Abstract
A distinct phosphodiesterasic activity (EC 3.1.4) was found in both mono- and dicotyledonous plants that catalyzes the hydrolytic breakdown of ADPglucose (ADPG) to produce equimolar amounts of glucose-1-phosphate and AMP. The enzyme responsible for this activity, referred to as ADPG pyrophosphatase (AGPPase), was purified over 1,100-fold from barley leaves and subjected to biochemical characterization. The calculated Keq′ (modified equilibrium constant) value for the ADPG hydrolytic reaction at pH 7.0 and 25°C is 110, and its standard-state free-energy change value (ΔG′) is −2.9 kcal/mol (1 kcal = 4.18 kJ). Kinetic analyses showed that, although AGPPase can hydrolyze several low-molecular weight phosphodiester bond-containing compounds, ADPG proved to be the best substrate (Km = 0.5 mM). Pi and phosphorylated compounds such as 3-phosphoglycerate, PPi, ATP, ADP, NADP+, and AMP are inhibitors of AGPPase. Subcellular localization studies revealed that AGPPase is localized exclusively in the plastidial compartment of cultured cells of sycamore (Acer pseudoplatanus L.), whereas it occurs both inside and outside the plastid in barley endosperm. In this paper, evidence is presented that shows that AGPPase, whose activity declines concomitantly with the accumulation of starch during development of sink organs, competes with starch synthase (ADPG:1,4-α-d-glucan 4-α-d-glucosyltransferase; EC 2.4.1.21) for ADPG, thus markedly blocking the starch biosynthesis.
Although the pyrophosphorolytic reactions leading to the production of gluconeogenic intermediates such as ADPglucose (ADPG) and UDPglucose (UDPG) are readily reversible, they mainly proceed toward the direction of nucleotide sugar synthesis (1). On the other hand, plant enzymes that irreversibly cleave nucleotide sugars have been described that may effectively interrupt the flow of glycosyl moieties toward the biosynthesis of end products such as starch, cell wall polysaccharides, or sucrose (2). It is conceivable that in conjunction with ADPG pyrophosphorylase (AGPase; EC 2.7.7.27), UDPG pyrophosphorylase, sucrose synthase, and starch synthase, among others, these enzymes will participate in controlling the levels of nucleotide sugars engaged in starch formation. Among a few enzymes reported to date that can hydrolyze nucleotide sugars in plants, ADPG phosphorylase is shown to catalyze the phosphorolytic breakdown of ADPG (3). UDPG phosphorylase is known to split UDPG in the presence of Pi, and its activities are greatly stimulated by some signal metabolites, indicating that it may play an important role in the control of photosynthate partitioning (4).
Based on experimental grounds, Pozueta-Romero et al. (5) have proposed the operation of synthesis/breakdown metabolic cycles controlling the rate of starch formation. According to this hypothesis, the balance between enzymatic activities catalyzing the synthesis of gluconeogenic intermediates and those activities catalyzing their breakdown can determine the net rate of starch synthesis. Because of the possibility that activities hydrolyzing ADPG, the universal starch precursor, may exist in plants that regulate the metabolism of this polyglucan, we have explored their possible occurrence in several plant species. As a result, we have now found a phosphodiesterasic activity that catalyzes the hydrolytic breakdown of ADPG. In this paper, we report the subcellular localization and biochemical characterization of the enzyme responsible for this activity, referred to as ADPG pyrophosphatase (AGPPase).† Based on the results presented in this work using different plant sources, we discuss that AGPPase may be involved in controlling the intracellular levels of ADPG linked to starch biosynthesis.
Materials and Methods
Plant Material.
Barley (Hordeum vulgare cv. Scarlett) and wheat (Triticum aestivum cv. Marius) plants were grown in the field. Seeds at different developmental stages and leaves were harvested and stored at −80°C until used. Bell pepper (Capsicum annuum cv. Yolo Wonder), tomato (Lycopersicon sculentum cv. Ailsa Craig), potato (Solanum tuberosum cv. Désirée), and Arabidopsis thaliana (cv. Columbia and Adg1) were grown in greenhouse conditions. Protoplasts and amyloplasts from the suspension-cultured cells of sycamore (Acer pseudoplatanus L.) were obtained as described by Pozueta-Romero et al. (6). Amyloplasts from young barley endosperms were isolated as described by Thorbjornsen et al. (7).
Protein Extraction and Purification.
Unless otherwise indicated, all steps were carried out at 4°C. For small-scale extractions, plant tissues were homogenized with 3-fold of extraction buffer (50 mM Mes, pH 6.0/1 mM EDTA/2 mM DTT) with a Waring Blender, and were filtered through four layers of Miracloth and desalted by ultrafiltration on Centricon YM-10 (Amicon, Bedford, MA). For purification of AGPPase, 600 ml of homogenate obtained from 200 g of young barley leaves were centrifuged at 100,000 × g for 30 min, and the supernatant was adjusted to 50% (NH4)2SO4. The precipitate obtained after 30 min of centrifugation at 30,000 × g (at 20°C) was resuspended in 520 ml of 50 mM Mes, pH 4.2/1 mM EDTA/2 mM DTT, heated in a water bath at 62°C for 20 min, cooled on ice, and centrifuged at 30,000 × g for 20 min. Proteins from the supernatant (520 ml) were precipitated with 50% (NH4)2SO4 and resuspended in 5.7 ml of extraction buffer. The sample was then subjected to gel filtration on a Superdex 200 column (Pharmacia LKB) preequilibrated with 50 mM Mes, pH 6.0/150 mM NaCl. The elution was carried out with the same buffer at a flow rate of 0.3 ml/min, and 150-μl fractions were collected. The purified enzyme preparation was subjected to electrophoresis in nondenaturating 7% acrylamide gels at pH 4.3, which were prepared as described by Reisfeld et al. (8).
The native molecular mass of AGPPase was determined from a plot of Kav (partition coefficient) versus log molecular mass of protein standards. Protein content was measured by the Bradford method using the Bio-Rad prepared reagent.
Enzyme Assays.
Unless otherwise indicated, all enzymatic reactions were performed at 37°C. Measurements of AGPase and AGPPase activities were performed by using the two-step spectrophotometric determination of glucose-1-phosphate (G1P) described by Sowokinos (9). In step one, the reaction mixture for AGPPase contained 50 mM Mes (pH 6.0), the specified amount of ADPG and protein extract, in a total volume of 50 μl. For AGPase assays, the reaction mixture contained 50 mM Hepes (pH 7.5), 5 mM MgCl2, 3 mM PPi, and 10 mM 3-phosphoglycerate (3-PGA). All assays were run with minus ADPG blanks. After 20 min of incubation, the reaction was stopped by boiling in a dry bath for 2 min. In step two, G1P was determined spectrophotometrically in a 300-μl mixture containing 50 mM Hepes (pH 7.0), 1 mM EDTA, 2 mM MgCl2, 15 mM KCl, 0.6 mM NAD+, 1 unit each of phosphoglucomutase and glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides, and 30 μl of the step-one reaction. After 20 min of incubation, the NADH production was monitored at 340 nm by using a Multiskan EX spectrophotometer (Labsystems, Chicago). Negligible NADH was produced by any protein extract in the absence of ADPG in step one. Hydrolytic breakdown of bis-p-nitrophenyl phosphate (bis-PNPP) was monitored by measuring the release of p-nitrophenol (PNP) at 405 nm as described by Nishimura and Beevers (10). Orthophosphate production from the hydrolysis of PPi, 3-PGA, G1P, and nucleoside mono-, di-, and triphosphates was determined with a colorimetric assay kit from Sigma. PNPP production from the hydrolysis of bis-PNPP was monitored by measuring the release of PNP at 405 nm in the presence of alkaline phosphatase (10). Production of nucleotides from the hydrolytic breakdown of 200 μg of linear, heat-denatured pSK plasmid DNA (Stratagene) was monitored by HPLC. Alkaline pyrophosphatase (PPase; EC 3.6.1.1) and alcohol dehydrogenase (EC 1.1.1.1) activities were measured as described by Thorbjornsen et al. (7). α-Mannosidase (EC 3.2.1.24) was determined as described by Nishimura and Beevers (10). Starch was determined by using a glucose oxidase-based test kit (Boehringer Mannheim). The unit is defined as the amount of enzyme that catalyzes the production of 1 μmol of product per minute.
HPLC Analyses.
Separations and quantitative analyses of nucleotides [ATP, ADP, AMP, cAMP, NAD, NMN, UTP, UDP, UMP, GTP, GDP, GMP, CTP, cytidine 5′-diphosphate (CDP), and CMP] and nucleotide sugars (ADPG, UDPG, CDPglucose, and ADPribose) were carried out as described by Shannon (11), using a Waters Associates' HPLC system fitted with a Partisil-10-SAX column (4.6 mm × 25 cm; Whatman).
Biochemical Calculations.
The modified equilibrium constant (Keq′) and the standard-state free-energy change (ΔG′) for the hydrolysis of ADPG catalyzed by AGPPase were calculated as described by Klotz (12). Toward this end, 100 mM ADPG was incubated at 25°C in a 100-μl reaction mixture containing 1 unit of AGPPase and 50 mM Hepes (pH 7.0). At different incubation times (ranging from 0 to 5 h), 20 μl of the reaction mixture was withdrawn, and the concentrations of ADPG, AMP, and G1P were measured. Kinetic parameters such as Km and Vmax were evaluated by Lineweaver–Burk plots. Hill coefficients for all substrates were calculated from Hill plots. The enzyme-inhibitor dissociation constants (Ki) as well as the pattern of inhibition were determined from Dixon plots and Lineweaver–Burk plots.
Starch Synthesis.
Starch granules were prepared from amyloplasts of 30-day-after-pollination barley endosperms according to the method of Thorbjornsen et al. (7). Ten milligrams of starch granules were resuspended in 250 μl of 50 mM Hepes (pH 7.0) and 2 mM MgCl2 and were incubated in Eppendorf tubes at 37°C with 0.5 mM ADP-(U-14C)-G under active shaking (1,400 rpm). Reactions were stopped at the indicated incubations times by adding 1 ml of 75% methanol/1% KCl to 50-μl aliquots of the starch granule suspension. Zero-time incubation was obtained by adding 1 ml of methanol/KCl solution to the starch granule suspension before the addition of ADP-(U-14C)-G. The methanol/KCl-insoluble fractions containing starch subsequently were washed thoroughly, and radioactivities were measured to estimate the starch formation as described by Hill and Smith (13).
Results
Hydrolytic Breakdown of ADPG Is Widely Present in Higher Plants.
The spectrophotometric measurement of the AGPase pyrophosphorolytic activities is a commonly used method that is based on the coupling of G1P production with an accompanying reduction of NAD+ (9, 14). Enzymatic assays are routinely initiated by the addition of PPi to the reaction mixture containing ADPG. Surprisingly, however, while monitoring the AGPase activities using the desalted protein extracts prepared from various mono- and dicotyledonous plant tissues as well as the cultured cells of sycamore, we were able to detect the production of G1P when PPi was not included in the reaction mixture (Table 1). This activity was found to be particularly high in barley and wheat leaves, whereas it was low in the endosperms from the same plants. In leaves, ADPG-dependent production of G1P was not affected or was even decreased when PPi was added to the assay mixture (Table 1); it was generally stimulated when both PPi and 3-PGA were added simultaneously to the reaction mixture (Table 1). This stimulation, ascribable to the allosteric effect of 3-PGA on AGPase catalyzing the pyrophosphorolytic breakdown of ADPG (1), was not observed in leaves of the starchless Adg1 mutant of A. thaliana, which lacks AGPase activity (15). These overall results thus show that enzymatic reaction(s) catalyzing the hydrolytic breakdown of ADPG are widely distributed in higher plants.
Table 1.
ADPG hydrolyzing enzyme activities in higher plants
| Plant tissues | Specific activity, milliunits/ mg of protein
|
||
|---|---|---|---|
| +ADPG | +ADPG+ PPi | +ADPG+ PPi+3-PGA | |
| Monocotyledonous | |||
| Barley leaf | 113.7 ± 3.5 | 72.2 ± 0.7 | 103.8 ± 5.0 |
| Barley leaf endosperm, 30 days after pollination | 2.0 ± 0.2 | 15.9 ± 1.2 | 21.8 ± 0.3 |
| Wheat leaf | 22.4 ± 2.5 | 15.1 ± 0.9 | 50.8 ± 0.6 |
| Wheat leaf endosperm, 30 days after pollination | 1.7 ± 0.2 | 48.3 ± 2.6 | 56.4 ± 1.4 |
| Dicotyledonous | |||
| A. thaliana leaf, Wt | 5.2 ± 0.6 | 5.1 ± 0.8 | 42.1 ± 3.3 |
| A. thaliana leaf, Adg | 5.9 ± 1.7 | 6.5 ± 1.3 | 7.2 ± 0.2 |
| Pepper leaf | 5.0 ± 0.6 | 7.3 ± 1.0 | 24.8 ± 0.8 |
| Tomato leaf | 5.6 ± 0.5 | 16.1 ± 0.1 | 82.4 ± 6.9 |
| Sycamore cultured cells | 16.5 ± 7.2 | 30.6 ± 5.4 | 137.4 ± 39.8 |
Data are presented as the mean ± SD obtained from five independent experiments. Details of enzyme assay methods to measure G1P production are described in Material and Methods.
Enzyme Purification.
We then attempted to purify the enzyme molecule responsible for these activities by using barley leaves. As presented in Table 2, the enzyme exists in a single soluble form and it is highly resistant to acid pH and high temperature treatment. By taking advantage of those characteristic behaviors, we were able to purify the enzyme over 1,100-fold to a specific activity of 23 units/mg of protein. The apparent molecular mass of the native enzyme measured by gel filtration was estimated to be 25–45 kDa. Electrophoretic separation of the purified enzyme preparation in nondenaturating gels and by subsequent staining revealed a single protein entity that, after elution, was shown to hydrolyze ADPG (data not shown). In addition to this finding, SDS/PAGE analysis of the purified protein preparation revealed a single band of about 35 kDa (data not shown). These results thus indicate that the enzyme responsible for the ADPG breakdown is a 35-kDa monomeric protein.
Table 2.
Purification of AGPPase from young barley leaves
| Step | Total volume, ml | Total protein, mg | Total activity, milliunits | Specific activity, milliunits/mg of protein | Purification, -fold | Yield, % |
|---|---|---|---|---|---|---|
| Crude extract | 560.0 | 5107.8 | 105,000 | 20.6 | — | 100.0 |
| Supernatant, 100,000 × g | 520.0 | 3436.7 | 100,500 | 29.2 | 1.4 | 95.7 |
| Ammonium sulfate, 50% | 520.0 | 748.6 | 97,500 | 130.2 | 6.3 | 92.8 |
| pH 4.2/62°C | 520.0 | 24.9 | 90,500 | 3,634.0 | 176.4 | 86.2 |
| Ammonium sulfate, 50% | 5.7 | 8.1 | 47,300 | 5,839.0 | 283.4 | 45.0 |
| Superdex 200 | 1.7 | 1.3 | 30,200 | 23,230.0 | 1127.6 | 28.7 |
Verification of Reaction Products and Enzyme Designation.
ADPG breakdown catalyzed by the purified enzyme preparation yields equimolar amounts of G1P and AMP (Table 3). Because of the similarity of this reaction with that of nucleoside diphosphate sugar pyrophosphatases (EC 3.6.1.13 and EC 3.6.1.21) occurring in mammalian tissues (16, 17), bacteria (18, 19), and yeast (20, 21), we have designated the enzyme responsible for the hydrolytic cleavage of ADPG as AGPPase.
Table 3.
Substrate specificity studies on barley AGPPase
| Substrates | Km, mM | Vmax, % with respect to ADPG | Products | Stoichiometry |
|---|---|---|---|---|
| ADPG | 0.5 | 100 | G1P + AMP | (1:1) |
| CDPglucose | 2.8 | 114 | G1P + CMP | (1:1) |
| UDPG | 2.1 | 114 | G1P + UMP | (1:1) |
| ADPribose | 2.4 | 100 | Ribose 5P + AMP | (1:1) |
| bis-PNPP | 0.3 | 100 | PNP + PNPP | (1:1) |
| NAD+ | 2.5 | 100 | AMP + NMN | (1:1) |
| NADP+ | NQ | NQ | — | — |
| cAMP | NQ | NQ | — | — |
| ATP | NQ | NQ | — | — |
| ADP | NQ | NQ | — | — |
Kinetic parameters (Km, Vmax, Ki, and stoichiometry) obtained are the mean values from five independent experiments. NQ, not quantifiable.
Keq′ and ΔG′ Calculations and Kinetic Analyses of AGPPase.
Time-course analyses of 100 mM ADPG incubated with 1 unit of AGPPase at pH 7 at 25°C revealed that at equilibrium, only 90 μM ADPG remained, whereas 99.9 mM G1P and 99.1 mM AMP were present in the assay mixture. It thus was calculated that the Keq′ of the reaction is 110 and its ΔG′ is −2.9 kcal/mol (1 kcal = 4.18 kJ) (12).
The substrate specificity of AGPPase was tested by using the purified enzyme preparation on a wide range of compounds at a concentration of 5 mM. These preliminary analyses showed that AGPPase is a phosphodiesterase that hydrolyzes some low-molecular weight phosphodiester bond-containing compounds, whereas it does not hydrolyze PPi, highly polymerized polynucleotides such as single-stranded DNA, and phosphomonoester bond-containing compounds such as G1P, AMP, UMP, GMP, or 3-PGA (data not shown). cAMP, a substrate for cyclic nucleotide phosphodiesterase (22), does not act as a favorable substrate for AGPPase (Table 3). Although having a phosphodiester bond, NADP+ as well as nucleoside di- and triphosphates are not hydrolyzed by AGPPase (Table 3).
Subsequently we performed kinetic analyses for those compounds that were shown to serve as AGPPase substrates (Table 3). The kinetics were hyperbolic (in every case the Hill coefficient was calculated to be n = 1) and the Vmax values were nearly the same, strongly indicating the presence of a unique active site in the enzyme molecule.
Among different types of substrates with possible physiological relevance tested, ADPG was shown to be the most favorable substrate, the apparent Km value being 0.5 mM. In terms of apparent Km, the affinity of AGPPase for NAD+, ADPribose, UDPG, and CDPglucose is about 4- to 5-fold lower than that for ADPG. In contrast to the case of nucleoside diphosphate sugar pyrophosphatases occurring in animals, yeast, and bacteria (16–21), AGPPase exhibits a high affinity to the synthetic substrate bis-PNPP, which is a reagent commonly used to measure phosphodiesterase activities (10).
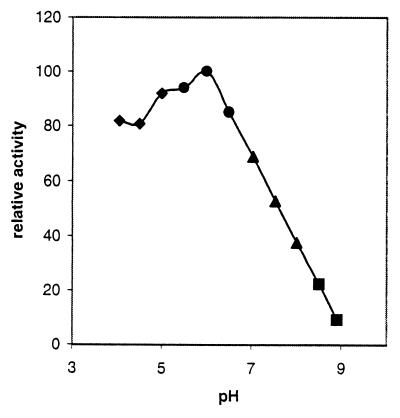
The ADPG breakdown catalyzed by AGPPase is inhibited in a competitive manner by phosphorylated compounds such as Pi, AMP, ADP, ATP, PPi, NADP+, and 3-PGA, whereas adenine, adenosine, NAD+, and glucose exhibit a poor or null effect (Table 4). Remarkably, the enzyme is not affected by typical phosphodiesterase and nucleoside diphosphate sugar pyrophosphatase effectors such as metal ions, fluoride, metal-binding reagents, sulfhydryl-group reacting agents, and reducing compounds (16–26). Also, in clear contrast to phosphodiesterases and nucleotide sugar hydrolases having basic optimum pH (16–26), AGPPase shows a sharp pH-activity profile with optimal activity occurring at pH 6.0, which dramatically decreases at basic pH values (Fig. 1).
Table 4.
Effect of different compounds on ADPG hydrolysis catalyzed by AGPPase
| Effectors | Ki, mM | Type of inhibition |
|---|---|---|
| Glucose, 100 mM | — | ND |
| Adenine, 5 mM | — | ND |
| Adenosine, 5 mM | — | ND |
| Metal ions: Co2+, Ca2+, Mg2+, and Mn2+, 1 mM | — | ND |
| NAD+ | 3.28 | Competitive |
| Phosphorylated compounds | ||
| Pi | 0.39 | Competitive |
| PPi | 0.82 | Competitive |
| 3-PGA | 1.21 | Competitive |
| ATP | 0.89 | Competitive |
| ADP | 0.43 | Competitive |
| AMP | 0.74 | Competitive |
| NADP+ | 0.67 | Competitive |
| Phosphodiesterase inhibitors | ||
| l-Cysteine, 5 mM | — | ND |
| Glutamate, 5 mM | — | ND |
| Reduced glutathione, 5 mM | — | ND |
| Aspartate, 5 mM | — | ND |
| Ascorbate, 1 mM | — | ND |
| Fluoride, 1 mM | — | ND |
| EDTA, 1 mM | — | ND |
| β-Mercaptoethanol, 1 mM | — | ND |
| DTT, 1 mM | — | ND |
Kinetic parameters (Km, Vmax, Ki, and stoichiometry) obtained are the mean values from five independent experiments. ND, not detectable.
Figure 1.
pH-activity curve of barley AGPPase. The buffers used were 50 mM sodium acetate-acetic acid (pH 3.0–5.5), 50 mM Mes/NaOH (pH 5.5–6.5), 50 mM Hepes/NaOH (pH 6.5–8.0), and 50 mM Tris⋅HCl (pH 8.0–9.0).
Subcellular Localization of AGPPase.
Employing the discontinuous Percoll gradient centrifugation technique (6), we have obtained highly purified, intact amyloplasts from the suspension-cultured sycamore cells. Judging by the activities of the soluble plastid marker alkaline pyrophosphatase (PPase), 1% of the amyloplasts originally present in the protoplast lysates were obtained in the final amyloplast preparation (Table 5). On the other hand, activities of cytosolic and vacuolar marker enzymes (i.e., alcohol dehydrogenase and α-mannosidase, respectively) were shown to be nearly undetectable. About 1% of the AGPPase activities initially present in the protoplast lysate was recovered in the final amyloplast preparations. This value showed a statistically significant difference (95% confidence limits) to the values of recovery of cytosolic and vacuolar marker enzymes. With a confidence limit of 95%, there was no significant difference between the values of AGPPase yields in the amyloplast preparations and those of the plastidial marker. Therefore, based on the percentages of the cytosolic, vacuolar, and plastidial marker enzymes found in the amyloplast preparation, we can conclude that practically all of the AGPPase activity in the sycamore cells is associated with amyloplasts. Independent analyses of protection against protease attack have further confirmed this conclusion (data not shown).
Table 5.
Subcellular localization of AGPPase in cultured sycamore cells
| Enzyme | Protopast lysate activity, milliunits/mg of protein | Stepped Percoll gradient centrifugation
|
||
|---|---|---|---|---|
| Amyloplast activity, milliunits/mg of protein | Amyloplast as % of lysate | Recovery range, % | ||
| ADH | 2563 ± 1331 | 0.13 ± 0.03 | 0.006 ± 0.01 | 110 ± 31 |
| α-Mannosidase | 325 ± 63 | 0.13 ± 0.09 | 0.042 ± 0.02 | 83 ± 26 |
| PPase | 4356 ± 1040 | 43.7 ± 8.71 | 1.0 ± 0.20 | 82 ± 14 |
| AGPPase | 16.5 ± 7.2 | 0.17 ± 0.06 | 1.0 ± 0.17 | 120 ± 39 |
Data are presented as the mean ± SD obtained from six separate preparations.
Employing the Nycodenz density gradient method described by Thorbjornsen et al. (7), we have obtained amyloplasts from young barley endosperms. The final yield of the vacuolar marker was marginally low (0.08%), and about 0.65% of the activity of the cytosolic marker was recovered in the final preparation, as compared with 7.6% of the activity of the plastidial marker enzyme (Table 6). About 3% of the activity of the AGPPase initially present in the homogenate was present in the amyloplast pellet. This value is significantly different (95% confidence limits) from the values of recovery of cytosolic, vacuolar, and plastid markers. Therefore, based on the percentages of cytosolic and plastidial markers and by using the equation used by Thorbjornsen et al. (7) to estimate the AGPase activity associated with amyloplasts, it could be concluded that ≈32% of the total AGPPase activity in the young barley endosperm is associated with amyloplasts.
Table 6.
Subcellular localization of AGPPase in barley endosperm
| Enzyme | Endosperm activity, milliunits/mg of protein | Nycodenz density gradient
|
||
|---|---|---|---|---|
| Amyloplast activity, milliunits/mg of protein | Amyloplast as % of endosperm | Recovery range, % | ||
| ADH | 3.61 ± 0.4 | 0.024 ± 0.0026 | 0.67 ± 0.07 | 95 ± 15 |
| α-Mannosidase | 2.48 ± 0.5 | 0.002 ± 0.0001 | 0.08 ± 0.01 | 101 ± 16 |
| PPase | 78.5 ± 1.6 | 5.98 ± 0.27 | 7.65 ± 0.50 | 98 ± 26 |
| AGPPase | 18.9 ± 0.8 | 0.55 ± 0.17 | 2.91 ± 0.47 | 112 ± 8 |
Data are presented as the mean ± SD obtained from six separate preparations.
AGPPase Prevents Starch Biosynthesis.
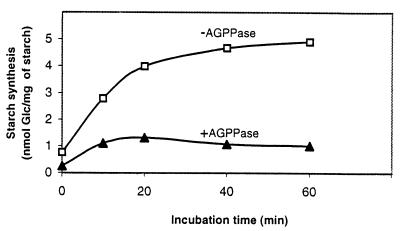
As a consequence of the overall experimental results obtained, it is conceivable that AGPPase and starch synthase can compete with each other for the same substrate and that AGPPase may effectively prevent the ADPG-dependent transglucosylation reaction catalyzed by starch synthase. For the purpose of testing this hypothesis, time-course analyses of ADP-(U-14C)-G-dependent starch biosynthesis were carried out by using starch granules incubated with or without purified AGPPase preparation (see Table 2). As presented in Fig. 2, a high transglucosylation rate was observed when starch granules were incubated with 0.5 mM ADP-(U-14C)-G. However, the reaction rate was markedly reduced when the AGPPase preparation was included in the assay mixture.
Figure 2.
Inhibitory effect of AGPPase on starch biosynthesis. Ten milligrams of starch granules isolated from barley amyloplasts was incubated at 37°C with 0.5 mM ADP-(U-14C)-G in the absence or in the presence of 0.2 units of AGPPase. At the indicated incubation times, samples were collected to measure the incorporated radioactivities as described in Materials and Methods.
Developmental Changes of AGPPase Activities and Starch Production in Sink Organs.
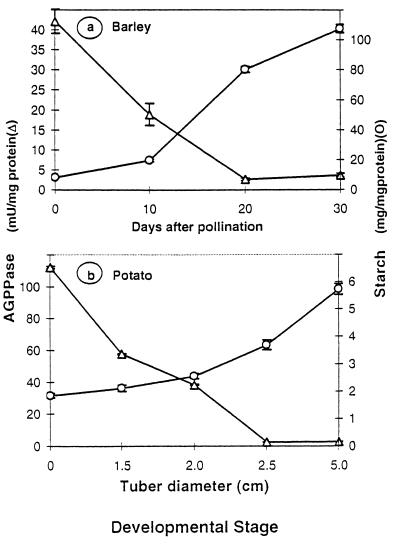
We have examined AGPPase activities and starch content at different developmental stages in sink organs, i.e., barley seeds and potato tubers. As shown in Fig. 3a, the starch content in the barley seed is low during the first 10 days after pollination, which are accompanied by high AGPPase activity. After a lag period, the starch accumulation pattern proceeds nearly at maximum rate until 30 days after pollination, at which time there occurs a dramatic decline of the AGPPase activity. Essentially a similar reciprocal pattern was observed during the developmental process of potato tuberization, which was characterized by a rapid increase in starch content and a dramatic decrease of the AGPPase activities (Fig. 3b).
Figure 3.
AGPPase activities and starch content at different developmental stages of sink organs. (a) Barley seeds. (b) Potato tubers. Pretuberized stolon samples, represented by zero tuber diameters, were collected by removing the terminal 3-cm section from their apical end. As tuberization occurred, tubers were removed, washed, and grated according to their diameter. Data are represented as mean ± SD (n = 4).
Discussion
The experimental results reported in this paper describe the wide occurrence of a distinct phosphodiesterase in plants (Table 1), designated as AGPPase, which catalyzes the hydrolytic conversion of ADPG, the universal glucosyl donor for starch biosynthesis, to AMP and G1P. Although its substrate specificity is not absolute for ADPG (Table 3), this nucleotide sugar was shown to serve as the best substrate among various substances of physiological relevance tested. AGPPase is clearly distinguishable from phosphodiesterases and nucleoside diphosphate sugar pyrophosphatases described to date (16–26) not only in its substrate specificity, but also in molecular weight, optimum pH (Fig. 1), inertness to reducing agents, and nonrequirement of metal ions (Table 4).
It is intriguing that AGPPase activities have escaped the attention of preceding investigators, especially considering that the utilization of phosphodiesterases from animal origin was a powerful and fundamental tool in identifying and characterizing the ADPG molecule in classic biochemical works (27). That AGPPase has not been noted until now is likely attributable to inadequate biochemical analyses performed on plant phosphodiesterases. Unfortunately, artificial substances such as bis-PNPP frequently have been used for analytical studies of plant phosphodiesterases (10), and few investigators have assessed Vmax and Km values for different substrates of possible physiological relevance. In many instances (23, 24, 28), putative physiological substrates have been assayed only at a single concentration, which makes meaningful kinetic comparisons among the different phosphodiesterases difficult.
Since the initial discovery showing that ADPG serves as the universal glucosyl donor for starch synthase (cf. 29), the mechanism of starch biosynthesis has generally been considered a unidirectional and vectorial process (1, 30, 31). Alternatively and basically analogous to the case of sucrose metabolism (32), Pozueta-Romero et al. (5) have proposed the possible operation of metabolic synthesis/breakdown cyclic turnover of starch, which may entail advantages such as sensitive regulation and channeling of excess gluconeogenic intermediates toward other metabolic pathways. In numerous plant tissues, for instance, starch synthase activity is generally severalfold lower than those of the ADPG-forming enzymes such as AGPase and sucrose synthase (5, 7, 33). Under conditions of very active ADPG production, it is highly conceivable that, unless some mechanism(s) for ADPG degradation exist, overaccumulation of ADPG will ensue in the cell. The question thus arises as to how the carbon units from excess ADPG can be effectively used by the starch-forming cell. The fact that AGPPase catalyzes the hydrolytic breakdown of ADPG strongly indicates that it may play a physiological significant role in cleaving excess ADPG. It is therefore highly conceivable that AGPPase plays a role in controlling the intracellular levels of ADPG and acts at a branchpoint, producing G1P and AMP for numerous metabolic pathways. In this context, it should be emphasized that nucleoside diphosphate sugar pyrophosphatases described in animals and bacteria are suggested to act as “housecleaning” enzymes whose role will be to cleanse the cell of potentially deleterious endogenous sugar nucleotides and to modulate the accumulation of metabolic intermediates by diverting them into alternative pathways in response to biochemical need (17, 21).
AGPPase is resistant to high temperature and low pH regimes, and it has relatively acid optimum pH (Fig. 1). Although these characteristics, also occurring in some chloroplastic enzymes (34), are suggestive of a vacuolar localization, subcellular fractionation studies shown in Tables 5 and 6 demonstrate that AGPPase is not confined to the vacuolar compartment. Most importantly, this study has shown that AGPPase is located in the plastid, which is the specific compartment where ADPG is produced by AGPase and is used by starch synthase to produce starch. In some instances, such as in the cultured cells of sycamore (Table 5), AGPPase occurs solely in the plastidial compartment. Exclusive plastidial localization of AGPPase has been further confirmed in our other investigations using barley leaves showing that AGPPase is a chloroplastic enzyme (unpublished data). It is remarkable that just like AGPase, AGPPase is shown to be located both inside and outside the amyloplast of developing barley endosperms (Table 6). Overall, our investigations on subcellular localization of AGPPase indicate that this enzyme is in the same compartment of the cell as are the established processes of ADPG synthesis and utilization.
AGPPase activities were found to be particularly high in tissues such as barley and wheat leaves (Table 1). In the cultured cells of sycamore, AGPPase activities (16.5 milliunits/mg of protein; Table 1) are exceedingly higher than those of starch synthase (1.3 milliunits/mg of protein; cf. 35). Because AGPPase is shown to exhibit relatively high affinity to ADPG (Table 3) and occurs inside the plastidial compartment (Tables 5 and 6), it can be readily predicted that, unless AGPPase is highly regulated, ADPG existing in the plastid will be instantaneously hydrolyzed to G1P and AMP, thus preventing starch synthesis (Fig. 2). However, as presented in Table 4, AGPPase is inhibited by nonsubstrate molecules. Moreover, regulation of AGPPase in photosynthetic tissues also may result from its optimum pH. The pH regime of the chloroplast is known to be alkaline in the light and essentially neutral in the dark, and indeed it is one of the most remarkable factors affecting the activities of various chloroplastic enzymes involved in photosynthesis and gluconeogenesis (34, 36). As presented in Fig. 1, AGPPase is shown to be potentially active in the hydrolysis of ADPG at neutral pH, but its activities are significantly lower at pH 8. Therefore, diurnal fluctuations of the pH in chloroplast stroma may result in the regulation of AGPPase activity in vivo.
The starch biosynthetic pathway in sink organs is known to be subjected to developmental control. In many cases, activities of enzymes involved in starch synthesis such as AGPase, sucrose synthase, and starch synthase, are shown to increase concomitantly with the accumulation of starch during the development of sink organs (37–39). Taking into account all of the limitations inherent in basing conclusions on in vitro activities (40), such a comparison nevertheless can be useful in pointing to potentially limiting steps in the starch biosynthetic process. Thus, we carried out time-course analyses of AGPPase activities and starch content in different sink tissues. As presented in Fig. 3, the clear reciprocal patterns observed in both developing potato tubers and barley endosperms suggest that AGPPase is subjected to developmental control and it may act as a potential limiting step of the gluconeogenic process.
Traditional criteria used to identify regulatory sites of a metabolic pathway lead to the conclusion that regulatory enzymes catalyze nonequilibrium reactions, and they are controlled by factors other than substrate concentration (30). However, recent reevaluation of these ideas (41) proved the distinction between “regulatability” of an enzyme (i.e., whether mechanisms exist to alter its activity) and its “regulatory capacity” (i.e., whether a change in the activity of the enzyme will lead to a change in flux through the pathway). AGPPase, which is now shown to prevent the ADPG-dependent transglucosylation reaction catalyzed by starch synthase (Fig. 2), is negatively affected by phosphorylated compounds that do not act as favorable substrates, and its activities are highly pH-dependent (Fig. 1). Furthermore, in view of the fact that the steady state concentrations of ADPG, G1P, and AMP in tissues such as rice and maize endosperms (11, 42) and potato tubers (43, 44) are substantially displaced from the equilibrium of the reaction catalyzed by AGPPase (Keq′ = 110; ΔG′ = −2.9 kcal/mol), it can be readily inferred that AGPPase is a “regulatable” enzyme. Although the overall results presented in this work pinpoint the step catalyzed by AGPPase as a potential point of regulation of the starch biosynthetic pathway, further investigations using transgenic plants with altered AGPPase activities will contribute to evaluating the importance of this enzyme in the control of the starch biosynthetic process.
Abbreviations
- ΔG′
standard-state free-energy change
- ADPG
ADPglucose
- AGPase
ADPG pyrophosphorylase
- AGPPase
ADPG pyrophosphatase
- G1P
glucose-1-phosphate
- 3-PGA
3-phosphoglycerate
- PNPP
p-nitrophenyl phosphate
- UDPG
UDPglucose
Footnotes
AGPPase also can be referred to as adenosine diphosphate glucose phosphodiesterase.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120168097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120168097
References
- 1.Preiss J. In: The Biochemistry of Plants. Preiss J, editor. Vol. 14. New York: Academic; 1988. pp. 181–254. [Google Scholar]
- 2.Feingold D S, Avigad G. In: The Biochemistry of Plants. Stumpf P K, Conn E E, editors. Vol. 3. New York: Academic; 1980. pp. 101–170. [Google Scholar]
- 3.Murata T. Agric Biol Chem. 1977;41:1995–2002. [Google Scholar]
- 4.Gibson D M, Shine W. Proc Natl Acad Sci USA. 1983;80:2491–2494. doi: 10.1073/pnas.80.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pozueta-Romero J, Perata P, Akazawa T. Crit Rev Plant Sci. 1999;18:489–525. [Google Scholar]
- 6.Pozueta-Romero J, Frehner M, Viale A M, Akazawa T. Proc Natl Acad Sci USA. 1991;88:5769–5773. doi: 10.1073/pnas.88.13.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorbjornsen T, Villand P, Denyer K, Olsen O-A, Smith A M. Plant J. 1996;10:243–250. [Google Scholar]
- 8.Reisfeld R A, Lewis U J, Williams D E. Nature (London) 1962;195:281–283. doi: 10.1038/195281a0. [DOI] [PubMed] [Google Scholar]
- 9.Sowokinos J R. Plant Physiol. 1981;68:924–929. doi: 10.1104/pp.68.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura M, Beevers H. Plant Physiol. 1978;62:44–48. doi: 10.1104/pp.62.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon J C, Pien F-M, Liu K-C. Plant Physiol. 1996;110:835–843. doi: 10.1104/pp.110.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klotz I M. Energy Changes in Biochemical Reactions. New York: Academic; 1967. [Google Scholar]
- 13.Hill L M, Smith A M. Planta. 1991;185:91–96. doi: 10.1007/BF00194519. [DOI] [PubMed] [Google Scholar]
- 14.Smith A M, Bettey M, Bedford I D. Plant Physiol. 1989;89:1279–1284. doi: 10.1104/pp.89.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin T-P, Caspar T, Somerville C R, Preiss J. Plant Physiol. 1988;86:1131–1135. doi: 10.1104/pp.86.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schliselfeld L H, van Eys J, Touster O. J Biol Chem. 1965;240:811–817. [PubMed] [Google Scholar]
- 17.Gasmi L, Cartwright J L, McLennan A G. Biochem J. 1999;344:331–337. [PMC free article] [PubMed] [Google Scholar]
- 18.Melo A, Glaser L. Biochem Biophys Res Commun. 1966;22:524–531. doi: 10.1016/0006-291x(66)90306-8. [DOI] [PubMed] [Google Scholar]
- 19.Sheikh S, O'Handley S F, Dunn C A, Bessaman M. J Biol Chem. 1998;273:20924–20928. doi: 10.1074/jbc.273.33.20924. [DOI] [PubMed] [Google Scholar]
- 20.Cabib E, Carminatti H. J Biol Chem. 1961;236:883–887. [PubMed] [Google Scholar]
- 21.Bessman M J, Frick D N, O'Handley S F. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 22.Gangwani L, Khurana J P, Maheshwari S C. Phytochemistry. 1994;35:857–861. [Google Scholar]
- 23.Harvey C L, Olson K C, Wright R. Biochemistry. 1971;9:921–925. doi: 10.1021/bi00806a030. [DOI] [PubMed] [Google Scholar]
- 24.Lerch B, Wolf G. Biochim Biophys Acta. 1972;258:206–218. doi: 10.1016/0005-2744(72)90979-5. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Yamamoto T, Minamiura N. J Biochem. 1987;102:359–367. doi: 10.1093/oxfordjournals.jbchem.a122062. [DOI] [PubMed] [Google Scholar]
- 26.Culver G M, Consaul S A, Tycowski K T, Filipowicz W, Phizicky E M. J Biol Chem. 1994;269:24928–24934. [PubMed] [Google Scholar]
- 27.Preiss J, Shen L, Greenberg E, Gentner N. Biochemistry. 1966;5:1833–1845. doi: 10.1021/bi00870a008. [DOI] [PubMed] [Google Scholar]
- 28.Shinshi H, Miwa M, Kato K, Noguchi M, Matsushima T, Sugimura T. Biochemistry. 1976;15:2185–2190. doi: 10.1021/bi00655a024. [DOI] [PubMed] [Google Scholar]
- 29.Akazawa T. Plant Mol Biol Rep. 1991;9:145–155. [Google Scholar]
- 30.ap Rees T. In: The Biochemistry of Plants. Stumpf P K, Conn E E, editors. Vol. 3. New York: Academic; 1980. pp. 1–42. [Google Scholar]
- 31.Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith A M. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geigenberger P, Stitt M. Planta. 1991;185:81–90. doi: 10.1007/BF00194518. [DOI] [PubMed] [Google Scholar]
- 33.Preiss J. Oxford Surv Plant Mol Cell Biol. 1991;7:59–114. [Google Scholar]
- 34.Randall D D, Tolbert N E. Plant Physiol. 1971;48:488–492. doi: 10.1104/pp.48.4.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pozueta-Romero J, Akazawa T. J Exp Bot. 1993;44,Suppl.:297–306. [Google Scholar]
- 36.Werdan K, Heldt H W, Milovancev M. Biochim Biophys Acta. 1975;396:276–292. doi: 10.1016/0005-2728(75)90041-9. [DOI] [PubMed] [Google Scholar]
- 37.Sowokinos J R. Plant Physiol. 1976;57:63–68. doi: 10.1104/pp.57.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prioul J-L, Jeannette E, Reyss A, Grégory N, Giroux M, Hannah L C, Causse M. Plant Physiol. 1994;104:179–187. doi: 10.1104/pp.104.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaffer A A, Petreikov M. Plant Physiol. 1997;113:739–746. doi: 10.1104/pp.113.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ap Rees T, Hill S A. Plant Cell Environ. 1994;17:587–589. [Google Scholar]
- 41.Stitt M, Sonnewald U. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:341–368. [Google Scholar]
- 42.Murata T, Sugiyama T, Minamikawa T, Akazawa T. Arch Biochem Biophys. 1966;113:34–44. doi: 10.1016/0003-9861(66)90153-6. [DOI] [PubMed] [Google Scholar]
- 43.Geigenberger P, Geiger M, Stitt M. Plant Physiol. 1998;117:1307–1316. doi: 10.1104/pp.117.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geigenberger P, Müller-Röber B, Stitt M. Planta. 1999;209:338–345. doi: 10.1007/s004250050641. [DOI] [PubMed] [Google Scholar]