Summary
The vertebrate retina is a tractable system in which to address the question of neuronal cell fate specification. Specification of retinal rod photoreceptors is determined by several different transcription factors that activate expression of rod-specific genes and repress expression of cone photoreceptor-specific genes. The mechanism by which this dual regulation occurs is unclear. We have found that Pias3, a transcriptional coregulator and E3 SUMO ligase that is selectively expressed in developing photoreceptors, promotes the differentiation of rod photoreceptors while preventing rods from adopting cone photoreceptor-like characteristics. Pias3 directly interacts with the photoreceptor-specific transcription factors Crx and Nr2e3 and is specifically targeted to the promoters of photoreceptor-specific genes. Pias3 SUMOylates Nr2e3, converting it into a potent repressor of cone-specific gene expression. Rod and cone-specific promoters are bound by hyperSUMOylated proteins in rod photoreceptors, and blocking SUMOylation in photoreceptors results in cells with morphological and molecular features of cones and an absence of rod-specific markers. Our data thus identifies Pias3-mediated SUMOylation of photoreceptor-specific transcription factors as a key mechanism of rod specification.
Introduction
The vertebrate retina is a well-established system for studying neuronal specification. Rod photoreceptors are the best studied retinal cell type at the molecular level. Several different transcription factors act to drive retinal precursors to a rod photoreceptor fate. Most of these genes are also expressed in mature rods, and regulate both photoreceptor survival and terminal differentiation. Among these are the paired-type homeodomain genes Crx and Otx2, which are also expressed in cone photoreceptors, the Maf-type bZip factor Nrl, and the nuclear hormone receptor Nr2e3 (Chen et al., 1997; Furukawa et al., 1997; Haider et al., 2000; Mears et al., 2001). Several of these factors not only promote rod differentiation, but also prevent rods from acquiring characteristics of other retinal cells. Among these are Otx2, which prevents rods from adopting an amacrine fate (Nishida et al., 2003), and Nrl and Nr2e3, which repress expression of cone-specific genes in rods (Akhmedov et al., 2000; Chen et al., 2005; Mears et al., 2001; Peng et al., 2005). Mutation of Nr2e3 in human patients leads to enhanced S-cone syndrome, and a spontaneous null mutation in Nr2e3 (the rd7/rd7 mouse line) results in rod photoreceptors that ectopically express S-cone markers (Haider et al., 2000; Jacobson et al., 2004). It is poorly understood how these photoreceptor-enriched transcription factors act as both repressors and activators. In other systems, however, this dual regulation is achieved by the action of transcriptional coregulators. Most coregulators are broadly expressed, and their activity is thought to be regulated by posttranslational modification and protein-protein interaction (Lonard and O'Malley, 2006).
Previously, we showed that Pias3 mRNA was selectively expressed in developing photoreceptors within the retina (Blackshaw et al., 2004). Pias3 (Protein Inhibitor of Activated Stat3) was first identified as an inhibitor of Stat3-dependent transcription (Chung et al., 1997). Later studies showed that Pias3 can act as both a transcriptional coactivator and corepressor, and regulates the activity of many classes of transcription factors in nonneuronal cells, including Smads (Long et al., 2004), nuclear hormone receptors (Junicho et al., 2000), and Oct4 (Tolkunova et al., 2007). Pias3 is also an E3 SUMO ligase, catalyzing covalent linkage of SUMO proteins, which are distant homologues of ubiquitin, to specific lysine residues on target proteins. SUMOylation, which in many ways mechanistically resembles ubiquitination, can reversibly modify the activity of a broad range of mostly nuclear proteins (Meulmeester and Melchior, 2008). Pias3 is known to catalyze SUMOylation of a number of different transcription factors, many of which are also bound and regulated by Pias3 by SUMOylation-independent mechanisms (Kotaja et al., 2002; Schmidt and Muller, 2002). The function of Pias3 in neural development has not been explored, however. Given the broad range of known Pias3 targets and its highly cell-specific expression pattern, we investigated whether Pias3 regulates rod photoreceptor development by regulating the activity of rod-specific transcription factors.
Results
Pias3 is selectively expressed in developing rod and cone photoreceptors
To determine if Pias3 protein was co-expressed with Pias3 mRNA in developing retina, we performed section immunostaining for Pias3. We confirmed the specificity of the antibody used by staining HEK293T cells transfected with a Pias3 overexpression construct (Figure S1A). We first observe weak expression of Pias3 in the outer region of the outer neuroblastic layer (ONBL) at postnatal day 0 (P0) (Figure 1A). Immunostaining with the rod precursor-specific transcription factor Nr2e3 revealed that Pias3 colocalized with Nr2e3. Coincident with previously reported in situ hybridization data, we observe that Pias3 protein expression increases in developing rods during the postnatal interval when rods are terminally specified. Protein expression peaks at P14, and then declines steadily. By P10, we also observe a subset of cells in the outer nuclear layer (ONL) that express Pias3 protein but not Nr2e3 (Figure 1A and B, arrowheads). We performed double immunohistochemistry for TRβ2, a transcription factor expressed in cone photoreceptors (Jones et al., 2007), and observed expression of Pias3 in a subset of TRβ2-positive cells (data not shown).
Figure 1.
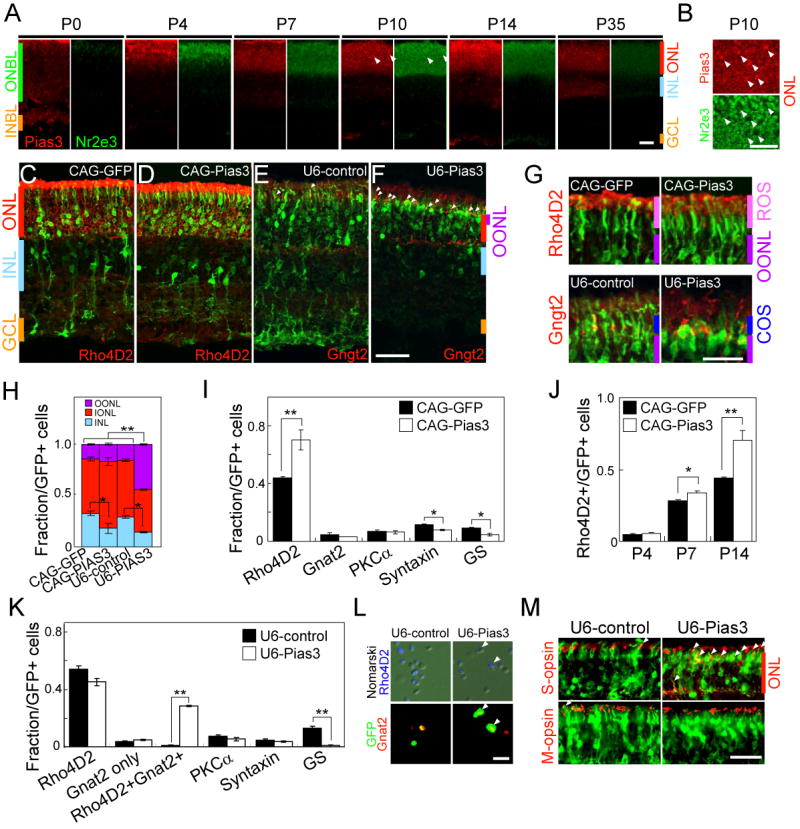
Pias3 overexpression and knockdown influence photoreceptor differentiation.
(A and B) Developmental expression of Pias3 (red) co-immunostained with Nr2e3 (green) from P0, 4, 7, 10, 14 and 35 (A), and higher magnificent image of the P10 retina focused on ONL (B). The arrowheads indicate Pias3-positive cells not co-labeled with Nr2e3.
(C-F) Section immunohistochemistry of the P14 mouse retinas electroporated in vivo at P0 with CAG-GFP (C), CAG-Pias3 (D), U6-control (E) and U6-Pias3 (F) labeled with Rho4D2 (C and D) or Gngt2 (E and F) antibodies. Arrowheads in (E) and (F) indicate GFP-positive cells labeled with Gngt2. OONL=outer region of outer nuclear layer, IONL=inner region of outer nuclear layer, INL=inner nuclear layer, GCL=ganglion cell layer.
(G) Higher magnification images of (C-F) focused on rod or cone outer segments (ROS or COS).
(H) Laminar position of electroporated cell bodies. Fraction of GFP-positive cells found in each retinal lamina is shown. OONL=outer region of outer nuclear layer, IONL=inner region of outer nuclear layer, INL=inner nuclear layer. Data are represented as mean +/- SD. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 6).
(I) Cell type composition of the P14 mouse retinas electroporated in vivo at P0 with CAG-GFP and CAG-Pias3 determined by immunostaining with retinal cell type-specific markers (Rho4D2 for rods, Gnat2 for cones, PKCα for rod bipolars, syntaxin for amacrines and GS for Muller glia). Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 3).
(J) Fraction of Rho4D2-positive cells harvested at P4, P7 and P14. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 3).
(K) Cell type composition of the P14 mouse retinas electroporated in vivo with U6-control and U6-PIAS3. The fraction of Rho4D2+ Gnat2+ indicates Rho4D2- and Gnat2-double positive cells shown in (L).
(L) Immunohistochemistry of the P14 dissociated retina eletroporated in vivo with U6-control and U6-Pias3 labeled with Rho4D2 (blue), GFP (green) and Gnat2 (red) antibodies. Arrowheads indicate triple-labeled cells.
(K) Immunostaining with M (red) or S (blue) opsin antibodies of ONL of retinas electroporated in vivo with U6-control and U6-Pias3. Arrowheads indicate electroporated cells co-labeled with the opsin antibodies.
Scale bar: A and C-F, 50μm; B, G, L and M, 20μm.
Pias3 promotes rod differentiation and represses expression of cone-specific genes
This selective expression of Pias3 in developing rods suggested that Pias3 regulate rod specification. We used in vivo electroporation of developing retina to determine the role of Pias3 in photoreceptor development (Matsuda and Cepko, 2004). After overexpressing full-length Pias3 under the ubiquitous CAG promoter (CAG-Pias3, Figures 1D and H) at P0, we observed by P14 a significant (p < 0.05) increase in the number of cells in the ONL with cell bodies in the inner outer nuclear layer (IONL) and rod-like morphology that labeled with an anti-rhodopsin antibody (Rho4D2), compared to cells electroporated with an empty vector control (CAG-GFP, Figures 1C and H).
To quantify the composition of cells electroporated with full-length Pias3, we performed dissociated cell immunocytochemistry with cell-type specific antibody markers for rods (Rho4D2), cones (cone transducin α subunit, Gnat2), rod bipolars (PKCα), amacrine cells (syntaxin) and Müller glia (glutamine synthetase, GS) (Matsuda and Cepko, 2004; Young et al., 2005). Significantly more Rho4D2-positive cells (p < 0.005) were observed following electroporation with full-length Pias3 than with empty vector (Figure 1I). Fewer syntaxin and GS-positive cells were also seen (p < 0.05) (Figure 1I). Since rhodopsin is not expressed until roughly six days after rod precursors exit mitosis (Belliveau et al., 2000), we tested if Pias3 overexpression altered the kinetics of rhodopsin expression (Figure 1J). No significant increase in the fraction of rhodopsin-positive cells was observed until P7, and we conclude that Pias3 does not induce precocious expression of rhodopsin. We observed that neither the fraction of proliferating cells (BrdU- positive cells pulsed at P0) nor of apotoptic cells (TUNEL-positive cells) changed significantly in electroporated cells following Pias3 overexpression at P7 and P10 (data not shown). These data suggest that Pias3 overexpression drives the electroporated retinal precursor cells to a rod cell fate at the expense of inner retinal cell types.
These data also suggest that Pias3 may directly enhance rhodopsin expression in transfected rod precursors after the first postnatal week. We tested the effects of Pias3 on expression of luciferase driven from a rhodopsin promoter construct in HEK293T cells. We found that Pias3 activates rhodopsin expression in a dose-dependent manner in the presence of the photoreceptor-specific transcription factors Crx, Nrl and Nr2e3 (Figure S2A). Since Nr2e3 is also known to be a strong transcriptional repressor when bound to its own consensus binding site, we tested whether Pias3 would have any effect on Nr2e3-mediated transcriptional repression. We found that Pias3 also significantly enhanced Nr2e3-depdendent repression (Figure S2B). Thus, Pias3 enhances both transcriptional activation and repression functions of Nr2e3, depending on the reporter tested and on other factors present.
We next tested the effect of reducing Pias3 expression in photoreceptor precursors. We used an shRNA expressed using the U6 promoter (U6-Pias3) that we confirmed to reduce Pias3 expression levels in retinal cells (Figure S1B and C). We observe an increase in the localization of cells electroporated with the Pias3 shRNA construct (U6-Pias3) in the ONL at P14 when compared to a negative control shRNA (U6-control) construct (Figure 1E and F). Significantly more transfected cells (p < 0.005) were found in the outer portion of the outer nuclear layer (OONL) than in controls (Figure 1H), and these cells had large cell soma and short outer segments, a pattern characteristic of cone photoreceptors. Cells in the ONL electroporated with control shRNAs rarely expressed the cone-specific markers Gngt2 (cone transducin γ subunit, Figure 1E), Gnat2 (cone transducin α subunit, Figure 1K), and S-cone opsin (Figure 1M). However, electroporation with Pias3-specific shRNA resulted in a dramatic increase (p < 0.005) in the fraction of cells expressing cone-specific markers (Figure 1F and K). Many photoreceptors electroporated with Pias3 shRNA expressed S-opsin, although the fraction expressing M-opsin was not increased (Figure 1M). We demonstrated that these shRNA effects are specific and can be rescued by shRNA-insensitive human PIAS3 (Figure S1D). We conclude that Pias3 is required to prevent developing rod photoreceptors from adopting a cone-like morphology and gene expression pattern.
When several rod-specific transcription factors including Nrl and Nr2e3 are mutated, cone-specific genes are ectopically expressed in rod photoreceptors, with S-opsin, but not M-opsin, being upregulated in these rods (Chen et al., 2005; Haider et al., 2000; Mears et al., 2001; Peng et al., 2005). In photoreceptors of mice mutant for Nr2e3, however, expression of rod-specific genes is only slightly reduced, and hybrid cells showing both rod and cone-like gene expression profiles are observed (Chen et al., 2005) (Corbo and Cepko, 2005). We next tested if cells electroporated with Pias3 shRNA expressed both rod-specific and cone-specific genes. Many rhodopsin-positive cells electroporated with Pias3 shRNA coexpressed Gnat2, though no cells expressing control shRNA did (Figure 1L). Quantifying these results (Figure 1K) revealed that over 60% of all rhodopsin-positive cells electroporated with the Pias3 shRNA expressed cone-specific markers. Though 35% of all electroporated cells expressed Gnat2, in comparison to only 4% in the control, roughly 30% of all electroporated cells expressed both Gnat2 and rhodopsin. The number of cells expressing only Gnat2 was not different between the two samples. Thus, we conclude that these cells, despite showing a cone-like morphology, are actually rods in which repression of cone-specific genes has been compromised – a phenotype like that seen with loss of Nr2e3 function.
We also measured expression of other cell-specific markers using dissociated cell immunohistochemistry in the same manner as for the overexpression studies shown in Figure 1I. We observe a slight reduction in the number of rhodopsin-positive cells following Pias3 knockdown (Figure 1K), along with a general reduction in the level of rhodopsin expression in each transfected cell (data not shown), consistent with a possible role for Pias3 in activation of rhodopsin expression (Figure S2A). We observe no changes in the fraction of PKCα or syntaxin-positive cells, but a significant decrease (p < 0.05) in the fraction of GS-positive Müller glia (Figure 1K), along with a reduction in the fraction of cells in the INL relative to controls (p < 0.05) (Figure 1H).
Pias3 interacts with Nr2e3 and Crx in vitro and in vivo
Since Pias3 is known to interact with multiple different classes of transcription factors in non-neuronal cells, we tested whether Pias3 interacted with known photoreceptor-specific transcription factors. We performed coimmunoprecipation using 35S-labeled in vitro translated proteins to determine whether Pias3 directly interacted with Crx and Nr2e3. Antibodies against both native Crx and Nr2e3 immunoprecipate radiolabeled Pias3 (Figure 2A, lanes 1-4). Antibodies to Nr2e3 were shown to immunoprecipitate radiolabeled Crx as previously reported (Peng et al., 2005), while antibodies to Crx did not precipitate Nr2e3 (Figure 2A, lanes 5 and 6). Antibodies to native Pias3 immunoprecipitated both Crx and Nr2e3 (Figure 2A, lanes 9-12).
Figure 2.
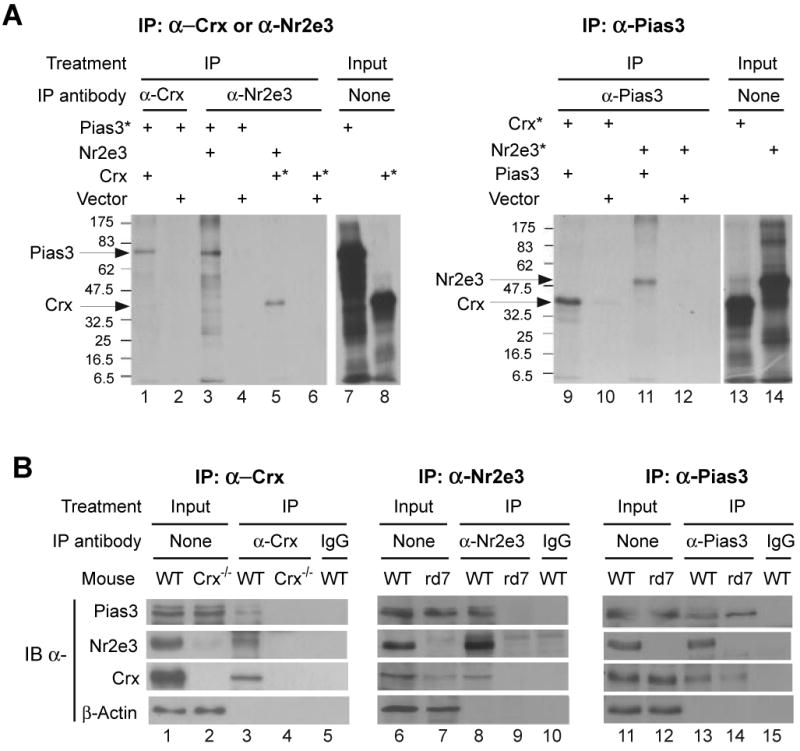
Pias3 interacts with Nr2e3 and Crx in vitro and in mouse retina.
(A) In vitro co-immunoprecipitation of Pias3 with Nr2e3 and Crx. Co-IP was performed using in vitro translated proteins with (*) or without 35S-labeling and the indicated IP antibodies (Lanes 1-6 and 9-12). Lanes 7, 8, 14 and 15 are input controls without IP.
(B) Co-IP of Pias3, Nr2e3 and Crx from postnatal day 10 retina of the indicated mouse strains. Proteins are detected via immunoblot with indicated antibodies. Nuclear extracts made from pooled retinal samples were immunoprecipitated (IP) using an antibody against one of the three proteins. Lanes 1-2, 6-7 and 11-12 are input controls without IP.
We next tested if Pias3 interacted with Nr2e3 and Crx in vivo. We performed co-IP using nuclear extracts of P10 retina from wildtype, Crx-/- and rd7/rd7 (Nr2e3 null) strains. Immunoprecipitations were performed using antibodies against one of each of these three proteins, and probed with all three antibodies. Immunoprecipitation using an anti-Crx antibody co-IPs Nr2e3, as previously reported (Chen et al., 2005; Cheng et al., 2004), as well as Pias3 from wildtype mice (Figure 2B, lane 3). Co-IP does not occur with a nonspecific IgG (Figure 2B, lane 5) or in Crx-/- mice where Crx protein is absent (Figure 2B, lane 4). Similarly, anti-Nr2e3 antibodies can immunoprecipitate Pias3 and Crx from wildtype retina (Figure 2B, lane 8) but not from rd7/rd7 mice (Figure 2B, lane 9). Co-IP did not occur when control IgG was used for wildtype retinal extracts (Figure 2B, lane 10). Finally, antibodies to native Pias3 can immunoprecipitate Crx and Nr2e3 from wildtype retina (Figure 2B, lane 13). However, no Nr2e3 protein can be immunoprecipitated with anti-Pias3 from rd7/rd7 retinas, while Crx protein can still be co-IPed (Figure 2B, lane 14). IgG controls do not precipitate any of these proteins (Figure 2B, lane 15), demonstrating the specificity of the anti-Pias3 antibody. Together, these data imply that Pias3 interacts directly with both Nr2e3 and Crx in vivo and in vitro, and that this interaction is independent of either Nr2e3 or Crx. Given that these three proteins are coexpressed in developing photoreceptors, and that Nr2e3 and Crx form a complex in vivo (Cheng et al., 2004; Peng et al., 2005), we conclude that Nr2e3 and Crx are physiological targets of Pias3.
We next tested whether the effects of Pias3 overexpression and knockdown were altered in rd7/rd7 animals. Overexpression of Pias3 in this background does not result in the increase in the cells in the inner outer nuclear layer (IONL), where the overwhelming majority of cells are rod photoreceptors (Figure S3). Significantly fewer cells with cone-like morphology and cell bodies in the outer outer nuclear layer (OONL) (p < 0.005) are seen in rd7/rd7 retinas electroporated with Pias3 shRNA compared to wildtype retinas electroporated with the same construct, though more are seen than in retinas electroporated with control shRNA. Finally, all rd7/rd7 retinas showed more electroporated GS-positive INL cells with Müller glia-like morphology than in wildtype. Neither CAG-Pias3 nor U6-Pias3 constructs repressed Müller glial development in rd7/rd7 retinas, implying a role for Nr2e3 in repression of glial development.
Pias3 is selectively present on the promoters of rod and cone-specific genes that are known targets of Crx and Nr2e3
Although the preceding data suggests that Pias3 interacts with Nr2e3 and Crx in vivo, we sought to confirm whether or not Pias3 is found together with Nr2e3 and Crx on the promoter regions of photoreceptor-specific genes. We first performed chromatin immunoprecipitation (ChIP) using antibodies to either Pias3 or Nr2e3 individually. We found that, as previously reported (Peng et al., 2005) antibodies to Nr2e3 selectively immunoprecipitate chromatin from promoters of rod and cone-specific genes such as rhodopsin, M- and S-cone opsin and rod-specific phosphodiesterase β subunit, while promoters of genes selectively expressed in non-photoreceptor cell types such as bipolar cells and retinal ganglion cells, or of genes not expressed in the retina such as albumin, are not immunoprecipitated with this antibody (Figure 3, lanes 1-3). We find that antibodies to Pias3 show the same pattern of immunoprecipitation as Nr2e3 (Figure 3, lanes 4-6). To determine whether Pias3 and Nr2e3 co-occupy the same promoters, we performed double ChIP (Figure 3, lanes 7-12) and the same pattern of immunoprecipitation was observed. Similar results were obtained using quantitative single and double ChIP for rhodopsin, M- and S-opsin and mGluR6 (Figure S4A). Promoter occupancy of each single ChIP experiment is approximately 40%, the values of which are in line with other previously reported ChIP studies (Masui et al., 2007; Miotto and Struhl, 2008). From this we conclude that Pias3 and Nr2e3 co-occupy the same sets of promoters in rod photoreceptor cells. However, this does not imply that Pias3 binding to these promoters requires both Nr2e3 and Crx. We performed ChIP assays for these same sequences from Crx-/- and rd7/rd7 mice and found that Pias3 is still immunoprecipitated from each photoreceptor promoter tested in either mutant background (Figure S4B), suggesting that neither protein is necessary to target Pias3 to these promoters.
Figure 3.
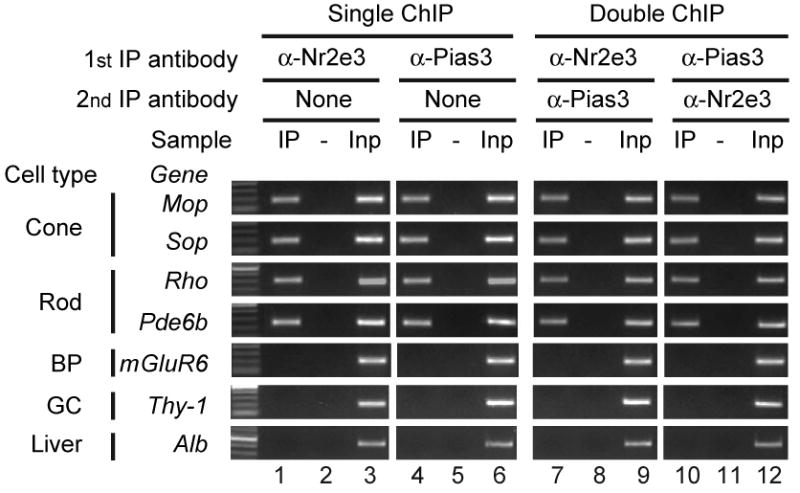
Pias3 and Nr2e3 selectively co-occupy the promoters of photoreceptor-specific genes.
Single and double chromatin immunoprecipitation (ChIP) assays were performed using P14 mouse retinas. Results are shown as gel images of PCR products representing promoter sequences of the indicated candidate genes: IP, single or double immunoprecipitation with the indicated antibodies; -, non-specific IgG controls (For double ChIP, the indicated specific antibody was used in 1st IP, and a control IgG was used in 2nd IP); Inp, Input without IP as a positive control for PCR. The candidate genes are shown based on the retinal cell type in which they are selectively expressed, with non Pias3/Nr2e3 target genes, which are not expressed in photoreceptor genes, is shown in the three bottom lanes (BP, bipolar; GC, ganglion cells).
Pias3 catalyzes SUMOylation of Nr2e3 and SUMOylation-deficient mutants of Pias3 no longer effectively promote rod differentiation
Since Pias3 can regulate target transcription factors both by direct protein-protein interaction and by its E3 SUMO ligase activity, we tested whether the E3 ligase activity is required for regulation of rod development by Pias3. Mutant forms of Pias3 tested included a double point mutation in the RING domain (C299S, H301A) that blocks the E3 SUMO ligase activity of Pias3 (Long et al., 2004). Pias3 also contains an LXXLL coregulator recruitment motif in its N-terminal domain (Jang et al., 2004), which can be inactivated by mutation of the terminal two leucines to alanines (Liu et al., 2001).
To characterize the interaction of Pias3 and Nr2e3, 35S-labeled-deletion mutants of Pias3 were tested for interaction with full-length Nr2e3 (Figure S5). We found both the N-terminal domain and the RING domains of Pias3 are sufficient for interaction with Nr2e3 in vitro. Since SUMO1 was predicted to be the most abundant retinal SUMO isoform based on our previous SAGE study (Blackshaw et al., 2004), we next determined if Pias3 can catalyze addition of SUMO1 to Nr2e3, and whether Pias3-Nr2e3 interaction was affected by the functional point mutations, first confirming the specificity of the SUMO1 antibody for immunblot and immunoprecipitation (Figure S6A). HEK293 cells were cotransfected with plasmids containing wildtype mouse Nr2e3, and wildtype and mutant forms of both Pias3 and Flag-tagged SUMO1. We observe bands with at ∼72 and ∼85 kD with both anti-Nr2e3 and anti-Flag indicating that Flag-tagged SUMO1 is conjugated to Nr2e3 in the presence of wildtype Pias3 (Figure 4A, lanes 1 and 9), corresponding to both di- and tri-SUMOylated forms of Nr2e3. The SUMOylation-deficient (ΔSUMO) RING domain mutant of Pias3, as expected, does not catalyze conjugation of Flag-SUMO1 to Nr2e3 to generate tri-SUMOylated Nr2e3 (Figure 4A, lane 5), although some di-SUMOylated Nr2e3 is observed with the anti-Flag antibody in both the ΔSUMO sample and the sample lacking transfected Pias3. We interpret this data to suggest that other E3 SUMO ligases in the HEK293 cells used may SUMOylate Nr2e3. Unexpectedly, mutation of the LXXLL domain (LXXAA) also blocks the ability of Pias3 to catalyze SUMOylation of Nr2e3 (Figure 4A, lane 3), with no di- or tri-SUMOylated Nr2e3 seen in the presence of this mutant using that anti-Flag antibody (Figure 4A, lane 11). We speculate that this mutant may, through mechanisms that are unclear, have a dominant-negative effect on the activity of endogenous E3 SUMO ligases that generate the di-SUMOylated form of Nr2e3. Interaction of Nr2e3 and Pias3, however, was unaffected in any of the mutants tested (Figure 4A, lanes 17-22).
Figure 4.
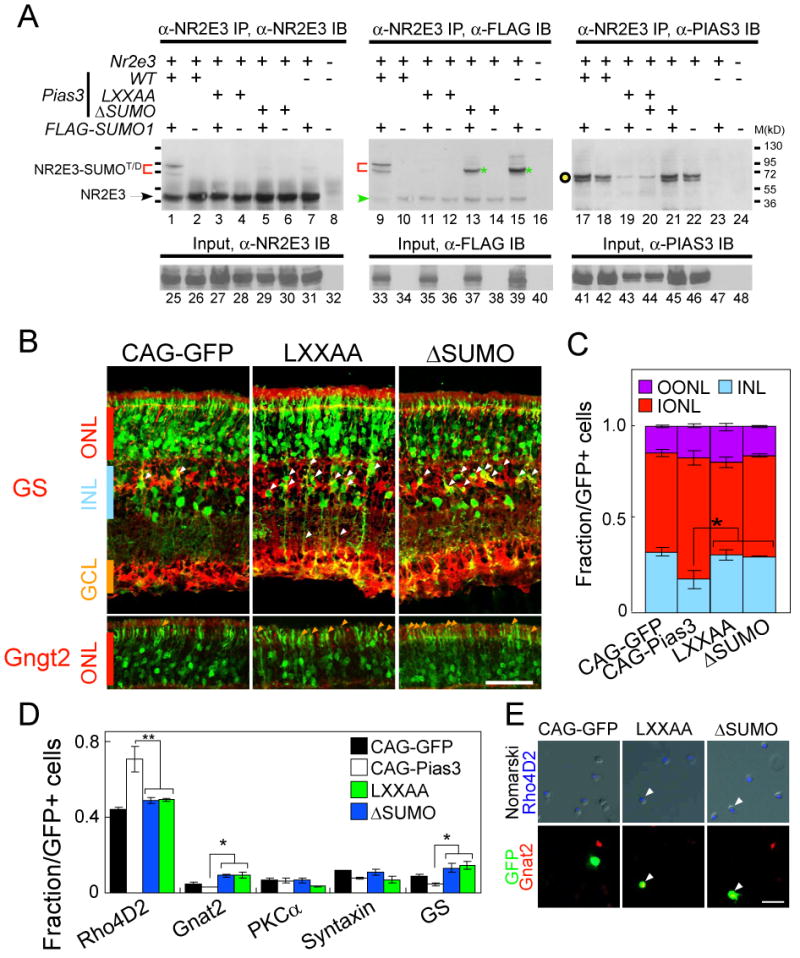
Nr2e3 is SUMOylated by Pias3, and the RING (E3 SUMO ligase) domain of Pias3 is necessary for its function in photoreceptor development.
(A) Pias3 catalyzes SUMOylation of Nr2e3 in an E3-ligase dependent manner. Whole cell extracts were prepared from HEK293 cells co-transfected with pCAG-Nr2e3, pCAG-Pias3 (wildtype, LXXAA or ΔSUMO mutant) and pTag-FLAG-SUMO1, or the respective empty vector controls “-”). The presence of SUMO1-modified forms of Nr2e3 and the interaction of Pias3 with Nr2e3 were analyzed by immunoprecipitation (IP) using anti-Nr2e3 antibody p183, and immunoblotting (IB) using antibodies to Nr2e3 (p183, lanes 1-8), FLAG-SUMO1 (anti-FLAG M2 monoclonal, Sigma; lanes 9-16) and Pias3 (Sigma, lanes 17-24). Red brackets indicate potential di- and tri- (T/D) SUMOylated Nr2e3 catalyzed by the recombinant PIAS3. Green asterisks mark di-SUMOylated Nr2e3, which may represent SUMOylation catalyzed by other endogenous E3 SUMO ligases. The green arrow in the FLAG IB blot points out background staining derived from Nr2e3 IB. The black open circle indicates the expected size for the Pias3. Input controls without IP were shown in lanes 25-48, demonstrating the expression of recombinant Nr2e3, FLAG-SUMO1 and Pias3 proteins by transfected 293 cells.
(B) Immunostaining of P14 retinas with GS (upper) or Gngt2 (lower) antibodies electroporated in vivo at P0 with CAG-GFP and Pias3 LXXAA and ΔSUMO mutants. White and orange arrowheads indicate electroporated cells labeled with the antibodies used.
(C) Fraction of GFP-positive cells found in the OONL, IONL and INL. Pias3 LXXAA and ΔSUMO mutants have significantly fewer cells in the IONL than Pias3 wildtype. All data are represented as mean +/- SD. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 6).
(D) Cell type composition of the P14 mouse retinas electroporated in vivo with CAG-GFP, CAG-Pias3, LXXAA and ΔSUMO determined by immunostaining with retinal cell type-specific markers. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 3).
(E) Immunohistochemistry of the P14 dissociated retina electroporated in vivo with CAG-GFP, LXXAA and ΔSUMO with Rho4D2 (blue), GFP (green) and Gnat2 (red) antibodies. Arrowheads indicate triple-labeled cells. Scale bar: B, 50μm; E, 20μm.
We next tested whether these mutant forms of Pias3 show altered ability to regulate rod photoreceptor development in vivo. We found that the morphology of cells electroporated with the ΔSUMO or LXXAA mutant forms of Pias3 were much like those electroporated with empty vector, but more GS-positive Müller glia were seen (Figure 4B, white arrowheads). We also observed more cells expressing Gngt2 in the ONL (Figure 4B, upper panel, orange arrowheads), along with Gnat2 and S-cone opsin (data not shown). We find that, unlike wildtype Pias3, overexpression of both mutant forms of Pias3 does not result in additional cells in the ONL (Figure 4C). We next performed dissociated cell immunostaining to quantify changes in cell composition resulting from overexpression of mutant forms of Pias3. Neither mutant produced an increase in rhodopsin-positive cells, unlike wildtype Pias3 (Figure 4D). Moreover, a SUMOylase-deficient mutant of the human orthologue of Pias3, unlike wildtype human PIAS3, did not rescue the reduction in rhodopsin seen following Pias3 knockdown (Figure S1D). More electroporated cells expressed the cone marker Gnat2 for both mutants relative than in either wildtype Pias3 or in vector control (p < 0.05). We observed that a number of electroporated cells coexpressed both rhodopsin and Gnat2 (Figure 4E). This phenotype is reminiscent of that seen with Pias3 knockdown (Figure 1L), although the increase in Gnat2-expressing cells is less dramatic. ΔSUMO mutants of Pias3 also failed to enhance Nr2e3-mediated transcriptional repression in HEK293T cells (Figure S2B). This suggests that these mutant forms of Pias3 act in vivo as partial dominant negative mutations. Both mutant forms of Pias3 also lead to an increase in the number of Müller glia (Figure 4B). The mechanism of this effect is unclear.
Nr2e3 is SUMOylated on multiple sites in vitro and in vivo and SUMOylation of Nr2e3 is necessary for its ability to repress cone-specific genes in vivo
Following up on the finding that Pias3 can SUMOylate Nr2e3 in vitro and in vivo, we sought to characterize the residues in Nr2e3 that can be SUMOylated. Analysis of mouse Nr2e3 sequence using SUMOplot (http://www.abgent.com/doc/sumoplot) identified three high probability sites for SUMOylation, at K178, K315 and K322 (Figure 5A), which are highly evolutionarily conserved. We tested whether these residues were necessary for SUMOylation of Nr2e3, constructing K to R point mutations for each of these sites individually, and also a construct in which all three lysines had been mutated. We first tested these using transfected HEK293T cells and found that wildtype forms of Nr2e3 show two SUMOylated products, corresponding in size to potentially di- and tri-SUMOylated isoforms, in the presence of Pias3 (Figure 5B, lane 1). Triple mutant forms of Nr2e3 are not SUMOylated (Figure 5B, lane 9). Finally, mutation of the K178 residue drastically affected the pattern and levels of SUMOylation of Nr2e3 (Figure 5B, lane 3). In this mutant isoform, the di-SUMOylated form of Nr2e3 was not observed in the presence of Pias3, although reduced levels of tri-SUMOylated Nr2e3 were still observed, presumably reflecting usage of a cryptic nonconsensus SUMOylation site. No change in SUMOylation levels or pattern was observed in either the K315R or K322R mutants (Figure 5B, lanes 5 and 7). Together, these results suggest that the observed di-SUMOylated form of Nr2e3 results from SUMOylation at K178 and at either K315 or K322.
Figure 5.
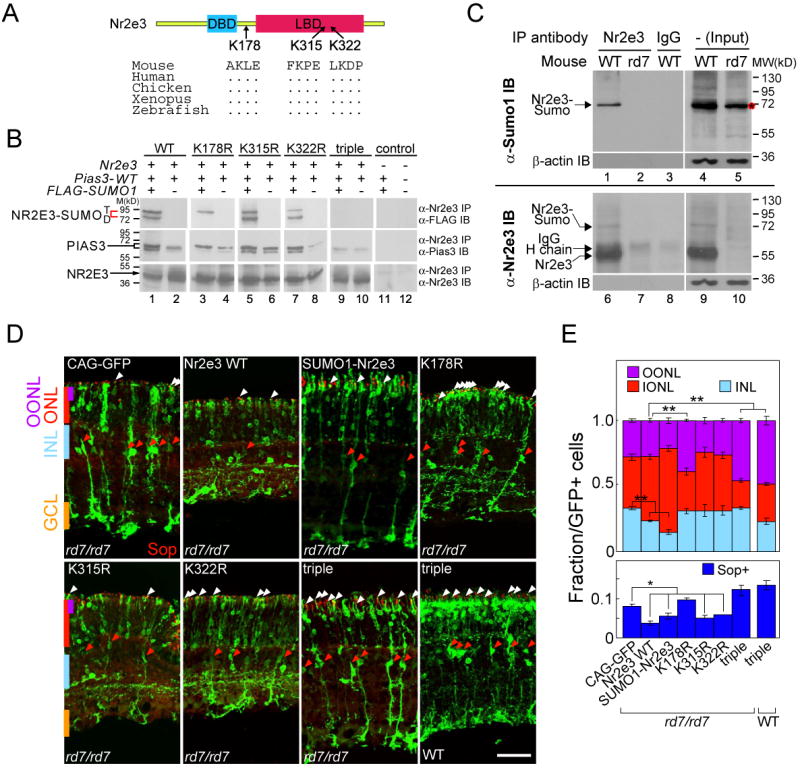
SUMOylation of Nr2e3 is required for repression of cone-specific genes in rod photoreceptors.
(A) Schematic structure of Nr2e3 is shown together with three predicted SUMOylation sites.
(B) Nr2e3 is SUMOylated by Pias3 at three potential sites. SUMOylation assays were performed using wildtype and mutant forms of Nr2e3 protein expressed in HEK293 cells. Whole cell extracts were prepared from HEK293 cells co-transfected with the wildtype and four mutant forms of pCAG-Nr2e3, pCAG-Pias3, and pTag-FLAG-SUMO1 with “-” as empty vector controls. Red brackets indicate the predicted di- or tri-SUMOylated forms of Nr2e3. Anti-Pias3 IB indicates that interaction of Nr2e3 and Pias3 is preserved in all combinations of Pias3 and Nr2e3 mutants. Anti-Nr2e3 IB is used to normalize loading.
(C) Nr2e3 is di-SUMOylated in the mouse retina. Immunoprecipitates from P10 wildtype or rd7/rd7 retina were analyzed for the presence of SUMOylated forms of Nr2e3 using immunoblotting with anti-SUMO1 (upper) and anti-Nr2e3 (lower). A red asterisk marks an unknown SUMOylated protein running at a same position as Nr2e3-SUMO in the input lane of the rd7/rd7 retinas.
(D) P14 rd7/rd7 or CD1 retinas electroporated in vivo with Nr2e3, SUMO1-fused Nr2e3 (SUMO1-Nr2e3) and the loss of SUMOylation mutants (K178R, K315R, K322R and triple mutant) immunostained with S-opsin antibody. White arrowheads indicate electroporated photoreceptors co-labeled with S-opsin, and red arrowheads indicate electroporated cells with Müller glial morphology. Scale bar: 50μm.
(E) Fraction of the GFP-positive cells localized at OONL, ONL and INL (upper graph) and S-opsin positive cells (lower graph) in rd7/rd7 mice electroporated with the indicated construct as shown in (D). Data are represented as mean +/- SD. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 6).
We next investigated whether Nr2e3 is indeed SUMOylated in developing retina. We performed immunoprecipitation assays from both wildtype and rd7/rd7 retina using antibodies directed against both Nr2e3 and SUMO1. Immunoprecipitation with anti-Nr2e3 followed by immunoblotting with anti-SUMO1 detects a band at ∼70 kD that corresponds to the size of di-SUMOylated Nr2e3 observed in transfected HEK293 cells (Figure 5C, lane 1). This band is not detectable in retinas of rd7/rd7 mice with either anti-SUMO1 (Figure 5C, lane 2) or with anti-Nr2e3 (Figure 5C, lane 7). The anti-SUMO1 antibody recognizes a protein that runs at roughly the same size as SUMOylated Nr2e3 in the input lane (Figure 5C, lane 5, asterisk). However, since this band is not immunoprecipitated with anti-Nr2e3 and is also present in retinas from rd7/rd7 animals, we conclude that it does not represent SUMOylated Nr2e3 but rather another SUMOylated protein that coincidentally has a similar electrophoretic mobility.
To determine whether SUMOylation of Nr2e3 is essential for its function in vivo, we tested if overexpression of SUMOylation-deficient mutants of Nr2e3 could rescue defects observed in rd7/rd7 animals. We electroporated wildtype Nr2e3 and SUMOylation-deficient mutants of Nr2e3, along with empty vector controls, into P0 rd7/rd7 animals and examined the phenotypes at P14. In retinas electroporated with empty vector, we observe many electroporated photoreceptors that are double-labeled with S-opsin, consistent with previously published data (Figure 5D) (Chen et al., 2005; Peng et al., 2005). We also observe more electroporated cells with Müller glial morphology that express GS than in wildtype animals. This may represent gliosis associated with the retinal rosettes seen in rd7/rd7 animals or else may represent an unreported function of Nr2e3 in repressing Müller glial differentiation. Electroporation of wildtype Nr2e3 results in a reduced number of cells expressing S-cone opsin (Figure 5D, white arrowheads), consistent with previous studies demonstrating a role of Nr2e3 in directly repressing S-cone opsin expression (Peng et al., 2005) along with a substantial reduction in the number of Müller glia (Figure 5D, red arrowheads) and other cells of the INL (Figure 5E), indicating that wildtype Nr2e3 partially rescues the phenotype observed in rd7/rd7 animals. Electroporation of single SUMOylation-deficient mutants reveals that the K178R mutant does not efficiently rescue the rd7/rd7 phenotype, as indicated by the large number of S-cone opsin positive photoreceptors and the increase in the number of cell with cone-like morphology in the outer ONL (p < 0.005) (Figure 5D and E). In fact, the number of cells with cone-like morphology (though not the number of cells expressing S-opsin) is greater in the K178R mutant than in vector control alone, suggesting that overexpression of K178R may disrupt the function of endogenous photoreceptor-specific transcription factors such as Crx and Nrl that are known to interact with Nr2e3 (Chen et al., 2005; Cheng et al., 2004). However, K315R and K322R mutants rescued both the morphology and S-opsin expression phenotypes at a level roughly equal to wildtype Nr2e3 (Figure 5D and E). These in vivo data are consistent with the pattern of SUMOylation observed in transfected cells (Figure 5B). Finally, we observed that the triple mutant form of Nr2e3 not only did not rescue the rd7/rd7 phenotype in electroporated photoreceptors, but appeared to enhance the mutant phenotype, generating a significantly higher fraction of cells located in the outer ONL compared to vector control and many more photoreceptors expressing S-cone opsin (p < 0.005) (Figure 5D and E). Similar results were observed in wildtype retinas electroporated with the triple mutant form of Nr2e3, suggesting that mutant Nr2e3 may interfere with the activity of wildtype Nr2e3 or associated proteins such as Nrl (Cheng et al., 2004). Triple mutant forms of Nr2e3 also failed to show any increase in transcriptional repression in the presence of Pias3 in HEK293T cells, while SUMO1-Nr2e3 fusion proteins repressed transcription more efficiently than wildtype Nr2e3, and that Pias3-dependent SUMOylation is needed for maximal repression of transcription by Nr2e3 (Figure S2B).
The promoters of rod and cone-specific genes are hyperSUMOylated and SUMOylation is essential for normal photoreceptor differentiation
To see if SUMOylation is associated with the promoters of photoreceptor-specific genes that are bound by Nr2e3, we performed single and double ChIP for Nr2e3 and SUMO1. Both single and double ChIP revealed similar results. We observed that the promoters of both rod and cone-specific genes are hyperSUMOylated relative to the promoters of genes selectively expressed in non-photoreceptor cell types, or not expressed in the retina (Figure 6A, Figure S6B). Since we observe hyperSUMOylation of rod and cone-specific promoters following initial immunoprecipitation with Nr2e3, this implies that rod and cone-specific promoters are hyperSUMOylated in rod photoreceptors, implying that SUMOylation is associated with both transcriptionally active and inactive photoreceptor-specific promoters in these cells.
Figure 6.
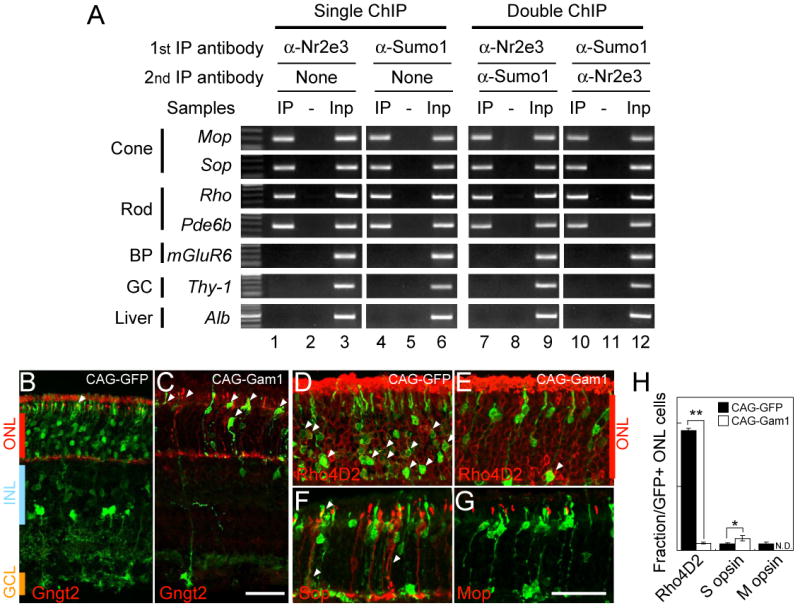
Photoreceptor-specific promotes are hyperSUMOylated and SUMOylation regulates rod photoreceptor development.
(A) Nr2e3 target promoters are hyperSUMOylated -Single and double ChIP assays were performed on P14 mouse retina using rabbit anti-Nr2e3 or/and mouse anti-SUMO1 antibodies. Results are shown as gel images of PCR products representing promoter sequences of the indicated candidate genes: IP, single or double immunoprecipitation with the indicated antibodies; -, non-specific IgG controls (For double ChIP, the indicated specific antibody was used in first IP, and a control IgG was used in second IP); Inp, Input without IP as positive controls for PCR. The candidate genes are shown based on the expressing cell types in the retina with non-Nr2e3 target genes in the three bottom lanes (BP; bipolar, GC; ganglion cells).
(B-G) Section immunohistochemistry of the P14 mouse retinas electroporated in vivo with CAG-GFP (B and D) and Gam1 (C, E, F and G) immunostained with Gngt2 (B and C), Rho4D2 (D and E), S-opsin (F) and M-opsin (G) antibodies. Scale bar: 50μm.
(H) Fraction of the GFP-positive cells localized at ONL labeled with Rho4D2, S-opsin or M-opsin. Data are represented as mean +/- SD. Asterisk, p < 0.05; double asterisk, p < 0.005 by Student's t-test (n = 3).
We next tested the effects of global inhibition of SUMOylation on photoreceptor development. The adenoviral protein Gam1 has been identified as a dominant negative inhibitor of SUMO E1 ligase (Boggio et al., 2004). Since all forms of SUMOylation use the same E1 ligase, overexpression of Gam1 abolishes all cellular SUMOylation. Overexpression of Gam1 in developing retina from P0 results in photoreceptors adopting a cone-like morphology and ectopically expressing Gngt2 (Figure 6C). We also observed more S-cone opsin positive cells compared to vector control (p < 0.05) (Figure 6F and H). This suggests that loss of SUMOylation results in a failure to repress cone-specific gene expression in rods, as is seen with knockdown of Pias3 and overexpression of SUMOylation-deficient mutants of Nr2e3. However, these cells also lack detectable rhodopsin expression (Figure 6E). We thus conclude that loss of SUMOylation not only results in ectopic expression of S-cone-specific genes in rods but is required for normal expression of rhodopsin.
Discussion
Pias3 promotes rod development, repressing expression of cone-specific genes in rods
Pias3 protein is first weakly detectable in developing photoreceptors at birth, with expression peaking in the postnatal week. This spatial and temporal expression pattern corresponds almost precisely with that of the known photoreceptor-specific transcription factors Crx, Nrl and Nr2e3, and closely matches the previously published Pias3 mRNA expression pattern (Blackshaw et al., 2004). Since Pias3 can regulate many different classes of transcription factors, we hypothesized that it might act in vivo to regulate the function of multiple photoreceptor-specific transcription factors. We tested this hypothesis using overexpression and knockdown of Pias3 and observed that Pias3 overexpression lead to an increased number of rhodopsin-positive, rod-like cells in the ONL and fewer inner retinal cells relative to controls, while Pias3 knockdown generates photoreceptors with cone-like morphology and molecular features but which still express rod-specific genes. Curiously, both overexpression and knockdown of Pias3 also lead to a reduced number of Müller glial cells, an effect that is dependent on the presence of Nr2e3. This may imply that an intermediate level of Pias3 expression in photoreceptor precursor cells may permit some to adopt a glial fate, while high or low levels of Pias3 may commit these cells to photoreceptor fate. Alternatively, Pias3 overexpression might mimic loss of function by titrating proteins that direct glial specification away from their endogenous targets.
Pias3 interacts with photoreceptor-specific transcription factors and regulates expression of photoreceptor-specific genes
The increase in rod photoreceptor number seen following Pias3 overexpression is like that seen following Crx overexpression (Furukawa et al., 1997). Likewise, the ectopic expression of S-cone specific transcripts seen in rods where Pias3 expression has been knocked down resembles that seen in mice mutant for Nr2e3. We found that Pias3 interacts with both these proteins in vivo and is selectively targeted to the promoters of photoreceptor-specific genes. It is likely that Crx and Nr2e3, however, do not represent the full set of Pias3 targets in rod photoreceptors. Crx and Nr2e3 can be found as part of a multiprotein complex along with Nrl,, Crx and the nuclear hormone receptor Nr1d1 in vivo (Chen et al., 2005; Cheng et al., 2004), We have observed that Pias3 can be immunoprecipitated from postnatal retina as part of a complex containing both Nrl and Otx2, and Pias3 can directly bind both these proteins in vitro (data not shown). It is thus possible that Pias3 may in part function as a bridging protein to facilitate the assembly of this transcription factor complex on photoreceptor promoters.
Pias3-mediated SUMOylation of Nr2e3 is required for repression of cone-specific genes in rods
Nr2e3, which is bound to the promoters of both rod and cone-specific genes in developing rod photoreceptors, is a transcriptional activator on the promoters of rod-specific genes but represses the expression of cone photoreceptor-specific genes in vivo. Since other identified photoreceptor-specific transcription factors such as Crx and Nrl are also present at each of these promoters (Peng et al., 2005), the mechanism of this divergent transcriptional regulation has been a mystery. We provide evidence that suggests that this may be mediated by selective SUMOylation of Nr2e3 by Pias3. We demonstrate that not only does Pias3 directly SUMOylate Nr2e3, and that Nr2e3 is SUMOylated in developing retina, but that SUMOylation-deficient mutants of Nr2e3 fail to rescue the mutant phenotype in rd7/rd7 mice. Pias3 enhances Nr2e3-mediated transcriptional repression in heterologous cells, and this effect is dependent on the presence of both the SUMO ligase of Pias3 and the SUMOylated lysine resides in Nr2e3. SUMOylation of Nr2e3 appears to be critical for repression of cone-specific genes, but it does not seem to be the only target of Pias3 in this pathway. Though Pias3 knockdown results in a less dramatic increase in cells with cone-like morphology and gene expression pattern in rd7/rd7 animals than in wildtype, we still observe an increase in cone-like cells over controls (Figure S3B). This suggests that other transcription factors that repress cone-specific gene expression, among them Nrl, may also be physiological targets of Pias3. While loss of function of Pias3 leads to an upregulation of S-opsin but M-opsin expression in rods, essentially phenocopying the rd7/rd7 in this respect (Chen et al., 2005; Corbo and Cepko, 2005; Peng et al., 2005), Pias3 protein is found associated with the promoters of both S- and M-opsin genes in rod photoreceptors, along with Nr2e3 (Figure 3). The reason why M-opsin is not upregulated following loss of function of either Pias3 or Nr2e3 is still unclear, although the presence of an additional cone-specific factor, such as the thyroid hormone receptor TRβ2 (Ng et al., 2001), may be necessary to allow expression of M-opsin.
SUMOylation of the promoters of photoreceptor-specific genes coordinates normal photoreceptor development
Many nuclear proteins are SUMOylated in vivo. SUMOylation has been traditionally thought of a signal for transcriptional repression and chromatin silencing (Gill, 2005; Girdwood et al., 2004). It has been proposed that SUMOylation may serve as a general signal for recruitment of histone deacetylases that trigger transcriptional silencing, or else may target SUMOylated DNA-protein complexes to regions of the nucleus associated with transcriptional silencing. Recent data, however, has suggested that SUMOylation of transcription factors may also lead to transcriptional activation (Lyst and Stancheva, 2007), though the mechanism by which this occurs is unclear. Our data suggests that hyperSUMOylation is a general property of both rod and cone-specific promoters in rod photoreceptor cells, but is not detectable in rods on promoters of genes that are selectively expressed in nonphotoreceptor cells of the retina. We hypothesize that this reflects the modification of substrate proteins, among them Nr2e3, by Pias3 targeted to these promoters.
Though we have shown that SUMOylated Nr2e3 represses cone-specific gene expression in rods, it is not clear if this represents the only SUMOylated protein found on the promoters of cone-specific genes (Figure 6A). Other than Nr2e3 itself, the identity of these SUMOylated proteins is unclear. Phosducin is the only SUMOylated mammalian photoreceptor-specific protein identified to date (Klenk et al., 2006). Phosducin is localized mainly in photoreceptor outer segments, and SUMOylation alters its stability and interaction with the beta and gamma subunits of transducin. It has been suggested that phosducin may also interact with Crx to regulate transcription of photoreceptor-specific genes (Zhu and Craft, 2000). If nuclear phosducin is indeed associated with photoreceptor promoters, it would represent a possible substrate for Pias3-dependent modification. One additional potential retinal target is phosphorylated Stat3, which was the first identified biochemical target of Pias3 (Chung et al., 1997), and can inhibit both inhibit rod photoreceptor development and promote Müller glial differentiation. However, the expression patterns of Pias3 protein and pStat3 overlap little in developing retina, with pStat3 expression undetectable in photoreceptor precursors by the time Pias3 expression is seen (Goureau et al., 2004; Ozawa et al., 2004; Zhang et al., 2004), and it is questionable whether pStat3 is a major retinal target of Pias3.
The function of SUMOylation in neural development is obscure. One recent study has demonstrated that Sox10 activity during neural crest development is modified by SUMOylation (Taylor and Labonne, 2005), while another has shown that SUMOylation of Mef2C by Pias1 can modulate dendrite development in cerebellar granule neurons (Shalizi et al., 2007; Shalizi et al., 2006). To our knowledge, however, this represents the first report in which SUMOylation has been directly linked to specification of a specific subtype of neuron.
Pias3 as central coordinator of rod photoreceptor development
Attention has historically focused on DNA-binding transcription factors as the key determinants of regional patterning and cell fate specification. Pias3 is one of a handful of transcriptional coregulators with highly cell-or tissue-specific expression patterns that have been identified. A few of these coregulators have been shown to play a critical role in developmental processes, including Dach1 in organogenesis (Li et al., 2003), and myocardin in smooth and cardiac muscle development (Wang et al., 2001; Wang et al., 2003). Pias3 represents the first protein in this category to play a clear role in neuronal cell fate specification.
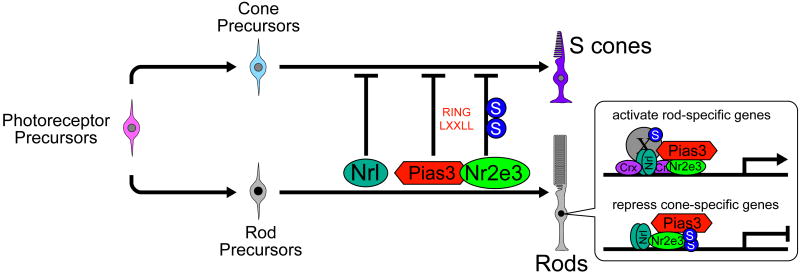
Based on these data, we conclude that Pias3 plays a multifunctional role in the regulation of photoreceptor development. Our findings suggest that Pias3 acts to promote rod photoreceptor development by modulating the activity of photoreceptor proteins, among them Nr2e3 and Crx (Figure 7). Though expression of rod-specific genes is dependent on Pias3-mediated SUMOylation, it is not yet clear which proteins must be SUMOylated for this effect. Whatever the relevant SUMOylated factors are for regulating expression of rod-specific genes, it is unlikely that these include Nr2e3, as both the ΔSUMO mutant of Pias3 and the triple SUMOylation-deficent mutant of Nr2e3 activate transcription of rhodopsin in heterologous cells equally as well as the wildtype forms of both proteins (Figure S2A). We interpret our results to show that Pias3 binds Nr2e3 when it is bound to the promoters of cone-specific genes and also SUMOylates it, converting it to a strong transcriptional repressor and ensuring that only rod-specific genes are expressed in rod photoreceptors. Pias3 thus acts to both enhance rod-promoting and confer cone-repressing functions on its target proteins, though the relevant targets of Pias3-dependent SUMOylation in activation of rod-specific genes remain to be determined (Figure 7). It is likely that the targets of Pias3 are not limited to Crx and Nr2e3, and further studies will clarify its mechanism of action.
Figure 7.
Model for action of Pias3 in photoreceptor differentiation. “S” indicates SUMOylated proteins, while “X” indicates an unidentified SUMOylated factor present on the promoters of rod-specific genes. Pias3 in rods binds and SUMOylates Nr2e3 to repress expression of S-cone-specific genes and simultaneously activates expression of rod-specific genes via a SUMOylation-dependent mechanism. Since Pias3-dependent activation of rod-specific genes does not require SUMOylation of Nr2e3 (see Figure S2), Pias3 must SUMOylate other transcription factors on rod-specific promoters (shown here as X).
Experimental Procedures
Animals
Timed pregnant CD1 mice were purchased from Charles River Breeding Laboratories. C57BL/6J and rd7/rd7 mice were purchased from the Jackson Laboratory. A Crx-/- mouse was provided by C. Cepko at Harvard Medical School. All experimental procedures were pre-approved by JHMI animal care and the Institutional Animal Care and Use Committee of Washington University School of Medicine, and conformed to the guidelines of the Association for Research in Vision and Ophthalmology for the use of live animals in vision research.
DNA Construction
Details for all cDNA constructs used in this study are shown in Table ST1.
In Vivo Electroporation
In vivo electroporation were performed at P0 as previously described (Matsuda and Cepko, 2004). ∼0.3 μl of DNA solutions (5 μg/μl) was injected into the subretinal space of P0 mouse pups, and square electric pulses (100 V; five 50-ms pulses with 950-ms intervals) were applied with tweezer-type electrodes (BTX, model 522). pCAGIG vector was coelectroporated (1 μg/μl) with both shRNA constructs to allow for visualization of transfected cells. The electroporated retinas were harvested at P4, P7 and P14 for immunohistochemistry.
Immunohistochemistry
For section immunohistochemistry, retinas were fixed with 4 % paraformaldehyde in PB for ∼1 hr at room temperature, cryoprotected in PBS containing 15% sucrose for overnight at 4 °C, and embedded in OCT compound (Sakura), and cryosectioned (20 μm). For dissociated cell immunohistochemistry, retinas were separated into single cells as described (Matsuda and Cepko, 2004), and plated on poly-D-lysine-coated glass slides, and fixed for 5 min at room temperature. Images were taken on a fluorescence light microscope (Axioskop2, Carl Zeiss) or a scanning confocal microscope (Meta 510, Carl Zeiss). Sources and dilutions of all antibodies used are listed in supplemental table ST2. A minimum of three retinas from two different litters were examined for each construct tested for section immunohistochemical analysis. For dissociated cell counting, a minimum of 100 cells that were positive for both GFP and the marker in question were counted from a minimum of three separate electroporation experiments.
Co-immunoprecipitation (co-IP), Western Blotting, and in vitro SUMOylation assays In vitro co-IP assays were performed using in vitro transcribed/translated recombinant proteins with or without 35S-labeling. Recombinant PIAS3, CRX, and NR2E3 proteins were made from the corresponding pcDNA3.1/HisC mammalian expression vectors using TnT T7 Quick Coupled Transcription/Translation Kit (Promega). Co-IPs were performed as described previously (La Spada et al., 2001) with minor modifications in the wash buffer [50 mM Tris-Cl (pH 7.4), 150∼400 mM NaCl, and 0.5% Triton X-100] and antibodies described in table ST2. In vivo co-IP assays were performed using retinal nuclear extracts made from P10 wild-type or mutant mice as described previously (Peng and Chen, 2007), with antibodies used detailed in supplemental Table ST2. The results were detected and visualized using the Millipore Western Blotting Detection System (Millipore, Billerica, MA) and Amersham Hyperfilm (GE Healthcare). Cell based co-IP and SUMOylation assays were performed as described in Supplemental Materials and Methods section.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation has performed as described (Peng et al., 2005). The results of ChIP assays were analyzed using candidate gene-based PCR with primers spanning the promoter region of each gene (listed in Peng et al., 2005). Results shown are representative of at least three separate experiments. Controls include the use of normal rabbit/mouse IgG (Santa Cruz) in immunoprecipitation reactions (negative controls) and input (without IP) as positive controls in PCR reactions. Double chromatin immunoprecipitation assays (double ChIP) were performed using a modification of a previously described protocol (Geisberg and Struhl, 2004). Details are described in the Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tomomi Shimogori, Heng Zhu, Jef Boeke, Jeremy Nathans, Don Zack and Geraldine Seydoux for comments on the manuscript; Connie Cepko for the Crx-/- mice; Eliazer Kopf and Michael Matunis for monoclonal antibodies; and Ke Shuai, Fang Liu and Albert La Spada for plasmids, Anne Hennig and Hui Wang and Belinda McMahan for technical assistance. This work was supported by R01EY017015, an Alfred P. Sloan Young Investigator Award, and a W. M. Keck Distinguished Young Scholar in Medical Research Award (to S.B.), research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to A.O.), R01 EY012543 (to S.C.), EY02687 and Research to Prevent Blindness (to WU-DOVS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci U S A. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau MJ, Young TL, Cepko CL. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J Neurosci. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Girdwood DW, Tatham MH, Hay RT. SUMO and transcriptional regulation. Semin Cell Dev Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Goureau O, Rhee KD, Yang XJ. Ciliary neurotrophic factor promotes muller glia differentiation from the postnatal retinal progenitor pool. Dev Neurosci. 2004;26:359–370. doi: 10.1159/000082278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Sumaroka A, Aleman TS, Cideciyan AV, Schwartz SB, Roman AJ, McInnes RR, Sheffield VC, Stone EM, Swaroop A, Wright AF. Nuclear receptor NR2E3 gene mutations distort human retinal laminar architecture and cause an unusual degeneration. Hum Mol Genet. 2004;13:1893–1902. doi: 10.1093/hmg/ddh198. [DOI] [PubMed] [Google Scholar]
- Jang HD, Yoon K, Shin YJ, Kim J, Lee SY. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J Biol Chem. 2004;279:24873–24880. doi: 10.1074/jbc.M313018200. [DOI] [PubMed] [Google Scholar]
- Jones I, Ng L, Liu H, Forrest D. An intron control region differentially regulates expression of thyroid hormone receptor beta2 in the cochlea, pituitary, and cone photoreceptors. Mol Endocrinol. 2007;21:1108–1119. doi: 10.1210/me.2007-0037. [DOI] [PubMed] [Google Scholar]
- Junicho A, Matsuda T, Yamamoto T, Kishi H, Korkmaz K, Saatcioglu F, Fuse H, Muraguchi A. Protein inhibitor of activated STAT3 regulates androgen receptor signaling in prostate carcinoma cells. Biochem Biophys Res Commun. 2000;278:9–13. doi: 10.1006/bbrc.2000.3753. [DOI] [PubMed] [Google Scholar]
- Klenk C, Humrich J, Quitterer U, Lohse MJ. SUMO-1 controls the protein stability and the biological function of phosducin. J Biol Chem. 2006;281:8357–8364. doi: 10.1074/jbc.M513703200. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Fu YH, Sopher BL, Libby RT, Wang X, Li LY, Einum DD, Huang J, Possin DE, Smith AC, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Liu B, Gross M, ten Hoeve J, Shuai K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc Natl Acad Sci U S A. 2001;98:3203–3207. doi: 10.1073/pnas.051489598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Long J, Wang G, Matsuura I, He D, Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3) Proc Natl Acad Sci U S A. 2004;101:99–104. doi: 10.1073/pnas.0307598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes & development. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes & development. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Ozawa Y, Nakao K, Shimazaki T, Takeda J, Akira S, Ishihara K, Hirano T, Oguchi Y, Okano H. Downregulation of STAT3 activation is required for presumptive rod photoreceptor cells to differentiate in the postnatal retina. Mol Cell Neurosci. 2004;26:258–270. doi: 10.1016/j.mcn.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Bilimoria PM, Stegmuller J, Gaudilliere B, Yang Y, Shuai K, Bonni A. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Tolkunova E, Malashicheva A, Parfenov VN, Sustmann C, Grosschedl R, Tomilin A. PIAS proteins as repressors of Oct4 function. J Mol Biol. 2007;374:1200–1212. doi: 10.1016/j.jmb.2007.09.081. [DOI] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Zhang SS, Wei J, Qin H, Zhang L, Xie B, Hui P, Deisseroth A, Barnstable CJ, Fu XY. STAT3-mediated signaling in the determination of rod photoreceptor cell fate in mouse retina. Invest Ophthalmol Vis Sci. 2004;45:2407–2412. doi: 10.1167/iovs.04-0003. [DOI] [PubMed] [Google Scholar]
- Zhu X, Craft CM. The carboxyl terminal domain of phosducin functions as a transcriptional activator. Biochem Biophys Res Commun. 2000;270:504–509. doi: 10.1006/bbrc.2000.2414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.