Abstract
Botulinum toxins (BoNT) are zinc proteases (serotypes A-G) which cause flaccid paralysis through the cleavage of SNARE proteins within motor neurons. BoNT/A was originally organized into two subtypes: BoNT/A1 and BoNT/A2, which are ~ 95 % homologous and possess similar catalytic activities. Subsequently, two additional subtypes were identified; BoNT/A3 (Loch Maree), and BoNT/A4 (657Ba), which have 81 and 88% homology with BoNT/A1, respectively. Alignment studies predicted that BoNT/A3 and BoNT/A4 were sufficiently different to BoNT/A1 to affect SNAP25 binding and cleavage. Recombinant Light Chain (LC) of BoNT/A3 (LC/A3) and BoNT/A4 (LC/A4) were subjected to biochemical analysis. LC/A3 cleaved SNAP25 at 50% the rate of LC/A1, but cleaved SNAPtide® at a faster rate than LC/A1, while LC/A4 cleaved SNAP25 and SNAPtide® at slower rates than LC/A1. LC/A3 and LC/A4 had similar Kms for SNAP25 relative to LC/A1, while the kcat for LC/A4 was 10- fold slower than LC/A1, suggesting a defect in substrate cleavage. Neither LC/A3 nor LC/A4 possessed autocatalytic activity, a property of LC/A1 and LC/A2. Thus, the four subtypes of BoNT/A bind SNAP25 with similar affinity but have different catalytic capacities for SNAP25 cleavage, SNAPtide® cleavage, and autocatalysis. The catalytic properties identified among the subtypes of LC/A may influence strategies for the development of small molecule- or peptide- inhibitors as therapies against botulism.
Keywords: Botulinum neurotoxin serotype A, Zinc metalloprotease, autocatalysis, SNAPtide®, SNAP25, SNARE proteins, botulism
The Clostridium botulinum neurotoxins (BoNT) are the most toxic proteins for humans and have been characterized as Category A agents by the Center for Disease Control and Prevention. BoNTs block neurotransmitter signal transduction in motor neurons, causing flaccid paralysis (1). This inhibition occurs through the cleaving of SNARE proteins that function in synaptic vesicle exocytosis (1, 2). Conversely, BoNTs are among the most useful reagents to treat neuromuscular afflictions (3).
Clostridia produce BoNTs as single-chain, 150-kDa proteins, which are cleaved into di-chain proteins that are linked by a disulfide bond with AB structure function properties (4). The N-terminal A domain (Light chain, LC) is an ~50 kDa zinc metalloprotease with the characteristic thermolysin-family zinc coordination motif (HExxH) (1). The C-terminal B domain (Heavy Chain, HC) is ~100 kDa and is composed of two functional domains that are involved in receptor recognition (HCR) and translocation of the LC across the endosomal membrane (HCT) (4). Antibody neutralization differentiates BoNTs into seven serotypes, A–G. BoNTs share ~65% primary amino acid similarity and ~35% amino acid identity with Tetanus toxin (5), and cleave different SNARE proteins: SNAP25 for A, E, and C; Syntaxin 1a for C; and VAMP-2 for B, D, F, G, and Tetanus toxin (2, 6, 7). Cleavage sites within each SNARE substrate differ among the various neurotoxins, suggesting specific residues on the LC are necessary for recognition of substrate. Recent studies have identified residues necessary for substrate recognition by LC/A, LC/B, LC/D, LC/F, and tetanus toxin (8–17), mechanistically supporting a crystal structure of a non-catalytic LC/A bound to SNAP25 through exosite and active site interactions (11, 12, 17). BoNT serotypes also comprise subtypes that can vary between 3 and 32% at the primary amino acid level (18). BoNT/A1 and BoNT/A2 show similar cleavage of SNAP25 and possess ~ 95% primary amino acid homology (19). Recent DNA sequencing analyses revealed two additional subtypes BoNT/A: BoNT/A3 and BoNT/A4 (19, 20). The bacterium producing BoNT/A3 was isolated from a botulism outbreak in Scotland in 1922, while the bacterium producing BoNT/A4 was isolated from a case of infant botulism in 1988 (21, 22). LC/A3 shows 81% identity to LC/A1, while LC/A4 shows 88% identity to LC/A1 (19). The HCs of the subtypes of BoNT/A are ~87% identical and polyclonal antisera to BoNT/A1 neutralizes BoNT/A2 (18, 23), but there are sufficient amino acid differences where neutralizing monoclonal antibodies against BoNT/A1 are not effective in neutralizing BoNT/A2 (18). Previous modeling studies suggested that LC/A3 and LC/A4 may possess different catalytic activities for SNAP25 relative to LC/A1 (19). This study reports the catalytic properties of LC/A3 and LC/A4.
Experimental Procedures
Plasmid Construction and protein expression
Plasmids containing full-length (1–448) Light Chain (LC) for LC/A1, LC/A2, LC/A3 and LC/A4 were obtained from Eric Johnson (Madison, WI). Primers were designed for PCR-amplification of DNA encoding amino acids 1–425 of LC/A2, LC/A3, and LC/A4, which has proved to be an optimal form of LC/A1 for structure-function studies (24). Amplified products were sub-cloned into pET-15b, to generate His6-LC/A(1–425). A point mutation was engineered into LC/A4 at residue 264 replacing Iso with Arg (I264R), using the Quikchange ™ reaction (Stratagene) as described by the manufacturer. The mutated plasmid was transformed into E. coli TG-1 and sequenced for mutation confirmation.
pET-15b containing LC/A1, LC/A2, LC/A3, LC/A4, or LC/A4-I264R was transformed into E. coli BL-21 RIL (DE3) (Stratagene). LC/A1, LC/A2, and LC/A3 were purified as previously described (24). Briefly, bacterial lawns were grown overnight on LB-agar with 100 μg/ml of ampicillin and 50 μg/ml of chloramphenicol and then inoculated into 400 ml cultures of LB (x6, 400 ml cultures). Cultures were incubated at 30°C for 2 hr at 250 rpm, induced with 0.25 mM IPTG and incubated at 16°C overnight at 250 rpm. Cultures were centrifuged at 5,000 ×g for 10 min, the pellet was suspended in binding buffer and bacteria were lysed using a French Press (two times). The lysate was centrifuged at 27,000 ×g for 20 min at 4°C (Percent soluble LC determined at this point), and purified as His6-fusion protein on a Ni2+-agarose (NiNTA) column (Qiagen), followed by size exclusion chromatography (Sephacryl S200 HR; 150 ml column equilibrated in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl). Peak fractions were subjected to anion exchange chromatography (DEAE–Sephacel; 5 ml column, equilibrated 10 ml in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl and eluted with a gradient (20 mM to 750 mM NaCl) (yield of LC determined). LC/A4 and LC/A4-I264R were purified as described above without utilizing the anion exchange chromatography (yield of LC determined). After dialysis, glycerol was added to a final concentration of 45% (vol/vol), and proteins were stored at −20°C.
Protease Activity Assays
Reactions were run in 20 μl volumes. Substrate Specificity, 0.2 μM LC/A was incubated with 3 μM of SNAP25(141–206)-HA or SNAP25(141–206)-R198A-HA in 10 mM Tris (pH 7.6) and 20 mM NaCl for 1 hr at 37°C. The reaction was stopped with an equal volume of protein loading buffer and the reaction product was separated from the substrate by 12 % SDS-PAGE and detected by Coomassie staining. Linear Velocity, Serially diluted LCs (2 μM–5nM) were incubated with 6 μM SNAP25(141–206) in 10 mM Tris (pH 7.6) and 20 mM NaCl at 37°C for 15 min. The reaction was analyzed as described above and quantified by densitometry (AlphaEaseFC, Alpha Innotech). Kinetics Constant, Kinetic constants for LC/As were determined in a reaction with increasing concentrations (0.015–120 μM) of SNAP25(141–206)-HA incubated for 15 min at 37°C. Reactions were run in accordance with Michaelis-Menten reaction kinetics. Kinetic parameters were determined utilizing non-linear regression equations using GraphPad Prism® 5.00 (GraphPad Software, Inc.) Reactions were run three or more times and averaged for statistical error. Auto-cleavage of LC/A, the subtypes of LC/A (120 μM) were incubated in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl for timed intervals at 37°C. The reaction was subjected to SDS-PAGE and cleavage products detected by Coomassie staining. SNAPtide® cleavage assay, LC/A was incubated with 5 μM SNAPtide® (List Biologics, CA) in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl (total reaction 100 μL). Reactions were run in 96-well, clear-bottom plates at 37°C in a Victor 3V© fluorescence plate reader (Perkin Elmer) with excitation at 488 nm and emission read at 523 nm at 2.5 min intervals. Fluorescent signals were subtracted from background fluorescence, and then plotted as fluorescent units (arbitrary units) vs. time for LC/A and recorded as relative fluorescent units/min/nmole LC.
Circular Dichroism (CD) Spectroscopy
LC/A subtypes were dialyzed into 10 mM NaPO4, 20 mM NaCl (pH 7.6), and subjected to CD analysis (10 mm cuvette, 250 nm-190 nm at a 0.5 nm data pitch, Jasco™ Circular Dichroism Spectrometer). Samples were scanned at least 10 times to obtain an average and data were plotted using GraphPad® 5.00.
Trypsin Digestion Assay
LC/A subtypes (amount or concentration) were incubated in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl (volume) for the indicated times(0–20 min) at 37°C alone or with 20 nM porcine Trypsin (Sigma-Aldrich). The reaction was stopped with SDS-PAGE sample buffer and subjected to SDS-PAGE. Cleavage products were detected by Coomassie staining.
Results
Production of LC/A3 and LC/A4 in E. coli
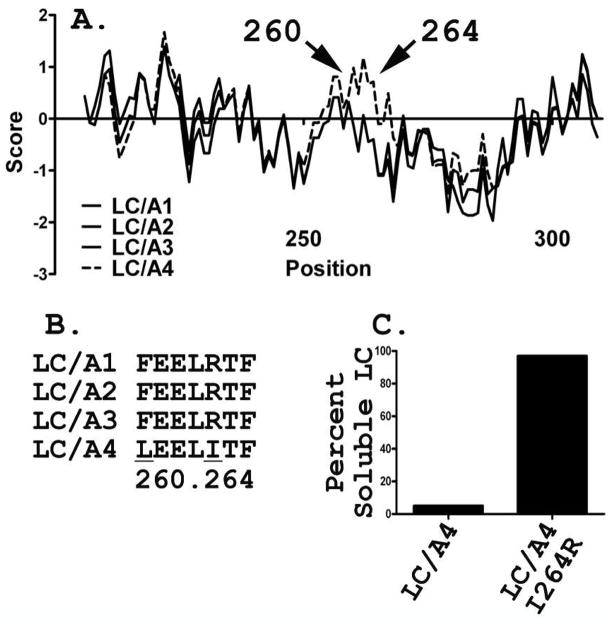
DNA encoding LC/A3(1–425) and LC/A4(1–425) were sub-cloned into pET-15b and expressed His6-fusion proteins, which migrated by SDS-PAGE as ~50kDa proteins. Approximately 85 % of LC/A3 was produced as a soluble protein, while <10% of LC/A4 was soluble. The amount of culture used for the LC/A4 purification was increased to obtain sufficient soluble LC/A4 for characterization. Purification of LC/A3 yielded ~1.5 mg and 79% purity, while LC/A4 yielded ~0.5 mg from the soluble fraction and 78% purity (Figure 1). The insolubility of LC/A4 was unexpected, since LC/A4 shared ~88% primary amino acid homology with LC/A1 (19). A Kyte-Doolittle plot of the primary amino acid sequences predicted 2 differences in hydrophobicity between of LC/A1 and LC/A4, which lay within the “250 loop”, where LC/A1 has an Leu at residue 260 and an Arg at residue 264 while LC/A4 has a Phe and Iso, respectively (Kyte and Doolittle Mean Hydrophobicity profile, Bioedit ©v. 7.0.4.1) (Figure 2A/B). A point mutation changing Leu 260 to Phe (LC/A4-L260F) did not change solubility (data not shown), while the Iso 264 to Arg (LC/A4-I264R) mutation was more soluble than LC/A4, approaching the solubility of LC/A1, when expressed in E. coli (Figure 2C).
Figure 1. Purification of LC/As.
LC/A subtype recombinant, purified protein (3μg) were subjected to 12% SDS-PAGE gel and visualized with Coomassie Brilliant Blue (shown).
Figure 2. Solubility of LC/A4.
(A) Kyte and Doolittle mean hydrophobicity profile of LC/A1-4. Arrow indicates residues substitutions causing a calculated increase in hydrophobicity locally: LC/A1, LC/A2, LC/A3 (—), and LC/A4 (---). Alignment and hydrophobicity plot calculated using BioEdit. LC/A1-A3 were plotted as solid lines due to their overlapping sequence. (B) LC/A subtype alignment of residues within solubility pocket of LC/A4 (260, 264). Underlined lettering indicates residue substitution. (C) Quantification of LC/A4 and LC/A4-I264R solubility upon induction, lysis, and centrifugation as measured by densitometry of a Coomassie-stained, 12% SDS-PAGE gel.
SNAP25 Cleavage by LC/A3 and LC/A4
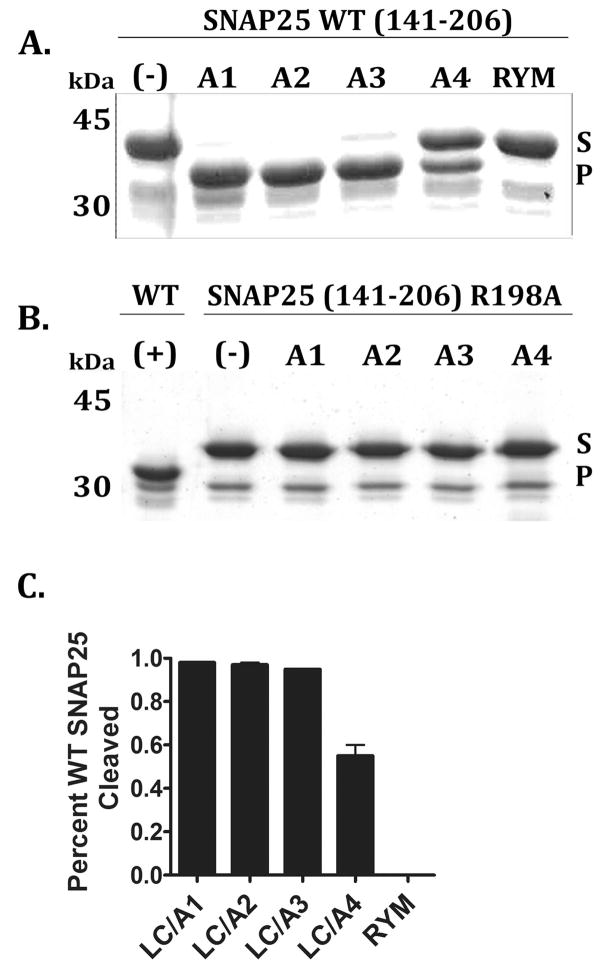
The four LC/A subtypes cleaved SNAP25 with LC/A4 less efficient than the other LC/A subtypes (Figure 3A/C). Neither LC/A3 nor LC/A4 cleaved SNAP25-R198A, a P1′ mutated form of SNAP25 that abrogates cleavage by LC/A1 (Figure 3B). This indicated that LC/A3 and LC/A4 possessed the same cleavage site specificity for SNAP25 as LC/A1. In a linear velocity assay (Table 1), LC/A2 had the same activity as LC/A1, while LC/A3 was ~2-fold less active than LC/A1 and LC/A4 was ~23-fold less active than LC/A1. LC/A4-I264R was ~ 10-fold more active than LC/A4, approaching the activity of LC/A3. Addition of 50 μM zinc did not increase activity of LC/A3 or LC/A4 for SNAP25 cleavage, indicating that the lower catalytic rate was not due to non-occupancy of zinc in the catalytic site (data not shown).
Figure 3. Cleavage site identification of LC/A subtypes.
GST-SNAP25 (141–206) (A) or GST-SNAP25(141-206-R198A) (B) (5 μM) was incubated with 0.2 μM of the indicated LC subtype (A1-A4) and a non-catalytic LC control (RYM, LC/A1-R363A, Y366F) for 1 hr at 37°C. Reaction were subjected to SDS-PAGE (shown) and the amount of cleaved GST-SNAP25(141–206) determined by densitometry of a Coomassie stained gel (C). Error bars represent the average of at least 3 independent experiments. In A and B, the SNAP25 (substrate) and cleaved product (product) are indicated with arrows, while “S” describes substrate and “P” describes the product band.
Table 1.
Linear velocity and SNAPtide® cleavage by LC/A derivatives
| LC/A | Linear Velocitya(Relative Velocity, 1=LC/A1) | SNAPtideb(Relative Velocity, 1=LC/A1) |
|---|---|---|
| LC/A1 | 1 | 1 |
| LC/A2 | 0.92 | 18 |
| LC/A3 | 0.52 | 49 |
| LC/A4 | 0.04 | 0.01 |
| LC/A4-I264R | 0.38 | 2.9 |
Linear Velocity assay. Two-fold dilutions of LC/A subtypes incubated with GST-SNAP25 as described in Methods. Percent cleavage was determined by densitometry. Comparative activity was determined by concentration of LC necessary for 20% substrate cleavage and expressed as a ratio of activity relative to LC/A1. Values are an average of at least 3 individual experiments with an error of +/− 4%.
LC/A subtype cleavage of a fluorescence energy transfer (FRET) synthetic peptide substrate, SNAPtide® (List Biologics, CA). Fluorescent signals were subtracted from background fluorescence, and then plotted as fluorescent units (arbitrary units) vs. time for LC/A subtypes. LC/A1 was determined to have relative initial velocity (fluorescence units (106)/min/nmoles LC). Values demonstrate fold change from LC/A1 initial velocity (LC/A1 initial velocity=1). Values were determined from a single representative experiment performed 3 independent times.
Experiments were also performed to determine the relative activity of native BoNT/A4. Although limited amount of BoNT/A4 in the culture supernatant of the dual toxin producing Clostridium strain 657 Ba has prevented purification of BoNT/A4 from BoNT/B, Western blotting with α-LC/A IgG determined that partially purified Ba toxin preparations contained 0.1 pmol of BoNT/A4/μl. In an overnight assay, 0.01 pmol of LC/A1 cleaved 25% of the available SNAP25, while 0.1 pmol of BoNT/A4 did not yield detectable cleavage of SNAP25 (< 2% cleavage, the limit of resolution) (data not shown). Controls showed that BoNT/B in the partially purified toxin preparation cleaved VAMP under standard assay conditions (25), indicating that the partial purification did not intrinsically inactivate the toxins. This is consistent with the native BoNT/A4 possessing a limited capacity to cleave SNAP25 relative to the A1 serotype.
Secondary Structure Analysis of LC/A4 and LC/A4-I264R
Improper protein folding could be responsible for the lower catalytic activity of LC/A4, which was addressed by measuring the CD spectra of LC/A4 and LC/A4-I264R relative to LC/A1. The spectra showed similar beta-strand content for the three proteins, while the alpha-helical content of LC/A4 was intermediate to LC/A4-I264R and LC/A1. The data indicated that the majority of LC/A4 was folded relative to LC/A4-I264R and LC/A1 and the difference in secondary structure content could not explain the low LC/A4 catalytic activity (Figure 4). Analysis of trypsin susceptibility showed that LC/A4, LC/A4-I264R, and LC/A1 had similar tryptic digestions patterns, also indicating that the overall structure of LC/A4 was similar to the catalytically active LCs (data not shown). This suggested that improper protein folding was not responsible for the observed differences in catalytic activity, and that more subtle changes in the active site of LC/A4 were responsible for the lower observed activity. This also indicated that the increased activity of the I264R mutation within LC/A4 had a more direct effect on catalysis than could be explained by global folding effects.
Figure 4. Structural folding of LC/A4.
Circular dichroism (CD) spectroscopy of LC/A4 derivatives. LC/A1 (Unbroken line), LC/A4 (Dashed line), and LC/A4-I264R (Dotted line) were examined by CD at equimolar concentrations. Data represents an average of at least 10 independent scans
Kinetic properties of the LC for SNAP25 cleavage
Kinetic constants of the LCs for SNAP25 cleavage were determined under Michaelis-Menten kinetic parameters. LC/A3 had a similar kcat and Km for SNAP25 relative to LC/A1, while LC/A4 had a 110-fold lower kcat and a two-fold higher Km for SNAP25 than LC/A1 (Table 2). Thus, LC/A3 and LC/A4 showed similar affinities for SNAP25, but LC/A4 was less efficient in the cleavage of SNAP25 relative to LC/A1. The I264R mutation in LC/A4 did not affect the Km for SNAP25 (difference < 2-fold), but increased in kcat to within 2-fold of LC/A1. This indicated that the I264R mutation influenced substrate cleavage to approach the kinetic parameters of LC/A1.
Table 2.
Kinetics of SNAP25 cleavage by LC/A deriviativesa
| Enzyme | Km (μM) | kcat (nmoles/min/nmoles LC) | kcat/Km |
|---|---|---|---|
| LC/A1 | 25 +/−7.5 | 10.7 +/− 1.4 | 0.44 |
| LC/A3 | 26 +/−6.8 | 9.3 +/− 1.0 | 0.34 |
| LC/A4 | 37 +/−9.4 | 0.13 +/− 0.01 | 0.003 |
| LC/A4-I264R | 46 +/−9.6 | 5.2 +/− 0.5 | 0.11 |
LC/A1, LC/A3, LC/A4 and LC/A4-I264R were incubated with SNAP25 (141–206) (1.25 μM–80 μM) for 10 min at 37°C. Reactions were then examined on SDS-PAGE, Coomassie-stained gels for cleavage, and quantified by densitometry. Kinetic parameters were determined utilizing non-linear regression equations on the GraphPad Prism® 5.00 (GraphPad Software, Inc). Values were determined where substrate cleavage was <10% and are the average of 3 independent experiments.
SNAPtide® cleavage by BoNT-LCs
The commercial BoNT peptide cleavage detection assay, SNAPtide®, was used as a second indication of the catalytic properties of the LC/As. SNAPtide® is a fluorescent energy resonance transfer (FRET) synthetic 12 amino acid peptide, comprised of SNAP25-residues that are adjacent to the P1′ cleavage site residue (R198) (26). LC/A2 was 18-fold more efficient in the cleavage of SNAPtide® than LC/A1 (Table1), while LC/A3 was ~50-fold more efficient than LC/A1. This suggests that LC/A2 and LC/A3 utilize different residues in the active site to recognize SNAPtide® relative to LC/A1. LC/A4 was 100-fold less efficient than LC/A1, while LC/A4-I264R had a similar efficiency for the cleavage of SNAPtide® relative to LC/A1. This indicated that the I264R mutation with LC/A4 influenced the active site conformation of LC/A4 making it a more efficient LC/A for SNAPtide® cleavage.
Auto-catalysis of LC/A subtypes
To further characterize the LC/A subtypes, LC/A3 and LC/A4 were examined for auto-catalysis using a procedure previously described (27). LC/A1 and LC/A2 stimulated ~35% and ~27% auto-cleavage by 6 hr respectively (Figure 5), while LC/A3, LC/A4, and LC/A4-I264R did not demonstrate auto-cleavage, even after 16 hr incubation (data not shown). To determine the effect of pH and salt concentrations, autocatalysis assays were performed at higher pH (8.6), lower pH (5.5), and higher salt (250 mM NaCl). In 12 h incubations, LC/A1 showed autocatalysis under each modified condition, while LC/A3 and LC/A4-I264R did not demonstrate autocatalysis (data not shown). Each LC/A subtype possessed the di-tyrosine auto-cleavage site (19), which indicated that residues in addition to the cleavage site were necessary for auto-cleavage. Control LC (LC/A1 DYM) contained mutations to cleavage site residues (Y250A, Y251A), which abrogated auto-catalysis in LC/A1 (Figure 5).
Figure 5. Auto-cleavage of LC/A subtypes.
LC/A1-A4 or LC/A1-Y250A, Y251A (LC/A1 DYM) (6 μM) were incubated at 37°C for 6 h in 10 mM Tris (pH 7.6), 20 mM NaCl. Reactions were subjected to SDS-PAGE and auto-cleavage was observed following staining with Coomassie Brilliant Blue. Asterisk (*) designates purification background that remains constant from 0–16 hr incubation.
Discussion
Each of the 7 serotypes of the botulinum neurotoxin contain subtypes that vary in primary amino acid identity by ~3–32% (18). Previous genetic analysis identified two new subtypes of BoNT/A (BoNT/A3,/A4) and demonstrated stronger homology between BoNT/A2 and/A3, than BoNT/A1, while BoNT/A4 shared a stronger homology to BoNT/A1 than BoNT/A2 and BoNT/A3 (20). These subtypes, while identified by sequence analysis, have had limited characterization for catalysis, neurotoxicity, or antibody neutralization. Single changes in residues involved in substrate recognition and catalysis may circumvent the development of inhibitor strategies (14, 18, 28), and must be taken into consideration for inhibitor development. Antibody recognition, utilized for intoxication prophylaxis and vaccination, varies between the highly conserved BoNT/A subtypes/A1 and A2 (18, 29). The goal of the current study was to determine the catalytic properties of the LC/A for subtypes A3-A4 to provide a greater understanding of substrate recognition and cleavage.
Earlier studies showed that LC/A1 use a pocket model for SNAP25 recognition. Breidenbach and Brunger (17), utilizing a crystal structure of a non-catalytic form of LC/A1 bound with SNAP25 peptide to develop a substrate recognition model for BoNT/A, proposing an a-exosite upstream of the cleavage site that affects Km, and a β-exosite downstream of the cleavage site, which primarily affects kcat. Analysis of this structure suggests a number of residues on the LC in potential interaction with SNAP25 peptide. Subsequent studies (12) utilized an alanine-scan mutation library of SNAP25 to determine kinetically-important residues on SNAP25 and point mutations on the LC to define pockets of residues on the LC necessary for interaction with SNAP25. Thus, the substrate recognition model has been refined to identify interaction sites on SNAP25 for binding (B) upstream of the active site (156–183), and an active site (AS) domain (187–203). The binding region for SNAP25 on LC/A1 is a band of residues wrapping around LC/A1, where residue interactions in SNAP25 with LC/A1 has been demonstrated along the a-helix as well as the unstructured loop of SNAP25. The AS of BoNT/A1 contained clusters of residues defined by the co-crystal of LC/A1 and SNAP25 by multiple groups to interact with SNAP25 (17, 30). Chen et al developed a “pocket” model for interactions between the LC/A1 and SNAP25 within the AS (12). The most important pocket described was the S1′ pocket, consisting of four residues (Phe163, Phe194, Thr220, Asp370). Asp370 has been shown, through an LC-inhibitor crystal, to be involved in stabilization of the catalytic intermediate of SNAP25 hydrolysis. Kinetic characterization by our laboratory (data not shown) found that LC/A2 contains identical catalytic activity and undergoes autocatalysis (31). In contrast, BoNT/A1 and BoNT/A2 demonstrate differences with neutralizing monoclonal antibody affinities (29), and an increase in SNAPtide® activity compared to LC/A1 (Table 1). This enzymatic difference is surprising, given LC/A1 and LC/A2 high residue identity (>95%), especially within known binding and catalysis residues.
LC/A3 contains the least residue identity to LC/A1 within the a-exosite, but is identical in the AS residues identified by Breidnebach et al (17) and Chen et al (12). Alternatively, LC/A4 contains similar residues to LC/A1 in the a-exosite binding site, with two exceptions; but contains three conserved, though sterically smaller changes at the S1′ site. Alignments of these primary protein sequences, taken with known structural and kinetic data, initially predicted a change in Km for LC/A3, and a change in kcat for LC/A4 (19). Kinetic data on LC/A3 did not exhibit a change in Km compared to LC/A1, demonstrating that the non-conserved residue changes within the binding exosite had little effect on SNAP25 recognition. The similarity of Km for SNAP25 by LC/A1 and LC/A3 suggests plasticity within the binding exosite for SNAP25, perhaps different interacting residues between the LC/A1 and LC/A3 for SNAP25. LC/A3 demonstrated a slightly lower activity in the linear velocity and catalytic efficiency (kcat/Km) for SNAP25. Unexpectedly, both LC/A2 and LC/A3 cleaved the synthetic substrate, SNAPtide®, more efficiently than LC/A1, demonstrating in congruency between WT SNAP25 and SNAPtide® cleavage among the LCs. The LC/A3 active site is identical in all residues recognized previously to be necessary cleavage of WT SNAP25. The unexpected difference between SNAP25 and SNAPtide® suggest the LCs recognize the two substrates differently. The binding exosite of SNAP25, which is not present in SNAPtide®, is identical between LC/A2 and LC/A1, suggesting that SNAPtide® activity differences of LC/A2 and LC/A3 compared to LC/A1 are not due to loss of the binding site. One explanation may be that in the SNAPtide® the P4′ residue, Lys201, is linked by the ε-amine of the R group to DABCYL, which may block interaction with the S4′ binding pocket. The S4′ pocket consists of a single residue (E257), shown to contribute to substrate recognition and catalysis in LC/A1 (12). Conversely, this also suggests that since there is a shared increase in activity between LC/A2 and LC/A3 compared to LC/A1 in SNAPtide® activity with the absence of S4′ pocket activation, residues that do not participate in the cleavage reaction of SNAP25 may now contribute to SNAPtide® recognition and catalysis for LC/A2 and LC/A3. Thus substrate recognition of SNAP25 and SNAPtide® may differ for specific LC subtypes. In contrast, SNAP25 recognition is the same for LC/A1, LC/A2, and LC/A3 which suggests that upon activation of the AS by the P4′-S4′ interaction, the LC ASs are comparable. Another possibility is that since LC/A3 is identical to LC/A1 within the catalytic site, regions outside the active site residues may change the fit of SNAP25 into the catalytic site, which could account for the observed differences in cleavage rate of the SNAPtide® by the different subtypes.
LC/A4 showed reduced cleavage for SNAPtide® and SNAP25. This was proposed to be due to conserved residue changes causing steric interaction differences within the putative S1′ pocket (12, 19). Unexpectedly, LC/A4 was inherently insoluble as a recombinant protein expressed in E. coli, and this property was examined as an explanation for the decrease in catalytic activity. CD spectroscopy of LC/A4 showed a similar α-helical pattern to LC/A1 and LC/A4-I264R, demonstrating a similar secondary structure to active proteins, which suggested that the change in activity was due to a subtle structural alteration via the residue substitution. A comparative hydrophobicity plot between LC/A1-4 showed two residue changes that were predicted to increase the hydrophobicity of LC/A4 related to LC/A1-A3. Examination of the LC/A1 crystal structure revealed a salt bridge between R264 and E262, which is apparently lacking in LC/A4. E263, directly adjacent to this salt-bridge stabilization in LC/A1, is a conserved residue within the LC/A subtypes and is necessary to stabilize the zinc atom within the active site during hydrolysis of SNAP25 (13). Previously published studies demonstrated that conserved residue substitutions of LC/A1 at E262 decreased the kcat 5-fold, while retaining CD-measured secondary structure and molar ratios of zinc (32). This suggests that the observed decrease in activity is not due to the alignment of the 264 residue that is required for optimal substrate cleavage. LC/A4-I264R demonstrated increased catalytic activity that approached LC/A1. One hypothesis is that substitution of Arg264 in LC/A1 to Iso in LC/A4 destabilizes the coordination activity of E262 in relation to the zinc atom in the active site. The solubility difference between LC/A4 and LC/A4-I264R may not be due to folding differences, but differences in protein interactions that occur after the folding process in an E. coli background compared to a Clostridia sp. background. BoNT/A4 was observed to have the I264 codon in multiple, independent overlapping DNA sequences confirming the substitution (M. Jacobson, unpublished data). The specific toxicity of purified BoNT/A4 has not yet been demonstrated in vivo due to the difficulty of purifying the 150-kDa zootoxin from a dual-neurotoxin producing Clostridium sp (W. TEPP, unpublished data).
Auto-cleavage is a characteristic of LC/A1 (31), where a di-tyrosine residue on the 250 loop is cleaved by active, trans-interacting LC/A1 and LC/A2. Crystal structures of the trans-interacting LC/A1 by Breidenbach and Brunger demonstrated non-canonical substrate binding and defined interacting residues on the 250 loop of both LC that were conserved within LC/A1-A4. The inability of LC/A3 and LC/A4 to undergo auto-cleavage was not predicted, since both subtypes are highly conserved for the 250 loop and possessed the di-tyrosine site of cleavage. Recently, Ahmed and Smith described a number of mutations to LC/A1 in which autocatalysis was abrogated, where the mutated enzyme lost both the capacity of auto-cleavage and cleavage of SNAP25, coupling the two activities (10). The observed properties of LC/A3 and LC/A4 suggest that the capacity to undergo auto-cleavage and SNAP25 cleavage can also be uncoupled. Alignment of LC/A1-A4 showed that the 250 loop region was conserved and did not explain the inability to recognize this self-substrate. LC/A3 contains a substitution to the loop region (N246K), which may cause a positively-charged microenvironment and a loss of accessibility of this loop to the AS, while LC/A4 loss of auto-cleavage may be explained through substitutions within the S1′ pocket of the AS.
Determining differences in substrate recognition between subtypes of botulinum neurotoxin provides information for vaccine and therapies against botulism. LC/A3 and LC/A4 cleaved SNAP25, but possessed differences in catalytic efficiency and autocatalysis compared to LC/A1. These data, as well as recent information on interacting residues of LC/A1 with SNAP25, provide a model by which LCs of other serotypes may be compared for substrate specificity and catalysis.
Acknowledgments
Funding: This work was sponsored by the NIH/NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (RCE) Program, Region V Great Lakes’ RCE (NIH award 1-U54-AI-057153).
We thank the members of the Barbieri laboratory for helpful discussion.
Abbreviations
- BoNT
Botulinum Neurotoxin
- LC
Light Chain
- HC
Heavy Chain
- FRET
Fluorescence resonance energy transfer
- CD
Circular dichroism
References
- 1.Shiva G, Rossetti O, Santucci A, DasGupta RR, Montecucco C. Botulinum Neurotoxins are Zinc Proteases. Journal of Biological Chemistry. 1992;267:23479–23483. [PubMed] [Google Scholar]
- 2.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Sudhof TC, Jahn R, Niemann H. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J Biol Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 3.Naumann M, So Y, Argoff CE, Childers MK, Dykstra DD, Gronseth GS, Jabbari B, Kaufmann HC, Schurch B, Silberstein SD, Simpson DM. Assessment: Botulinum neurotoxin in the treatment of autonomic disorders and pain (an evidence-based review): Reports of the Therapeutics and Technology Assessment Subcommittee of the American Academy of Neurology. Neurology. 2008;70:1707–1714. doi: 10.1212/01.wnl.0000311390.87642.d8. [DOI] [PubMed] [Google Scholar]
- 4.Bandyopadhyay S, Clark AW, DasGupta BR, Sathyamoorthy V. Role of the heavy and light chains of botulinum neurotoxin in neuromuscular paralysis. J Biol Chem. 1989;262:2660–2663. [PubMed] [Google Scholar]
- 5.Lacy DB, Stevens RC. Sequence homology and structural analysis of the clostridial neurotoxins. J Mol Biol. 1999;291:1091–1104. doi: 10.1006/jmbi.1999.2945. [DOI] [PubMed] [Google Scholar]
- 6.Foran P, Lawrence GW, Shone CC, Foster KA, Dolly JO. Botulinum neurotoxin C1 cleaves both Syntaxin and SNAP-25 in intact and permeabilized Chromaffin cells: Correlation with its blockade of Catecholamine release. Biochemistry. 1996;35:2630–2636. doi: 10.1021/bi9519009. [DOI] [PubMed] [Google Scholar]
- 7.Schiavo G, Shone CC, Bennett MK, Scheller RH, Montecucco C. Botulinum neurotoxin type C cleaves a single Lys-Ala bond within the carboxyl-terminal region of syntaxins. J Biol Chem. 1995;270:10566–10570. doi: 10.1074/jbc.270.18.10566. [DOI] [PubMed] [Google Scholar]
- 8.Arndt JW, Yu W, Bi F, Stevens RC. Crystal structure of botulinum neurotoxin type G light chain: serotype divergence in substrate recognition. Biochemistry. 2005;44:9574–9580. doi: 10.1021/bi0505924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Hall C, Barbieri JT. Substrate recognition of VAMP-2 by botulinum neurotoxin B and tetanus neurotoxin. J Biol Chem. 2008;283:21153–21159. doi: 10.1074/jbc.M800611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed SAOM, Ludivico ML, Gilsdorf J, Smith LA. Identification of Residues Surrounding the Active Site of Type A Botulinum Neurotoxin Important for Substrate Recognition and Catalytic Activity. Protein J. 2008;27:151–162. doi: 10.1007/s10930-007-9118-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Barbieri JT. Unique substrate recognition by botulinum neurotoxins serotypes A and E. Journal of Biological Chemistry. 2006;281:10906–10911. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Kim JP, Barbieri JT. Mechanism of Substrate Recognition by Botulinum Neurotoxin Serotype A. J Biol Chem. 2007;282:9621–9627. doi: 10.1074/jbc.M611211200. [DOI] [PubMed] [Google Scholar]
- 13.Rigoni M, Caccin P, Johnson EA, Montecucco C, Rossetto O. Site-Directed Mutagenesis Identifies Active-Site Residues of the Light Chain of Botulinum Neurotoxin Type A. Biochemical and Biophysical Research Communications. 2001;288:1231–1237. doi: 10.1006/bbrc.2001.5911. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt JJ, Stafford RG. Botulinum neurotoxin serotype F: identification of substrate recognition requirements and development of inhibitors with low nanomolar affinity. Biochemistry. 2005;44:4067–4073. doi: 10.1021/bi0477642. [DOI] [PubMed] [Google Scholar]
- 15.Sikorra S, Henke T, Swaminathan S, Galli T, Binz T. Identification of the Amino Acid Residues Rendering TI-VAMP Insensitive toward Botulinum Neurotoxin B. J Mol Biol. 2006;357:574–582. doi: 10.1016/j.jmb.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 16.Arndt JW, Chai Q, Christian T, Stevens R. Structure of Botulinum Neurotoxin Type D Light Chain at 1.65 A Resolution: Repercussions for VAMP-2 Substrate Specificity. Biochemistry. 2006;45:3255–3262. doi: 10.1021/bi052518r. [DOI] [PubMed] [Google Scholar]
- 17.Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 18.Smith TJ, Lou J, Geren IN, Forsyth CM, Tsai R, LaPorte SL, Tepp WH, Bradshaw M, Johnson EA, Smith LA, Marks JD. Sequence Variation within Botulinum Neurotoxin Serotypes Impacts Antibody Binding and Neutralization. Infect Immun. 2005;73:5450–5457. doi: 10.1128/IAI.73.9.5450-5457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arndt JW, Jacobson MJ, Abola EE, Forsyth CM, Tepp WH, Marks JD, Johnson EA, Stevens RC. A structural perspective of the sequence variability within botulinum neurotoxin subtypes A1-A4. J Mol Biol. 2006;362:733–742. doi: 10.1016/j.jmb.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 20.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. Genetic diversity among Botulinum Neurotoxin-producing clostridial strains. J Bacteriol. 2007;189:818–832. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edmond BJ, Guerra FA, Blake J, Hempler S. Case of infant botulism in Texas. Tex Med. 1977;73:85–88. [PubMed] [Google Scholar]
- 22.Leighton GR. HM Stationery Office. London: 1923. [Google Scholar]
- 23.Baldwin MR, Tepp WH, Przedpelski A, Pier CL, Bradshaw M, Johnson EA, Barbieri JT. Subunit Vaccine against the Seven Serotypes of Botulism. Infect Immun. 2008;76:1314–1318. doi: 10.1128/IAI.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldwin MR, Bradshaw M, Johnson EA, Barbieri JT. The C-terminus of botulinum neurotoxin type A light chain contributes to solubility, catalysis, and stability. Protein Expression & Purification. 2004;27:187–195. doi: 10.1016/j.pep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Fang H, Luo W, Henkel J, Barbieri J, Green N. A yeast assay probes the interaction between botulinum neurotoxin serotype B and its SNARE substrate. PNAS. 2006;103:6958–6963. doi: 10.1073/pnas.0510816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boldt GEKJP, Hixon MS, McAllister LA, Barbieri JT, Tzipori S, Janda KD. Synthesis, characterization and development of a high-throughput methodology for the discovery of botulinum neurotoxin A inhibitors. J Comb Chem. 2006;8:513–521. doi: 10.1021/cc060010h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed SABM, Jensen M, Hines HB, Brueggemann E, Smith LA. Enzymatic autocataylsis of botulinum A neurotoxin light chain. J Prot Chem. 2001;20:221–231. doi: 10.1023/a:1010952025677. [DOI] [PubMed] [Google Scholar]
- 28.Boldt GEELM, Janda KD. Identification of a botulinum neurotoxin A protease inhibitor displaying efficacy in a cellular model. Chem Commun. 2006;29:3063–3065. doi: 10.1039/b603099h. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Rodriquez CLR, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 30.Lacy DBTW, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 31.Ahmed SA, Byrne MP, Jensen M, Hines HB, Brueggemann E, Smith LA. Enzymatic autocatalysis of Botulinum A Neurotoxin Light Chain. J Prot Chem. 2005;20:221–231. doi: 10.1023/a:1010952025677. [DOI] [PubMed] [Google Scholar]
- 32.Kukreja RV, Sharma S, Cai S, Singh BR. Role of two active site Glu residues in the molecular action of botulinum neurotoxin endopeptidase. Biochim Biophys Acta. 2007;1774:213–222. doi: 10.1016/j.bbapap.2006.11.007. [DOI] [PubMed] [Google Scholar]