Abstract
PURPOSE
The inability to predict clinical outcome of prostate cancer is a major impediment to effective treatment decisions and patient counseling. New markers of recurrence are needed to improve the accuracy of risk assessment and treatment of prostate cancer. Our previous studies identified a mismatch repair protein, PMS2, to be elevated in prostate cancer; here we investigate the prognostic potential of this marker. We hypothesized that the elevation of PMS2 would correlate with disease outcome.
EXPERIMENTAL DESIGN
Retrospective quantitative immunohistochemistry was performed to measure PMS2 in high grade cancers of 166 men treated by radical prostatectomy with a biochemical recurrence rate of 56%. Associations between PMS2 levels, pathological variables and biochemical recurrence over time were determined.
RESULTS
The mean level of PMS2 protein was consistently higher in both cancer-associated benign epithelium and cancer cells of patients who recurred, compared to non-recurrent patients. PMS2 was an independent predictor of time-to-recurrence in Cox multivariate analyses and significantly stratified patients based on outcome. PMS2 was able to improve the sensitivity of total percent Gleason 4/5 as a risk factor for recurrence in this cohort.
CONCLUSIONS
PMS2 protein levels were shown to be a predictor of time-to-recurrence after surgery. This study is the first to document that the elevation of a mismatch repair protein negatively correlates with prognosis and has implications in patient diagnosis and molecular profiling.
Keywords: prostate cancer, clinical biomarker, mismatch repair, PMS2, risk factor
INTRODUCTION
Prostate cancer risk assessment for recurrence is performed with the help of two well-established clinical prognostic markers: blood serum prostate specific antigen (PSA) and pathological Gleason grading of the cancer (1-4). These markers are useful for patients that present with either benign (low risk) or aggressive (high risk) cancer; however they are uninformative for patients with less severe disease presentations (5,6). These intermediate risk patients, characterized by localized cancer, a Gleason score of 7 and a pre-operative prostate specific antigen (PSA) level between 2.1 and 10.0 ng/mL, may develop an aggressive form of the disease during their lifetime. A lack of reliable markers for these patients impairs the clinician’s ability to plan appropriate treatment regimens and novel biomarkers are needed.
Generally, only mismatch repair (MMR) defects or loss have been described in association with cancer (7-12). Our recent discovery that a MMR protein, PMS2, is specifically elevated in prostate cancer was unexpected. The PMS2 protein is part of the MMR machinery and forms a heterodimeric complex with its homolog MLH1 (13). This complex is thought to promote the recruitment of downstream proteins after the recognition of mismatches or damage in DNA. PMS2 overexpression has been shown to result in an increased mutation frequency (14), a resistance to methylation-induced apoptosis (14) and correlate with microsatellite instability (15).
The purpose of this study was to define the prognostic potential of PMS2 elevation in a cohort of intermediate risk prostate cancer patients. We hypothesized that patients with biochemical recurrence would have significantly higher levels of PMS2 than non-recurrent patients. Here we report that PMS2 elevation is a predictor of biochemical recurrence after surgery in intermediate risk patients and that it enhances the sensitivity of Gleason grading in prostatectomy samples. This finding identifies a potential new marker for recurrent prostate cancer and uncovers a potentially novel pathway for targeted cancer therapeutics.
MATERIALS AND METHODS
Patients
Retrospective analysis was performed on 166 of 379 prostatectomy samples collected at Stanford University Medical Center (Stanford, CA) between 1983 and 1992 (16). All patients were treated by prostatectomy alone and all tumors were extensively analyzed for total tumor volume, the volume of the largest tumor, the total percent Gleason grade represented by all cancer in the prostate, serum PSA, age, tumor location (peripheral zone versus transition zone), nodal involvement, prostate weight and 5-year follow-up (16). Disease recurrence was defined by biochemical failure, which was described as two consecutive PSA values above a cut-off point of 0.07 ng/ml for PSA measured by the Tosoh method, and 0.2 ng/ml for measurements by less sensitive methods.
From this larger cohort, patients were excluded that had cancer consisting of only Gleason pattern 3, as these patients have less than a 5% chance of recurrence (17). Our final cohort consisted of 166 cancers from men who had localized peripheral zone cancer with at least 2% Gleason grade 4/5 cancer. Within this cohort and despite surgical treatment, 56% of patients developed biochemical recurrence with an average PSA value of 0.72 ng/mL at time of recurrence (Table 1).
Table 1. Pathological descriptors and patient demographics of prostate cancer patient cohort.
The cohort used in this study consisted of 166 cancers from men who had localized peripheral zone cancer. Their pathological descriptors and demographics are listed here.
| Total number of patients | 166 | ||
|---|---|---|---|
| % Recurrencea | 56 | ||
| Median | Range | Average | |
| Age Distribution (years) | 66 | 35 – 76 | 64 |
| Time to Recurrence (days) | 256 | 0 – 3048 | 1797 |
| PSA values | |||
| pre-surgical (ng/ml) | 16.4 | 2 – 266 | 22.4 |
| at time of recurrencea (ng/ml) | 0.2 | 0.07 – 12 | 0.72 |
| last PSAb (ng/ml) | 0.1 | 0 – 1366 | 33.24 |
| Vol. of Largest Cancer (cc) | 4.37 | 0.4 – 45 | 6.12 |
| Total Cancer Volume (cc) | 4.8 | 0.4 – 45 | 6.58 |
| Gleason Grade | |||
| % Grade 3 | 60 | 0 – 98 | 55.7 |
| % Grade 4 | 30 | 2 – 100 | 36.1 |
| % Grade 5 | 0 | 0 – 90 | 3.9 |
| % Intraductal | 0 | 0 – 60 | 4.3 |
| % high grade (4/5) tumor | 35 | 2 – 100 | 40 |
| Prostate weight (g) | 46 | 20 – 366 | 51.5 |
recurrence as determined by PSA >0.05 ng/mL after prostatectomy
last PSA: either at time of death or at the last 5 year follow-up visit
Histopathological Evaluation
Surgically removed prostates had been subjected to a comprehensive histopathologic review according to the method described previously(18). Briefly, each specimen was fixed in formalin, serially blocked at 3-mm intervals, and embedded in paraffin. A 5 μm-thick section was cut from each block and stained with hematoxylin and eosin for histologic assessment. The cancer volume was calculated by tracing the exact tumor outline on each slide, determining the area of tumor at each level of section with a digitizing pad, summing the tumor areas at different levels multiplied by the section thickness, and correcting the volume for tissue shrinkage during processing. Tumor grade was determined according to the Gleason system (1). The Stanford modified Gleason scale was used to estimate the proportion of the largest cancer in each case that was poorly differentiated (grades 4 and 5) or well-differentiated (grades 1, 2 and 3) (19). The percentage of each cancer occupied by Gleason grades 4 and 5 (% Gleason grade 4/5) was estimated by a single pathologist (J.E. McNeal). This measurement of tumor grade was shown to be the strongest predictor of PSA failure(20).
Immunohistochemistry
Four-micron sections were cut using a microtome and dried onto charged glass slides at least overnight. These sections were used for subsequent immunohistochemical analysis as previously described (15). The PMS2 antibodies used were from BD Biosciences at 1:100 dilution. To verify specificity of staining, various PMS2 antibodies (Sigma, BD Biosciences, Invitrogen) were used on near-identical serial-sections. Patterns and staining levels were highly repeatable in all samples tested.
Microscopy & Quantitation
Images were captured using a Nikon Eclipse 50i inverted microscope attached to a Nikon Micropublisher 3.3 RTV camera. The pictures were captured at 300x magnification and analyzed using the NIH shareware program, ImageJ. For the quantification of staining, a modified “H score” method was used as described previously (15). Briefly, the modified H-score specified that a score of “zero” designates no stain and a score of “three” designates darkest stain. The scores of “one” and “two” reflect cells that have intermediate staining. Staining and quantification were performed blindly to the patient outcome or disease status.
Statistical Analysis
A mixed model approach was used to examine predictors of PMS2 levels. This model accounted for multiple measurements taken from each patient (e.g. PMS2 in cancer and benign tissue) and was used to examine the impact of other patient level characteristics such as tumor grade, age, PSA, tumor volume, etc.
where Yijk is the outcome (i.e., PMS2 level) measured on the ith individual in the jth tissue type (benign/cancer) in the kth replicate sample (1st or 2nd field for each tissue type); μ is the grand mean; αj is the fixed cancer effect; βk(i) is the random effect of the kth replicate nested within the ith patient; and eijk is the error term.
To examine the correlation between PMS2 and other variables, a non-parametric Spearman correlation was used to account for the fact that several of the measurements were not normally distributed. A Stepwise Cox Proportional Hazards regression model was fit to determine if study variables were significant predictors of the time to recurrence. Only significant (p≤0.05) predictors are reported. For each of the individuals in this study, data concerning the time to relapse (if relapse occurred) was available. Survival curves and the Log-Rank test were performed to examine whether PMS2 levels were predictive of recurrence. A 4-level stratification variable was also created, based on both PMS2 levels and total percent Gleason 4/5.
SAS version 9.1 for Windows was used to perform all statistical analyses. For all statistical tests performed we used two-tailed tests and considered an alpha level of 0.05 for statistical significance. We report actual p-values associated with each significant test unless the observed p-value was less than 0.001 where it would be reported as <0.001.
RESULTS
Study Population
As described in the methods, a cohort consisting of 166 patients with cancers containing more than 2% Gleason grade 4/5 and treated by radical prostatectomy were included in this study. This group of patients was selected because while patients with only Gleason grade 3 (score of 6) tumor are not likely to recur, patients with only Gleason grade 5 (score of 10) are extremely likely to recur. Therefore, we chose to study patients with an uncertain risk of recurrence; those patients typically have a combination of Gleason grade 4 and 5 tumor (score of 7-9). Accordingly, this group had a 56% rate of recurrence and average PSA value of 0.72 ng/mL at time of recurrence. Thus, for each individual, recurrence was uncertain and additional prognostic factors could be useful. PSA levels were measured by the sensitive Tosoh assay, which has been previously shown to successfully detect PSA recurrence at 0.07ng/mL (21,22). We defined biochemical failure as having consecutive PSA values above 0.07 ng/ml, a definition that has been used extensively in previous studies (23-27). Table 1 includes the descriptive statistics on the clinical and pathological variables collected for this cohort.
PMS2 protein levels in human prostate cancer
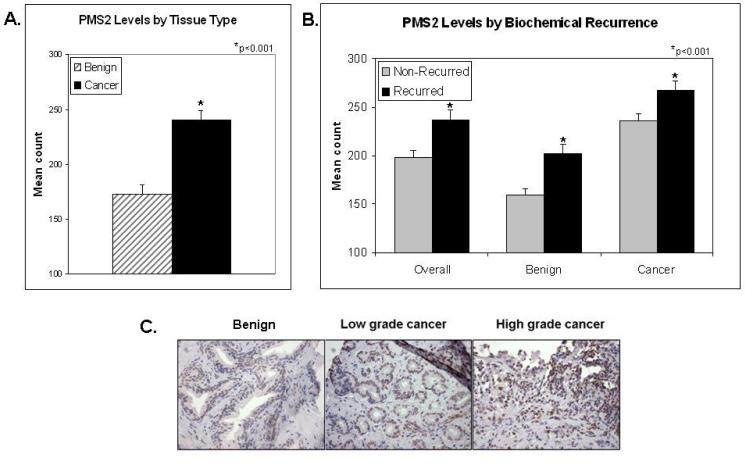
We determined the PMS2 protein levels of cancerous and cancer-associated benign tissue immunohistochemically. The intensity of PMS2 stain was quantitated using an H-score semi-quantitative method as performed previously (15). The variability for this method (calculated for the same sample analyzed at different time points) was approximately 10 H-score counts. Within this cohort, PMS2 protein levels were elevated in prostate cancer (Figure 1A). PMS2 elevation was elevated in 67% (111/166) of patients with the mean cancer PMS2 count of 240 compared to the mean normal PMS2 count of 173 (p<0.001). This finding validated our previous results (15) and confirmed that PMS2 elevation associates with prostate cancer.
Figure 1. PMS2 protein levels are elevated in recurrent prostate cancer patients.
(A) Mean and standard deviation of PMS2 counts in cancer-surrounding benign (hatched bar) and cancer tissue (black bar), *p <0.001;(B) Mean and standard deviation of PMS2 counts of patients with biochemical recurrence (gray bars) and no recurrence (black bars), stratified by tissue type (e.g. benign, cancer or the average of both “overall”), *p<0.001;(C) Representative immunohistochemical staining of PMS2 in benign cancer-associated or cancer tissue of a patient with non-recurrent disease (Patient 1) or recurrent disease (Patient 2). Notice the difference in PMS2 staining in cancer-associated benign cells and the similarity in the cancer cells. Twenty-micron scales are shown by the black bars. Pictures taken under 300x magnification.
We next stratified patients based on biochemical recurrence (Figure 1B). As seen in the left-most pair of bars, recurrent patients had statistically higher levels of PMS2 than non-recurrent patients (p<0.001). When further stratified by tissue type (e.g. benign or cancer) we show that cancer PMS2 levels are significantly higher in cancerous tissue than in patient-matched cancer-associated benign tissue. However in both cases, PMS2 levels were elevated in patients with biochemical recurrence. Characteristic staining of PMS2 in a recurrent versus non-recurrent patient is shown in Figure 1C.
PMS2 Correlations with Pathological Variables
Next, PMS2 elevation was correlated with pathological variables. The results from the Spearman correlation analyses are shown in Table 2. PMS2 levels in cancer (left column) marginally correlated with volume of the largest cancer (p=0.05) and total tumor volume (p=0.01), but not with age, pre-operative serum PSA, prostate weight or total percent Gleason 4/5. PMS2 in cancer-surrounding benign tissue (right column) marginally correlated with volume of the largest cancer (p=0.05), prostate weight (p=0.01) and total percent Gleason 4/5 but not age, PSA or total tumor volume (Table 2).
Table 2. Spearman correlation analyses between PMS2 levels in either benign or cancer tissue and pathological variables.
To determine the correlation between PMS2 elevation and the pathological variables in Table 1, a Spearman analyses was performed. The resulting coefficients and p-values are listed here. Bolded numbers indicate those variables which significantly correlated with PMS2 elevation.
| PMS2 in cancer | PMS2 in benign | |
|---|---|---|
| Variable | Coeffic. Pr>│r│ | Coeffic. Pr>│r│ |
| Age | 0.04 0.60 | -0.08 0.34 |
| PSA | 0.20 0.01 | 0.02 0.77 |
| largest cancer volume |
0.22 0.05 | 0.15 0.05 |
| total tumor volume | 0.19 0.01 | 0.12 0.12 |
| prostate weight | -0.11 0.18 | -0.20 0.01 |
| % Gleason 4/5 | 0.09 0.22 | 0.15 0.05 |
PMS2 Protein Levels and Biochemical Recurrence
We next determined if elevation of PMS2 is predictive of biochemical recurrence in this cohort. Receiver operating characteristic analyses were used to determine a cut-point H-score of 200 for PMS2. This number best corresponded to the highest sensitivity/specificity ratio and was also approximately the median value in this cohort. It should be noted that the cutpoint used here was the most effective for this cohort and analysis, but may not be as effective in a larger or different population. Further studies are needed to specifically address this aspect.
A multivariate analysis was performed to determine which pathological variables, if any, predicted time-to-recurrence. As shown in Table 3, only preoperative PSA (p<0.001), percent Gleason grade 4/5 (p<0.001) and PMS2 in tumor-associated benign tissue (p=0.002) were independent predictors of biochemical recurrence. For all variables, the parameter estimates were positive indicating that the higher PSA, higher percent Gleason 4/5 and higher PMS2 counts all predicted time-to-recurrence. The fact that all three variables were highly significant in the same regression model indicates that each contributes independent information in the prediction of time-to-recurrence.
Table 3. Stepwise Cox Proportional Hazards regression model analyses for time to biochemical recurrence.
A multivariate stepwise Cox Proportional Hazards regression model analysis was performed to determine which pathological variables, if any, predicted time-to-recurrence. This table displays those variables that were significant. PMS2 (B): PMS2 levels in benign tissue
| Variable | DF | Parameter Estimate |
Standard Error |
Chi- Square |
Pr>ChiSq | Hazard Ratio |
|---|---|---|---|---|---|---|
| Pre-op PSA | 1 | 0.015 | 0.002 | 35.215 | <0.001 | 1.015 |
| % Gleason 4/5 | 1 | 1.010 | 0.211 | 23.022 | <0.001 | 2.747 |
| PMS2(B) | 1 | 0.003 | 0.001 | 9.3311 | 0.002 | 1.004 |
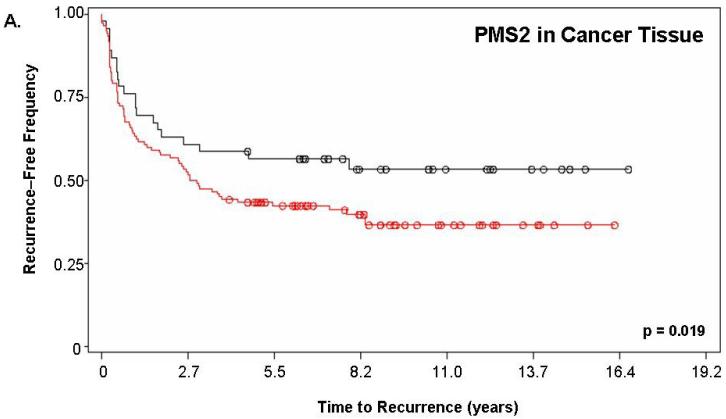
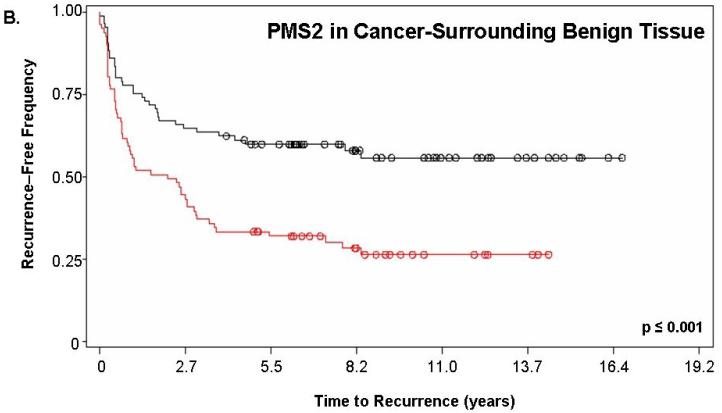
Figure 2 shows the Kaplan-Meier estimate of recurrence-free survival stratified by PMS2. For this analysis, “PMS2 high” patients corresponded to those having a PMS2 score ≥200 while “PMS2 low” corresponded to patients with a score <200. Figure 2a depicts the curves for PMS2 levels in cancer tissue alone (p=0.019) while Figure 2b depicts the curves resulting from PMS2 levels in cancer-surrounding benign tissue alone (p<0.001). In both cases, patients with low PMS2 (black lines; line 1) have higher recurrence-free survival when compared to patients with high PMS2 (red lines; line 2) and according to the p-value, the level of PMS2 in cancer-surrounding benign tissue was better able to stratify patients based on biochemical recurrence. For PMS2 in cancer tissue (Figure 2a), the median time-to-recurrence for PMS2 low patients (1999 ± 143 days) was longer than the PMS2 high patients (1264 ± 141 days). Similar results were found for the cancer-associated benign tissue (Figure 2b), where the median time-to-recurrence for PMS2 low patients (1797 ± 188 days) was equally longer than the PMS2 high patients (1542 ± 121 days).
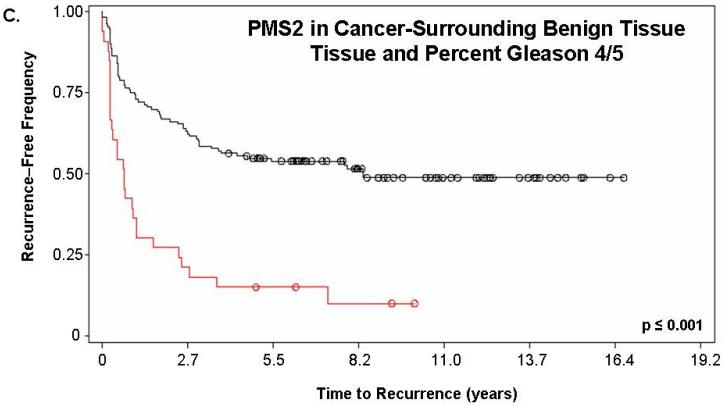
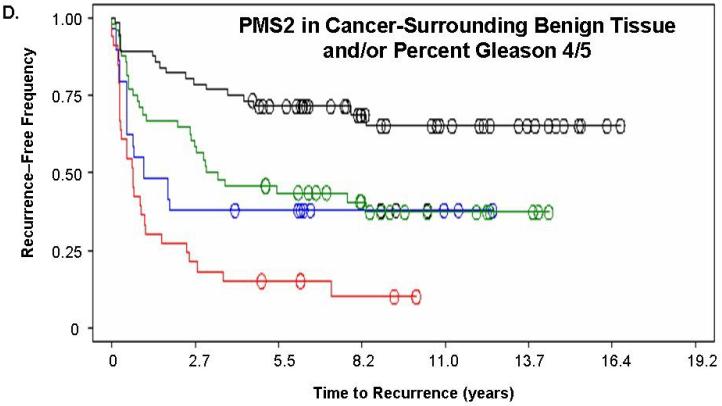
Figure 2. PMS2 protein levels in cancer-associated benign tissue stratifies patients based on recurrence and enhances the predictive power of Gleason grading.
Kaplan-Meier analysis of time to biochemical recurrence using total percent Gleason 4/5 and PMS2 measurements. Analyses were performed using (A) only PMS2 in cancer tissue (p=0.019), (B) only PMS2 in cancer-surrounding benign tissue (p<0.001) or (C-D) a combination of PMS2 in cancer-surrounding benign tissue and total percent Gleason 4/5. (A) Low PMS2 (1999 ± 143 days; black; line 1) versus high PMS2 (1264 ± 141 days; red; line 2); (B) Low PMS2 (1797 ± 188 days; black; line 1) versus high PMS2 (1542 ± 121 days; red; line 3) (C) Low PMS2 or low percent Gleason 4/5 (1874 ± 114 days; black; line 1) versus high PMS2 or high percent Gleason 4/5 (657 ± 158 days; red; line 2); (D) Low PMS2 and low percent Gleason 4/5 (2355 ± 154 days; black; line 1), high PMS2 and low percent Gleason 4/5 (1651 ± 188 days; green; line 2), low PMS2 and high percent Gleason 4/5 (418 ± 51 days; blue; line 3), or high PMS2 and high percent Gleason 4/5 (657 ± 158 days; red; line 4). Median time-to-recurrence values are reflected in the parentheses. Each drop is a recurrence event and each circle is end-of-data for a patient. P-values were computed from Log-Rank test.
We next determined if a combination of prognostic markers would improve the predictive value of either alone. PMS2 levels in cancer-surrounding benign tissue were combined with total percent Gleason 4/5. Kaplan-Meier analyses of the combination of PMS2 and percent Gleason 4/5 is shown in Figure 2c-d. For these analyses, we assigned patients that had ≥50% Gleason grade 4/5 tumor as “high percent Gleason” and patients with <50% Gleason grade 4/5 tumor as “low percent Gleason”. Total percent of Gleason 4/5 measurements were used in place of Gleason score, since they were previously shown to be the best predictor of outcome with this population (16). As seen in Figure 2c, patients who had low PMS2 or low percent Gleason 4/5 (black; line 1) had a higher recurrence-free survival (1874 ± 114 days versus 657 ± 158 days, respectively) when compared to patients with high PMS2 or high percent Gleason 4/5 (red; line 2; p<0.001). Figure 2d depicts patients that were further stratified into low PMS2 and low percent Gleason 4/5 (black; line 1), high PMS2 and high percent Gleason 4/5 (red; line 4), high PMS2 with low percent Gleason 4/5 (green; line 2) or low PMS2 with high percent Gleason 4/5 (blue; line 3).
Of these four sub-groups, patients that had high PMS2 and high percent Gleason 4/5 had the fastest time-to-recurrence (418 ± 51 days) when compared to all other groups (low PMS2 and low percent Gleason 4/5 was 2355 ± 154 days; high PMS2 and low percent Gleason 4/5 was 1651 ± 188 days; low PMS2 and high percent Gleason 4/5 was 657 ± 158 days). The Kaplan-Meier curves for the low PMS2 and high percent Gleason 4/5 (blue; line 3) and the high PMS2 and low percent Gleason 4/5 (green; line 2) become virtually identical over time indicating that both groups have the same frequency of biochemical recurrence approximately 4 years post-surgery (Figure 2d).
We further used two-by-two table analyses to determine the sensitivity (true positive) and specificity (true negative) of PMS2 alone and in combination with percent Gleason 4/5. As shown in Table 4, these parameters for PMS2 levels in cancer-surrounding benign tissue (72%, 58%) are better than PMS2 levels in cancer (61%, 54%) and are approximately equivalent to percent Gleason 4/5 (76%, 55%). When percent Gleason 4/5 was combined with PMS2 in cancer-surrounding benign tissue, the sensitivity was increased to 88% at some expense to the specificity (48% versus 55%). These analyses indicate that PMS2 provides prognostic benefits alone and in combination with traditional clinical indicators.
Table 4. Two by two table analyses of PMS2 levels (in benign and cancer tissue), of total percent Gleason 4/5 and of both combined.
To determine the sensitivity (true positive) and specificity (true negative) of PMS2 alone and in combination with percent Gleason 4/5, a two-by-two table analyses was performed. The resulting percentages are shown above. PMS2 (B): PMS2 levels in benign tissue
| Group | Sensitivity | Specificity |
|---|---|---|
| PMS2 in Cancer | 61% | 54% |
| PMS2 in Benign | 72% | 58% |
| % Gleason 4/5 | 76% | 55% |
| % Gleason 4/5 + PMS2 (B) | 88% | 48% |
DISCUSSION
Currently in the diagnosis of prostate cancer, in patients with Gleason score 7 cancer, no prognostic markers are able to distinguish between those whose cancer will and will not recur. Unfortunately, this group represents the largest number of patients at presentation. The consequences of ineffective risk assessment are under-diagnosis (increasing the number of deaths from cancer) or over-diagnosis (increasing the treatment-associated morbidity and life-altering side effects). Thus, new markers that can improve predictions of disease progression and treatment planning of patients with prostate cancer are needed.
Here we show that PMS2, a MMR protein that is elevated in prostate cancer, predicts biochemical recurrence in a population of patients containing Gleason score 7 prostate cancer treated by prostatectomy. The levels of PMS2 were consistently higher in recurrent individuals (Figure 1). The levels of PMS2 in both cancer and the cancer-surrounding benign epithelium were able to stratify patients based on recurrence (Figure 2), while PMS2 levels in the benign epithelium surrounding the cancer were actually predictive of disease recurrence (Table 3). When used in combination, PMS2 was able to improve the prognostic sensitivity of total percent Gleason 4/5 by 12% (Table 4). The presence of PMS2 in benign tissue is reminiscent of previous reports on risk markers that were found to be present in cancer-associated, benign tissue (28,29) and suggests that PMS2 elevation is an early event in prostate tumorigenesis.
The search for molecular prognostic indicators has produced a number of markers that can be detected immunohistochemically and that correlate with prostate cancer progression. Well-characterizedmarkers include Bcl-2 family proteins (30-34), CDK1/p34 (35,36), E-cadherin(37-40), insulin-like growth factor binding proteins (41,42), Ki67 (30,32,43,44), p27(45-47) and PTEN (48,49). Many more potential biomarkers are the subject of further investigation and verification (50,51). Like PMS2, many of these markers are detectable in tissue samples and correlate with patient outcome. PMS2 is unique, however, in that its quantitation in histologically benign epithelium adjacent to the tumor is informative for biochemical relapse. Also, while the well-characterized markers mentioned above are primarily indicators and effectors of cellular proliferation or apoptotic activation, PMS2 elevation is associated with genomic instability and mutagenesis.
Elevation of PMS2 has previously been shown to have biological consequences including: increased mutation frequency (14), increased resistance to methylation-induced apoptosis (14), reduced mismatch repair (52) and microsatellite instability in tumors (15). PMS2 elevation even in early, pre-neoplastic PIN lesions (15) indicates that it may be an early event in tumorigenesis. The discovery that PMS2 levels in cancer-surrounding benign epithelium are predictive of biochemical recurrence suggests that PMS2 may be a consequence of early changes in the tumor microenvironment that contribute to aggressive tumor formation. At this point, the exact role of PMS2 elevation in tumorigenesis is still speculative but may involve aberrant and improper binding to inactivate the MMR system. This mechanism would be consistent with the biological consequences already associated with PMS2 elevation.
As this study was retrospective and would be defined as a Phase 2 biomarker study(53), further studies are required to 1.) validate these findings in an independent patient population, 2.) to investigate the effect of treatment (e.g. radiation, adjunct chemotherapy) and, 3.) to determine a standardized scale of PMS2 quantitation. Overall, this study confirms previous findings of a MMR protein, PMS2, being elevated in a significant proportion of prostate cancer (15) and demonstrates that PMS2, is a novel prognostic indicator for biochemical recurrence after surgery.
ACKNOWLEDGMENTS
We thank Drs. Chen and Torti for helpful discussions and to the granting agencies for funding.
Supported by DoD grants W81XWH-04-1-0057 and W81XWH-07-1-0047 and by an American Federation of Aging Research student award
Standard Abbreviations
- (MMR)
mismatch repair
- (DNA)
deoxyribonucleic acid
- (PIN)
prostatic intraepithelial neoplasia
REFERENCES
- 1.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 1974;111(1):58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 2.Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep. 1966;50(3):125–8. [PubMed] [Google Scholar]
- 3.Cookson MM. Prostate cancer: screening and early detection. Cancer Control. 2001;8(2):133–40. doi: 10.1177/107327480100800203. [DOI] [PubMed] [Google Scholar]
- 4.Catalona WJ, Richie JP, Ahmann FR, et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6,630 men. J Urol. 1994;151(5):1283–90. doi: 10.1016/s0022-5347(17)35233-3. [DOI] [PubMed] [Google Scholar]
- 5.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004;17(3):292–306. doi: 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 6.Fall K, Garmo H, Andren O, et al. Prostate-specific antigen levels as a predictor of lethal prostate cancer. J Natl Cancer Inst. 2007;99(7):526–32. doi: 10.1093/jnci/djk110. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Wang J, Fraig MM, et al. Alterations in PMS2, MSH2 and MLH1 expression in human prostate cancer. Int J Oncol. 2003;22(5):1033–43. [PubMed] [Google Scholar]
- 8.Leach FS. Microsatellite instability and prostate cancer: clinical and pathological implications. Curr Opin Urol. 2002;12(5):407–11. doi: 10.1097/00042307-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84(1):27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 10.Murata H, Khattar NH, Kang Y, Gu L, Li GM. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002;21(37):5696–703. doi: 10.1038/sj.onc.1205683. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61(20):7369–74. [PubMed] [Google Scholar]
- 12.Chintamani, Jha BP, Bhandari V, Bansal A, Saxena S, Bhatnagar D. The expression of mismatched repair genes and their correlation with clinicopathological parameters and response to neo-adjuvant chemotherapy in breast cancer. Int Semin Surg Oncol. 2007;4:5. doi: 10.1186/1477-7800-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 14.Gibson SL, Narayanan L, Hegan DC, Buermeyer AB, Liskay RM, Glazer PM. Overexpression of the DNA mismatch repair factor, PMS2, confers hypermutability and DNA damage tolerance. Cancer Lett. 2006;244(2):195–202. doi: 10.1016/j.canlet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Norris AM, Woodruff RD, D’Agostino RB, Jr., Clodfelter JE, Scarpinato KD. Elevated levels of the mismatch repair protein PMS2 are associated with prostate cancer. Prostate. 2007;67(2):214–25. doi: 10.1002/pros.20522. [DOI] [PubMed] [Google Scholar]
- 16.Stamey TA, McNeal JE, Yemoto CM, Sigal BM, Johnstone IM. Biological determinants of cancer progression in men with prostate cancer. JAMA. 1999;281(15):1395–400. doi: 10.1001/jama.281.15.1395. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez DJ, Nielsen ME, Han M, et al. Natural History of Pathologically Organ-Confined (pT2), Gleason Score 6 or Less, Prostate Cancer After Radical Prostatectomy. Urology. 2008 doi: 10.1016/j.urology.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamey TA, McNeal JE, Freiha FS, Redwine E. Morphometric and clinical studies on 68 consecutive radical prostatectomies. J Urol. 1988;139(6):1235–41. doi: 10.1016/s0022-5347(17)42876-x. [DOI] [PubMed] [Google Scholar]
- 19.McNeal JE, Villers AA, Redwine EA, Freiha FS, Stamey TA. Histologic differentiation, cancer volume, and pelvic lymph node metastasis in adenocarcinoma of the prostate. Cancer. 1990;66(6):1225–33. doi: 10.1002/1097-0142(19900915)66:6<1225::aid-cncr2820660624>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 20.Stamey TA, Yemoto CM, McNeal JE, Sigal BM, Johnstone IM. Prostate cancer is highly predictable: a prognostic equation based on all morphological variables in radical prostatectomy specimens. J Urol. 2000;163(4):1155–60. doi: 10.1016/s0022-5347(05)67713-0. [DOI] [PubMed] [Google Scholar]
- 21.Prestigiacomo AF, Stamey TA. A comparison of 4 ultrasensitive prostate specific antigen assays for early detection of residual cancer after radical prostatectomy. J Urol. 1994;152(5 Pt 1):1515–9. doi: 10.1016/s0022-5347(17)32459-x. [DOI] [PubMed] [Google Scholar]
- 22.Pruthi RS, Haese A, Huland E, Stamey TA. Use of serum concentration techniques to enhance early detection of recurrent prostate cancer after radical prostatectomy. Urology. 1997;49(3):404–10. doi: 10.1016/S0090-4295(96)00500-6. [DOI] [PubMed] [Google Scholar]
- 23.McNeal JE, Yemoto CE. Significance of demonstrable vascular space invasion for the progression of prostatic adenocarcinoma. Am J Surg Pathol. 1996;20(11):1351–60. doi: 10.1097/00000478-199611000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Shinghal R, Yemoto C, McNeal JE, Brooks JD. Biochemical recurrence without PSA progression characterizes a subset of patients after radical prostatectomy. Prostate-specific antigen. Urology. 2003;61(2):380–5. doi: 10.1016/s0090-4295(02)02254-9. [DOI] [PubMed] [Google Scholar]
- 25.Risbridger GP, Shibata A, Ferguson KL, Stamey TA, McNeal JE, Peehl DM. Elevated expression of inhibin alpha in prostate cancer. J Urol. 2004;171(1):192–6. doi: 10.1097/01.ju.0000100048.98807.b7. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Shibata A, McNeal JE, Stamey TA, Feldman D, Peehl DM. Vitamin D receptor start codon polymorphism (FokI) and prostate cancer progression. Cancer Epidemiol Biomarkers Prev. 2003;12(1):23–7. [PubMed] [Google Scholar]
- 27.Shibata A, Garcia MI, Cheng I, et al. Polymorphisms in the androgen receptor and type II 5 alpha-reductase genes and prostate cancer prognosis. Prostate. 2002;52(4):269–78. doi: 10.1002/pros.10119. [DOI] [PubMed] [Google Scholar]
- 28.Malins DC, Gilman NK, Green VM, Wheeler TM, Barker EA, Anderson KM. A cancer DNA phenotype in healthy prostates, conserved in tumors and adjacent normal cells, implies a relationship to carcinogenesis. Proc Natl Acad Sci U S A. 2005;102(52):19093–6. doi: 10.1073/pnas.0509630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malins DC, Gilman NK, Green VM, et al. Metastatic cancer DNA phenotype identified in normal tissues surrounding metastasizing prostate carcinomas. Proc Natl Acad Sci U S A. 2004;101(31):11428–31. doi: 10.1073/pnas.0404572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bubendorf L, Sauter G, Moch H, et al. Prognostic significance of Bcl-2 in clinically localized prostate cancer. Am J Pathol. 1996;148(5):1557–65. [PMC free article] [PubMed] [Google Scholar]
- 31.Scherr DS, Vaughan ED, Jr., Wei J, et al. BCL-2 and p53 expression in clinically localized prostate cancer predicts response to external beam radiotherapy. J Urol. 1999;162(1):12–6. doi: 10.1097/00005392-199907000-00003. discussion 6-7. [DOI] [PubMed] [Google Scholar]
- 32.Pollack A, Cowen D, Troncoso P, et al. Molecular markers of outcome after radiotherapy in patients with prostate carcinoma: Ki-67, bcl-2, bax, and bcl-x. Cancer. 2003;97(7):1630–8. doi: 10.1002/cncr.11230. [DOI] [PubMed] [Google Scholar]
- 33.Keshgegian AA, Johnston E, Cnaan A. Bcl-2 oncoprotein positivity and high MIB-1 (Ki-67) proliferative rate are independent predictive markers for recurrence in prostate carcinoma. Am J Clin Pathol. 1998;110(4):443–9. doi: 10.1093/ajcp/110.4.443. [DOI] [PubMed] [Google Scholar]
- 34.Mackey TJ, Borkowski A, Amin P, Jacobs SC, Kyprianou N. bcl-2/bax ratio as a predictive marker for therapeutic response to radiotherapy in patients with prostate cancer. Urology. 1998;52(6):1085–90. doi: 10.1016/s0090-4295(98)00360-4. [DOI] [PubMed] [Google Scholar]
- 35.Kallakury BV, Sheehan CE, Ambros RA, Fisher HA, Kaufman RP, Jr., Ross JS. The prognostic significance of p34cdc2 and cyclin D1 protein expression inprostate adenocarcinoma. Cancer. 1997;80(4):753–63. doi: 10.1002/(sici)1097-0142(19970815)80:4<753::aid-cncr15>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 36.Kallakury BV, Sheehan CE, Rhee SJ, et al. The prognostic significance of proliferation-associated nucleolar protein p120 expression in prostate adenocarcinoma: a comparison with cyclins A and B1, Ki-67, proliferating cell nuclear antigen, and p34cdc2. Cancer. 1999;85(7):1569–76. doi: 10.1002/(sici)1097-0142(19990401)85:7<1569::aid-cncr19>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 37.Brewster SF, Oxley JD, Trivella M, Abbott CD, Gillatt DA. Preoperative p53, bcl-2, CD44 and E-cadherin immunohistochemistry as predictors of biochemical relapse after radical prostatectomy. J Urol. 1999;161(4):1238–43. [PubMed] [Google Scholar]
- 38.Umbas R, Isaacs WB, Bringuier PP, et al. Decreased E-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res. 1994;54(14):3929–33. [PubMed] [Google Scholar]
- 39.Kallakury BV, Sheehan CE, Ross JS. Co-downregulation of cell adhesion proteins alpha- and beta-catenins, p120CTN, E-cadherin, and CD44 in prostatic adenocarcinomas. Hum Pathol. 2001;32(8):849–55. doi: 10.1053/hupa.2001.26463. [DOI] [PubMed] [Google Scholar]
- 40.Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA. Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. J Natl Cancer Inst. 2003;95(9):661–8. doi: 10.1093/jnci/95.9.661. [DOI] [PubMed] [Google Scholar]
- 41.Miyata Y, Sakai H, Hayashi T, Kanetake H. Serum insulin-like growth factor binding protein-3/prostate-specific antigen ratio is a useful predictive marker in patients with advanced prostate cancer. Prostate. 2003;54(2):125–32. doi: 10.1002/pros.10175. [DOI] [PubMed] [Google Scholar]
- 42.Thrasher JB, Tennant MK, Twomey PA, Hansberry KL, Wettlaufer JN, Plymate SR. Immunohistochemical localization of insulin-like growth factor binding proteins 2 and 3 in prostate tissue: clinical correlations. J Urol. 1996;155(3):999–1003. [PubMed] [Google Scholar]
- 43.Bubendorf L, Tapia C, Gasser TC, et al. Ki67 labeling index in core needle biopsies independently predicts tumor-specific survival in prostate cancer. Hum Pathol. 1998;29(9):949–54. doi: 10.1016/s0046-8177(98)90199-x. [DOI] [PubMed] [Google Scholar]
- 44.Khoo VS, Pollack A, Cowen D, et al. Relationship of Ki-67 labeling index to DNA-ploidy, S-phase fraction, and outcome in prostate cancer treated with radiotherapy. Prostate. 1999;41(3):166–72. doi: 10.1002/(sici)1097-0045(19991101)41:3<166::aid-pros3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 45.Yang RM, Naitoh J, Murphy M, et al. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159(3):941–5. [PubMed] [Google Scholar]
- 46.Freedland SJ, deGregorio F, Sacoolidge JC, et al. Preoperative p27 status is an independent predictor of prostate specific antigen failure following radical prostatectomy. J Urol. 2003;169(4):1325–30. doi: 10.1097/01.ju.0000054004.08958.f3. [DOI] [PubMed] [Google Scholar]
- 47.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9(4):1474–9. [PubMed] [Google Scholar]
- 48.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59(17):4291–6. [PubMed] [Google Scholar]
- 49.Whang YE, Wu X, Suzuki H, et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A. 1998;95(9):5246–50. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar-Sinha C, Chinnaiyan AM. Molecular markers to identify patients at risk for recurrence after primary treatment for prostate cancer. Urology. 2003;62(Suppl 1):19–35. doi: 10.1016/j.urology.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Wright JL, Lange PH. Newer potential biomarkers in prostate cancer. Rev Urol. 2007;9(4):207–13. [PMC free article] [PubMed] [Google Scholar]
- 52.Shcherbakova PV, Hall MC, Lewis MS, et al. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21(3):940–51. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]