a molecular signature that might distinguish familial EE from sporadic EE, we retrieved gene expression data in these two groups from microarray analysis on esophageal biopsies.21 In the biopsies from familial (n=6) and sporadic (n=10) EE patients, the expression of the 54,681 genes represented in the genome-wide microarrays was subjected to Welch t test with false discovery rate (FDR). Significantly different gene expression was not found in familial EE compared to sporadic EE patients (including expression of eotaxin-3). To identify genes that were possibly truly differently expressed between the groups but eliminated from analysis by the FDR correction, the same analysis was performed without correction. The number of genes that were thus obtained was always fewer than the number of genes expected to be obtained by chance, and the expression of only 15 genes was significantly different (P<0.001, no FDR) in familial EE compared to sporadic EE patients. Of these 15 genes, the expression of only 5 genes differed by more than 2-fold in familial EE compared to sporadic EE patients (Figure 3). The expression of eotaxin-3 gene did not differ between the groups. The relative average expression of the genes of the EE transcript signature (574 genes) in familial compared to sporadic EE patients is presented in Figure 3.
Figure 3. Gene expression in familial EE patients vs. sporadic EE patients.

A. The peak eosinophil number in distal esophageal biopsies from familial EE and sporadic EE patients are presented in a dot plot; the line is the mean. B. The expression of the EE transcript consisting of 574 genes in familial EE and sporadic patients is presented in a heat diagram. Up regulated genes are in red, down regulated genes are in blue, and yellow corresponds to gene expression unchanged compared to controls. Peak eosinophil number in distal esophageal biopsies of each patient is shown at the right. C. The average expression of the EE transcript signature in familial EE and sporadic EE is presented in a heat diagram. Each line corresponds to the average of the patents in each group. Up regulated genes are in red, down regulated genes are in blue, and yellow corresponds to gene expression unchanged compared to controls.
DISCUSSION
This is the largest series of EE patients who have other affected family members, and includes both pediatric and adult patients. Major findings include: all patients in this study whose race was known were Caucasian; patients who had dysphagia at diagnosis were significantly older than patients who complained of emesis or abdominal pain;10 mucosal furrows, thick mucosa, and mucosal exudates were common esophageal findings in the diagnostic endoscopy; most patients had positive skin prick test reactions to food and aeroallergens; and distal esophageal biopsies were diagnostic in 90% of patients in the study. One member of each family was matched for age and gender to EE patients who did not have other affected family members: familial EE patients who had furrows at diagnostic endoscopy had significantly lower peak eosinophil count in distal esophageal biopsies compared to sporadic patients who had mucosal furrows at diagnostic endoscopy. We report comparable degrees of eosinophilic inflammation in esophageal biopsies and a comparable prevalence of positive skin prick tests in older compared to younger patients. To our knowledge, our study represents the largest analysis of EE across a wide age range (pediatric and adult), and although the number of adult patients is small (only five individuals were older than 20 years), our data provide evidence that adult and pediatric EE are likely to involve the same primary disease processes.
We report members of two successive generations in multiple families who have biopsy-proven EE. However, several of these families reported one or more members of a preceding third generation who exhibited signs and symptoms suggestive of EE. We have not yet had the opportunity to examine slides of biopsies from those family members to confirm the diagnosis, and they are therefore not included in this report. Other investigators have recently reported that esophageal biopsies of symptomatic grandparents of EE probands did not contain sufficient numbers of intraepithelial eosinophils to diagnose EE, including biopsies from older patients who had Schatzki rings that may be found in EE.19 The number of intraepithelial eosinophils in adults who have EE has been reported to diminish over time in one study;25 however, our cohort analysis of esophageal eosinophil levels as a function of age at diagnosis (Figure 1) did not reveal statistical differences. The significance of a relatively low number of intraepithelial eosinophils in esophageal biopsies from older members of a proband’s family remains unclear.
The probability that three male siblings would develop EE if there were not a familial component has been estimated as 10−12.17 In one of the families we report, five male siblings are affected (Table 1), and the probability that a disease devoid of a familial component would occur in five siblings must be considerably greater than 10−12. These findings do not eliminate the possibility that disease in multiple family members is in some manner related to their environment, but our findings enhance the view that there is a familial form of EE that includes an inherited predisposition to the disease. Notably, the estimated recurrence risk for siblings of affected EE patients is much larger than the estimate for other atopic diseases,21,26 suggesting a strong role for genetics in EE. The results of genome-wide DNA microarray analyses of esophageal biopsies from EE patients and controls suggest that the eotaxin-3 gene may be important in disease pathogenesis.22 A single nucleotide polymorphism at the 3′ untranslated region of the eotaxin-3 gene contains a GG allele that is preferentially transmitted from heterozygous parents to their children who have EE in a manner that appears to be independent of atopic status.22 This finding further supports genetic transmission as an important component of EE pathogenesis, and suggests that inheritance of EE occurs independently of inheritance of atopy.
The occurrence of EE exclusively in Caucasian patients in this study is intriguing, and has not been reported in other atopic diseases. We postulated that genes with large allele frequency differences between Caucasian and African populations might contribute to the apparently virtual restriction of EE to Caucasian people.21 From the HapMap database (http://www.hapmap.org) we obtained the allele frequencies of each identified single nucleotide polymorphism in each of 34 genes that were over/under expressed (>10 fold change) in EE patients compared with controls identified in our genome-wide DNA microarray analysis.22 Five of those genes (EPB41L3, TMEM71, CES4, FLG, CDH26) have a significantly greater allele frequency difference between Caucasians and Africans (>0.6) compared to the average allele frequency difference between these groups (Table 3). The FLG gene encodes the protein filaggrin, which is essential for skin barrier integrity, and loss-of-function variants of FLG predispose to atopic dermatitis.27 Notably, in mice, epicutaneous antigen exposure predisposes to the development of EE,28 further highlighting the potential importance of skin and barrier function in EE pathogenesis.
TABLE 3.
LIST OF GENES WITH >10 FOLD EXPRESSION CHANGE IN EE PATIENTS AND A LARGE ALLELE FREQUENCY DIFFERENCE BETWEEN CAUCASIAN AND AFRICAN POPULATIONS
| Gene Symbol | Gene Description | Fold Change£ | Largest allele frequency difference§ | |
|---|---|---|---|---|
| SNP rs# | allele freq. difference | |||
| EPB41L3 | Erythrocyte membrane protein band 4.1-like 3 | 0.097 | rs2874686 | 0.869 |
| TMEM71 (FLJ33069) | Transmembrane protein 71 | 14.03 | rs2280871 | 0.680 |
| CES4 | Carboxyl esterase 4-like | 0.088 | rs7188256 | 0.636 |
| FLG | Filaggrin | 0.097 | rs2184953 | 0.619 |
| CDH26 | Cadherin-like 26 | 23.64 | rs2426865 | 0.608 |
fold change was obtained by comparing the gene expression level between patients with EE and controls;
largest allele frequency difference between Caucasian and African was obtained from HapMap database (http://www.hapmap.org) by comparing the allele frequency of each identified SNP of the gene between CEU (Utah residents with ancestry from northern and western Europe) and YRI (Yoruba in Ibadan, Nigeria).
Statistically significant differences in signs and symptoms, esophageal mucosal appearance at endoscopy, or atopic status were not found between a subset of familial patients compared to patients with sporadic EE. Although we report on a relatively large number of EE patients, our study is retrospective and therefore we do not have complete information on all the patients in the study; it is possible that differences would be uncovered with larger numbers of patients in each group. In addition, we cannot eliminate the possibility that the sporadic cohort includes patients whose family members do not currently manifest disease, but who will in the future.
In this study, biopsies from the distal esophagus were more commonly diagnostic compared to biopsies from other esophageal sites, generally the proximal esophagus. However, in seven cases with nondiagnostic distal esophageal biopsies, biopsies from other esophageal sites established the diagnosis, underscoring the patchy nature of the disease, and the need to obtain biopsies from multiple sites. In a study of adult EE patients, biopsies from sites other than the distal esophagus were diagnostic in a patient whose distal biopsies were nondiagnostic.29 In our study, eosinophil density tended to be greater in biopsies from the distal esophagus compared to biopsies from other sites, which was also observed in adult EE patients.29, 30
There was wide variation in eosinophil number among and within biopsies at all esophageal sites (Table 1), and therefore lack of statistical differences between the familial and nonfamilial pairs is not surprising. The large standard deviations result from the varying intensity of inflammation within biopsies obtained from any site, a fact that also emphasizes the need to obtain multiple biopsies. In a study of biopsies from adult patients using a threshold number of 15/hpf, the sensitivity to detect EE was 55% with one esophageal biopsy, but sensitivity was 100% with five esophageal biopsies.30
Among all 85 patients for whom the presence or absence of furrowed esophageal mucosa was recorded (n=59), peak eosinophil numbers in biopsies from proximal or distal esophagus were not significantly different for patients who had furrows compared to those who did not. However, among familial and sporadic patient pairs who had furrowed esophageal mucosa, the peak number of intraepithelial eosinophils in biopsies from the distal esophagus was significantly lower in familial patients compared to sporadic patients. The cause of furrowed mucosa is unclear, but likely involves alterations of the mucosa, lamina propria, and possibly deeper layers of the esophageal wall, that could be related to inflammation. Furrows are reversible following proper therapy for EE, but the natural history if left untreated is unknown. We cannot rule out the possibility that the difference between familial and nonfamilial patients is related to recording bias, but it is also possible that the alterations that lead to mucosal furrows occur with less inflammation in familial compared to nonfamilial patients. This finding may indicate a fundamental difference between familial and nonfamilial patients, and requires further investigation. Currently, it should provide greater impetus to obtain detailed family histories of patients, and to thoroughly evaluate symptomatic family members of affected patients.
Gender-related analysis uncovered significant differences among all patients and sporadic patients, but not familial EE patients. EE is much more prevalent among males than females, and the number of females in the sporadic group is small (n=7). Our data nonetheless suggests that thickened mucosa and chest pain are more common among males and females respectively, and these findings should be confirmed in larger studies.
We did not require absence of eosinophilic inflammation in biopsies from extra esophageal sites for inclusion in this study. Our practice is to obtain biopsies from stomach and duodenum, and the lower gastrointestinal tract if indicated, in patients suspected to have EE.13 However, some of the initially diagnostic biopsies of our patients were obtained at other institutions that do not routinely biopsy extra esophageal sites in patients suspected of having esophageal disease. The biopsies from extra esophageal sites generally were normal, and those that showed eosinophilic inflammation were usually not histologically impressive. However, twice as many familial as sporadic patients had any eosinophilic inflammation in extra esophageal biopsies, indicating that familial EE is more likely part of eosinophilic gastroenteritis than sporadic EE. This also may indicate a fundamental difference between familial and sporadic patients, and requires further study. This finding also provides support for obtaining careful family histories from EE patients, and strongly suspecting EE in symptomatic family members, particularly if eosinophilic inflammation is found in extra esophageal sites of the proband.
It is remarkable that global transcript expression profile analysis of esophageal tissue from sporadic EE and familial EE patients were largely similar even in the absence of false discovery correction. It is important to note that the EE transcriptome is also conserved across gender, age, and atopic background.22 The EE transcriptome consists of a common set of genes that include Th2-associated genes and effector pathways (e.g. eotaxin-3) that is remarkably similar between individuals despite their age, gender, and atopic status.20,21 The largely similar transcript expression profile between EE patients indicates that the effector phase of tissue disease is largely conserved despite the underlying upstream trigger (e.g. genetics).
Acknowledgments
This work was funded in part by the NIH U19 AI070235 (M.E.R.), the Food Allergy and Anaphylaxis Network FAAN (M.E.R.), Campaign Urging Research for Eosinophil Disorders (CURED), the Buckeye Foundation (M.E.R.), the Food Allergy Project (M.E.R.), and the American Heart Association (C.B.).
Abbreviations
- EE
eosinophilic esophagitis
- EGID
eosinophilic gastrointestinal disease
- FDR
false discovery rate
- GERD
gastroesophageal reflux disease
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID) J Allergy Clin Immunol. 2004;113:11–28. doi: 10.1016/j.jaci.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg ME, Mishra A, Collins MH, et al. Pathogenesis and clinical features of eosinophilic esophagitis. J Allergy Clin Immunol. 2001;108:891–894. doi: 10.1067/mai.2001.120095. [DOI] [PubMed] [Google Scholar]
- 3.Gupta SK, Fitzgerald JF, Chong SKF, et al. Vertical lines in distal esophageal mucosa (VLEM): a true endoscopic manifestation of esophagitis in children? Gastrointest Endosc. 1997;45:485–489. doi: 10.1016/s0016-5107(97)70178-0. [DOI] [PubMed] [Google Scholar]
- 4.Lim JR, Gupta SK, Croffie JM, et al. White specks in the esophageal mucosa: an endoscopic manifestation of non-reflux eosinophilic esophagitis in children. Gastrointest Endosc. 2004;59:835–838. doi: 10.1016/s0016-5107(04)00364-5. [DOI] [PubMed] [Google Scholar]
- 5.Ngo P, Furuta GT, Antonioli DA, et al. Eosinophils in the esophagus—peptic or allergic eosinophilic esophagitis? Case series of three patients with esophageal eosinophilia. Am J Gastroenterol. 2006;101:1666–1670. doi: 10.1111/j.1572-0241.2006.00562.x. [DOI] [PubMed] [Google Scholar]
- 6.Kelly KJ, Lazenby AJ, Rowe PC, et al. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–1512. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 7.Ruchelli E, Wenner W, Voytek T, et al. Severity of esophageal eosinophilia predicts response to conventional gastroesophageal reflux therapy. Pediatr Develop Pathol. 1999;2:15–18. doi: 10.1007/s100249900084. [DOI] [PubMed] [Google Scholar]
- 8.Walsh SV, Antonioli DA, Goldman H, et al. Allergic esophagitis in children. A clinicopathological entity. Am J Surg Pathol. 1999;23:390–396. doi: 10.1097/00000478-199904000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Markowitz JE, Spergel JM, Ruchelli E, et al. Elemental diet is an effective treatment for eosinophilic esophagitis in children and adolescents. Am J Gastroenterol. 2003;98:777–782. doi: 10.1111/j.1572-0241.2003.07390.x. [DOI] [PubMed] [Google Scholar]
- 10.Noel RJ, Putman PE, Collins MH, et al. Clinical and immunopathologic effects of swallowed fluticasone for eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2004;2:568–575. doi: 10.1016/s1542-3565(04)00240-x. [DOI] [PubMed] [Google Scholar]
- 11.Konikoff MR, Noel RJ, Blanchard C, et al. A randomized, double blind, placebo-controlled trial of fluticasone propionate for pediatric eosinophilic esophagitis. Gastroenterology. 2006;131:1381–1391. doi: 10.1053/j.gastro.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Stein ML, Collins MH, Villaneuva JM, et al. Anti-IL-5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol. 2006;118:1312–1319. doi: 10.1016/j.jaci.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: A systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Spergel JM, Beausoleil JL, Mascarenhas M, et al. The use of skin prick tests and patch tests to identify causative foods in eosinophilic esophagitis. J Allergy Clin Immunol. 2002;109:363–368. doi: 10.1067/mai.2002.121458. [DOI] [PubMed] [Google Scholar]
- 15.Ober C. Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol. 2005;116:274–278. doi: 10.1016/j.jaci.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 16.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis (Letter to the Editor) New Engl J Med. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 17.Patel SM, Falchuk KR. Three brothers with dysphagia caused by eosinophilic esophagitis. Gastrointest Endosc. 2005;61:165–167. doi: 10.1016/s0016-5107(04)02459-9. [DOI] [PubMed] [Google Scholar]
- 18.Meyer GW. Eosinophilic esophagitis in a father and a daughter (Letter to the editor) Gastrointest Endosc. 2005;61:932. doi: 10.1016/s0016-5107(05)00508-0. [DOI] [PubMed] [Google Scholar]
- 19.Zink DA, Amin M, Gebara S, Desai TK. Familial dysphagia and eosinophilia. Gastrointest Endosc. 2007;65:330–334. doi: 10.1016/j.gie.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Straumann A, Simon H-U. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–419. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–1059. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 22.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assa’ad AH, Putnam PE, Collins MH, et al. Pediatric patients with eosinophilic esophagitis: An 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–738. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 24.DeBrosse CW, Case JW, Putnam PE, et al. Quantity and distribution of eosinophils in the gastrointestinal tract of children. Pediatr Develop Pathol. 2006;9:210–218. doi: 10.2350/11-05-0130.1. [DOI] [PubMed] [Google Scholar]
- 25.Straumann A, Spichtin H-P, Grize L, et al. Natural history of primary eosinophilic esophagitis: A follow-up of 30 adult patients for up to 11.5 years. Gastroenterology. 2003;125:1660–1669. doi: 10.1053/j.gastro.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Malerba G, Lauciello MC, Scherpbier J, et al. Linkage analysis of chromosome 12 markers in Italian families with atopic asthmatic children. Am J Respir Crit Care Med. 2000;162:1587–1590. doi: 10.1164/ajrccm.162.4.9909031. [DOI] [PubMed] [Google Scholar]
- 27.Palmer CAN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 28.Akei HS, Mishra A, Blanchard C, et al. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–994. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Remedios M, Campbell C, Jones DM, et al. Eosinophilic esophagitis in adults: clinical, endoscopic, histological findings, and response to treatment with fluticasone propionate. Gastrointest Endosc. 2006;63:3–12. doi: 10.1016/j.gie.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 30.Gonsalves N, Policarpio-Nicolas M, Zhang Q, et al. Histopathological variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc. 2006;64:313–319. doi: 10.1016/j.gie.2006.04.037. [DOI] [PubMed] [Google Scholar]


