Abstract
The aim of the study was to identify chromosomal regions containing putative genetic variants influencing age-at-onset in familial late-onset Alzheimer’s disease. Data from a genome-wide scan that included genotyping of APOE was analyzed in 1,161 individuals from 209 families of Caribbean Hispanic ancestry with a mean age-at-onset of 73.3 years multiply affected by late-onset Alzheimer’s disease. Two-point and multipoint analyses were conducted using variance component methods from 376 microsatellite markers with an average inter-marker distance of 9.3 cM. Family-based test of association were also conducted for the same set of markers. Age-at-onset of symptoms among affected individuals was used as the quantitative trait. Our results showed that the presence of APOE-ε4 lowered the age-at-onset by three years. Using linkage analysis strategy, the highest LOD scores were obtained using a conservative definition of LOAD at 5q15 (LOD 3.1) 17q25.1 (LOD=2.94) and 14q32.12 (LOD=2.36) and 7q36.3 (LOD=2.29) in covariate adjusted models that included APOE-ε4. Both linkage and family-based association identified 17p13 as a candidate region. In addition, family-based association analysis showed markers at 12q13 (p=0.00002), 13q (p=0.00043) and 14q23 (p=0.00046) to be significantly associated with age at onset. The current study supports the hypothesis that there are additional genetic loci that could influence age-at-onset of late onset Alzheimer’s disease. The novel loci at 5q15, 17q25.1, 13q and 17p13, and the previously reported loci at 7q36.3, 12q13, 14q23 and 14q32 need further investigation.
Keywords: Alzheimer’s disease, age-at-onset, linkage analysis, family-based association analysis, APOE
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of dementia and the most frequent degenerative brain disorder encountered in old age. The risk of developing AD substantially increases after 65 years of age [1]. While the cause of this disease remains unknown, there is evidence for substantial genetic influence [2, 3]. Mutations in three genes: amyloid precursor protein (APP), presenilin 1 and 2 (PSEN 1, PSEN2) result in an early onset, autosomal dominant form of the disease beginning in the third or fourth decade. The ε4 allele of apolipoprotein E (APOE) increases the risk of both sporadic and familial AD occurring later in life around the sixth decade. Each of these genes is involved in the production or processing of the amyloid β peptide, which is deposited in the brain as dense plaques that are characteristic of the disease [2].
Over the past two decades, genetic studies of AD have relied on linkage analysis in multiply affected families, or allelic association studies of candidate genes in cohorts of unrelated AD patients and healthy, elderly controls. It is well known that gene identification for complex genetic traits, such as late-onset Alzheimer’s disease (LOAD), requires far greater efforts than for Mendelian traits [4]. Several factors contribute to the difficulty, including locus heterogeneity, allelic heterogeneity, phenotypic variability as well as influence of non-hereditary factors such as age and sex. Because of the steep increase in prevalence of LOAD with advancing age, studies have used age-at-onset of symptoms as a quantitative trait to identify chromosomal loci related to LOAD [5]. Covariates approaches may be useful in identifying loci in the presence of locus heterogeneity. Variants in APOE provide an example. The number of APOE-ε4 alleles consistently influences age-at-onset of LOAD [6–11].
Daw et al. [12] estimated that there may be as many as four additional loci contributing to the variance in age-at-onset of LOAD, and that the magnitude of the effect from one of these genes could be similar to APOE. Indeed, analyses using age-at-onset of disease as a quantitative trait have yielded multiple putative loci on chromosomes 2, 7, 9, 14, 15, 20 and 21, and some of these loci were also found to be significant in the analyses of AD phenotype [13–19]. However, most of these analyses lack independent confirmation, with the exception of the locus at 14q32[16] which is of interest because of its location near PSEN 1. In the current study, we used variance component (VC) linkage analysis to attempt to confirm previous studies and to identify candidate genetic loci related to age-at-onset of disease in a unique cohort of families of Caribbean Hispanic ancestry multiply affected by LOAD [20, 21].
MATERIALS AND METHODS
Families
We included data from 1,161 individuals from 209 Caribbean Hispanic families genotyped in a family study of Alzheimer’s disease (AD) (Table 1). These families represent 101 families from our first report of the AD genome scan [21], and 108 additional families used in the expanded second genome scan [20]. The recruitment strategy and detailed characteristics of the participants have been described previously [22]. Briefly, multiple sources were used to recruit families. We recruited from clinics in the Dominican Republic and Puerto Rico, as well as the Alzheimer’s Disease Research Center Memory Disorders Clinic at Columbia University in the New York City, which includes a large number of Hispanic patients. To augment family recruitment, we also advertised in local newspapers and media in Dominican Republic, Puerto Rico and New York. In addition, we recruited probands identified in an epidemiological study in northern Manhattan when the informant reported family members with dementia. The pedigrees were of variable complexity. The majority of families consisted of probands with AD and their siblings. However, when more distant relatives of probands -- primarily aunts, uncles or cousins -- were affected with AD, all available relatives from that branch as well as individuals linking the two branches of the pedigree were studied.
Table 1.
Characteristics of Families and Family Members.
| Characteristics | |
|---|---|
| Number of Families | 209 |
| Number of family members examined | 1,161 |
| Number of genotyped individuals per family (range) | 5.6 (2–25) |
| Proportion of women (%) | 65.9 |
| Mean age of onset of AD (years) | |
| Probable and Possible | 73.3 (±10.48) |
| Probable only | 73.6 (±10.46) |
| Affection status | |
| Probable and Possible LOAD (n) | 50.4% (585) |
| Unaffected | 42.2% (490) |
| Unknown (ambiguous) | 7.4% (86) |
| Probable LOAD Only | 38.7% (449) |
| Unaffected | 42.3% (491) |
| Unknown (ambiguous) | 19.0% (221) |
| Number of Families | |
| Total (Probable & Possible) | 209 |
| Families with ≥ 5 affecteds | 20 |
| Families with 4 affecteds | 22 |
| Families with 3 affecteds | 47 |
| Families with 2 affecteds | 98 |
| Families with 1 affected | 22 |
| Total (Probable Only)3 | 202 |
| Families with ≥ 5 affecteds | 11 |
| Families with 4 affecteds | 10 |
| Families with 3 affecteds | 33 |
| Families with 2 affecteds | 97 |
| Families with 1 affected | 51 |
| APOE - ε4 genotype frequency | |
| LOAD (probable and possible) | 27.0% |
| Unaffected | 19.0% |
| Age-at-onset APOE – ε4 (years) | |
| Homozygous – ε4 | 67.5 ± 8.9 |
| Heterozygous – ε4 | 72.7 ± 8.9 |
| No ε4 | 75.3 ± 11.0 |
The Institutional Review Board of Columbia Presbyterian Medical Center and Columbia University Health Sciences and the Bioethics National Committee for Research in the Dominican Republic approved the study. We obtained informed consent from the participant directly or from a family member (surrogate) when the individual had dementia.
Diagnosis
A physician, typically a gerontologist, internist or neurologist (RL, MM, RM), examined all patients and participating family members and obtained blood at the time of examination. To be included in the study, the proband and a living sibling were required to meet National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association criteria for probable or possible AD [23]. These two definitions differ only by the allowance of features in persons with the diagnosis of possible AD that could have contributed to the dementia but were deemed not to be the primary cause (e.g. thyroid disease). The Clinical Dementia Rating Scale (CDR) was used to rate disease severity [24]. Brain imaging and other laboratory studies were reviewed when they were available, and offered when medically required for diagnosis. The battery of neuropsychological tests used has been developed and evaluated extensively [25–27]. All diagnoses were established by a group of physicians and neuropsychologists at a consensus conference. Eleven patients died during the course of the study and subsequent autopsies confirmed the clinical diagnoses in each person.
We used two definitions of affection status for analysis. First, we considered patients with the diagnosis of either possible or probable LOAD as “affected”. In subsequent analyses we attempted to reduce phenotypic heterogeneity by considering only the patients with the diagnosis of probable AD as “affected”. For this analysis, individuals with possible AD were classified as unknown, which reduced the number of affected individuals by approximately 20%. For both definitions, individuals with other forms of dementia or mild cognitive impairment were also considered unknown for the purposes of analysis.
Age-at-Onset
For the current study, we conducted linkage analysis using age-at-onset of symptoms of dementia. This variable was derived from a semi-structured interview with the patient’s informant. The spouse or offspring of the proband, usually the most knowledgeable concerning the proband or affected relative, was chosen as the informant and asked which of the following symptoms first appeared: changes in memory, language, behavior or personality. When the information was available, we used the reported age at which the symptoms first appeared. When the informant was unavailable, we used the age at the date of examination when the diagnosis of AD was made. To be conservative, we used the age at onset phenotype of affected individuals only and treated the age at onset phenotype of unaffected individuals as unknown; however, we used the genotype data of all family members regardless of their affection status to maximize the genotype information.
Covariates
Sex was included as a covariate because of the higher frequency of women than men surviving into old age. Education, as determined by the number of years of formal education including college, technical or graduate school, was also included because low education is associated with AD [28].
Genotyping
A total of 376 autosomal microsatellite markers from the Marshfield Screening Set 13 with an average inter-marker distance of 9.3 cM were genotyped at the Center for Medical Genetics, Marshfield Medical Research Foundation. Marker heterozygosity ranged from 0.53 to 0.92 with an average of 0.77. The Marshfield Medical Research Foundation (http://research.marshfieldclinic.org/genetics/) provided chromosomal locations for the microsatellite markers. We confirmed locus order and inter-marker distance by comparing with either Ensembl (http://www.ensembl.org/index.html) or the UCSC Genome Browser (http://genome.ucsc.edu/). APOE genotyping was carried out as previously described with slight modifications [29, 30]. Genotyping for the PSEN1 Gly206Ala mutation, which we previously described as a founder mutation in this population, was performed as in Athan et al. [31].
Statistical and Genetic Analysis
To determine the effect of APOE genotype on the age-at-onset, we contrasted individuals by the number of the APOE-ε4 alleles (2, 1 or 0) using generalized estimating equations [32, 33]. This method allowed each family to be analyzed as a cluster and circumvented the problem of non-independence. We then adjusted for education and sex as covariates. Quality control procedures (i.e., non-paternity check, Mendelian errors, Hardy-Weinberg equilibrium, etc.) were described in detail in our earlier study [34]
Two-point and multipoint analyses of microsatellite markers were conducted using the variance component (VC) method implemented in SOLAR (version 2.1.4). In SOLAR, heritability is first estimated in a polygenic model, including covariates followed by the general pedigree VC approach developed by Amos [35] extended to use multipoint identity-by-descent (IBD) estimates to localize quantitative trait locus (QTL) [36] . The VC model compares the likelihood under the null hypothesis of no additive genetic variance due to a QTL (σ2 q) against the alternative hypothesis that estimates the variance due to the QTL. Twice the difference in the natural log likelihood of these two models yields a test statistic that is asymptotically distributed as a 50:50 mixture of a χ2 variable and a point mass of zero [36]. The maximum likelihood estimates of the allele frequencies were calculated using all marker data from the Caribbean Hispanic families. We corrected for ascertainment by designating probands and computing likelihoods conditional on the proband. Correction by exclusion of the initial proband has been found to be an effective means to correct ascertainment bias, and can improve power to detect linkage [37].
Age-at-onset of dementia was analyzed as a continuous variable under two definitions as described above: either probable or possible AD as affected; then probable AD only. Genome-wide linkage analyses were limited to affected individuals and adjusted by education and sex as well as their product term. We then performed the analysis also allowing the incorporation of APOE ε4 as an additional covariate.
For markers with a LOD score of one or higher in the two-point analysis, we computed empirical P values to limit the possibility of false positives, using the genotyping information and marker allele frequencies from the data to simulate genotypes under the assumption of no linkage. Each simulation consisted of 10,000 replicates. In each replicate, a fully-informative marker, completely unlinked to the trait, was simulated, and the actual trait (age-at-onset) was then tested against the simulated genotype for the marker to estimate the likelihood of a false positive finding for the observed LOD score. All computations for empirical p-values were conducted using SOLAR.
Single-point family-based association was assessed using an additive genetic model with the null hypothesis of no linkage and no association, as implemented in FBAT version 1.7.3 [38, 39]. Allele frequencies were estimated by FBAT using the EM algorithm.
RESULTS
Demographics
Among the 1,161 family members in the 209 families, 65.9% were women (Table 1). There were approximately 6 individuals per family. The average age of onset was 73.3 years (SD=10.6; range=30–98 years), and 50.4% (n=585) of the participants met criteria for either probable or possible LOAD [23]. The majority (77%) of affected individuals met criteria for probable LOAD. In most families, there were two affected individuals but 43% of the families had three or more affected. In 7.4% of family members, no consensus diagnosis could be made or other forms of dementia were present. For these individuals, the phenotype was coded as unknown, but their genetic data were used to estimate allele frequencies and to establish identity by descent.
APOE
The presence of APOE-ε4 had clear effects on the age-at-onset of LOAD (Table 1). Individuals homozygous for APOE-ε4 had onset of symptoms on average six years earlier than those without this allele (69.1 years vs. 75.0 years). Thus, separate analyses were performed taking into account the presence of APOE-ε4 in the VC linkage analyses.
Linkage analysis
The most significant two-point LOD of 2.70 (pempirical=0.0025) for probable and possible AD was obtained at chromosome 7q36 with APOE adjustment (Table 2). Without APOE adjustment, the LOD score at this position significantly lowered to 1.95. Through all the analyzed markers we observed that inclusion of APOE as covariate in the two-point analysis resulted in increased LOD scores. Particularly, markers at 17q25, 14q32 and 5q15 yielded LOD scores of 2.58, 2.46 and 2.38, respectively (0.0016<pempirical<0.0064) under the APOE adjusted model.
Table 2.
LOD scores from the two-point variance component linkage analysis*
| chr | Marker | Cytogenetic Location | cM | Probable & Possible | Probable only | ||
|---|---|---|---|---|---|---|---|
| APOE Unadjusted | APOE ε4+ | APOE Unadjusted | APOE ε4+ | ||||
| 1 | TATC028 | 1q24 | 184.6 | 1.84 | 1.56 | 1.61 | 1.36 |
| 2 | D2S1394 | 2p13 | 90.8 | 1.53 | 1.18 | 1.42 | 1.43 |
| 2 | D2S1328 | 2q14 | 132.6 | 1.57 | 1.28 | ||
| 2 | D2S1391 | 2q32 | 186.2 | 1.36 | 1.22 | ||
| 3 | D3S1768 | 3p22 | 62 | 1.43 | 1.04 | 1.23 | 1.07 |
| 4 | GATA70E01 | 4p15 | 35 | 1.79 | 1.76 | 1.39 | 1.28 |
| 5 | D5S1501 | 5q14 | 85 | 1.06 | 1.17 | 1.33 | 1.69 |
| 5 | D5S1462 | 5q15 | 105 | 1.80 | 2.38 | 2.72 | 3.11 |
| 5 | D5S2501 | 5q22 | 117 | 1.55 | 1.35 | 1.95 | 2.18 |
| 5 | D5S1480 | 5q32 | 147.0 | 1.05 | 1.07 | ||
| 6 | ATTT030 | 6p24 | 17.9 | 0.84 | 1.03 | ||
| 6 | GATA184A08 | 6q24 | 146.0 | 0.99 | 1.23 | ||
| 7 | GATA104 | 7q35 | 155.0 | 1.55 | 1.24 | ||
| 7 | D7S3070 | 7q36 | 163 | 1.85 | 1.36 | 2.07 | 1.57 |
| 7 | D7S3058 | 7q36 | 174 | 1.38 | 1.18 | 1.87 | 1.67 |
| 7 | MFD442GTTT002 | 7q36 | 178.6 | 1.95 | 2.70 | 1.77 | 2.29 |
| 13 | D13S787 | 13q12 | 9.0 | 1.30 | 1.26 | ||
| 14 | D14S617 | 14q32 | 106 | 2.25 | 2.46 | 2.60 | 2.36 |
| 16 | TTAT023Z | 16p12 | 38.4 | 1.25 | 1.27 | ||
| 16 | D16S3253 | 16q12 | 72 | 1.60 | 1.04 | ||
| 16 | AAT107 | 16q21 | 81.1 | 1.08 | 1.04 | ||
| 17 | G10693 | 17p13 | 14.7 | 1.67 | 1.49 | ||
| 17 | D17S1301 | 17q25 | 100 | 2.63 | 2.58 | 2.71 | 2.94 |
| 17 | AAT095 | 17q26 | 119.5 | 1.27 | 1.05 | ||
| 18 | D18S542 | 18p11 | 41 | 1.87 | 1.55 | 1.36 | 1.43 |
| 21 | D21S1446 | 21q22 | 57.8 | 0.82 | 1.08 | ||
Individuals with probable AD and probable or possible AD were considered affected. Only markers with LOD score above threshold of 1 are shown.
We then repeated the two-point analysis using probable AD only (Table 2). As with the results from the previous analysis of probable and possible AD, the same four loci showed support for linkage. Under the model without APOE adjustment, the most significant linkage was observed at 5q15 with a LOD score of 2.72. Additional three loci, namely 17q25, 14q32, and 7q36, showed suggestive linkage with LOD scores of 2.71, 2.60, and 1.77, respectively. With the APOE adjustment, the most significant LOD score of 3.11 was observed for 5q15, with a flanking marker at 5q22 reaching a LOD score of 2.18. The remaining three loci had elevated LOD scores of 2.94 for 17q25, 2.36 for 14q32 and 2.29 for 7q36.
Because the marker at 14q32 yielded a significant LOD and is proximal to PSEN, we screened one affected person from each family for a PSEN1 mutation, Gly206Ala, specific to Caribbean Hispanics [31]. From the screen, we identified six individuals with this mutation who had an average age-at-onset of 57 years. We then re-analyzed this region after removing the families with the Gly206Ala mutation to determine whether the linkage signal at 14q32 can be explained by these families with the mutation. For the probable and possible AD definition, LOD score at 14q32 increased from 2.46 to 2.93 for an APOE-ε4 adjusted analysis. When restricted to probable AD, the LOD score increased from 2.36 to 2.83 for an APOE adjusted analysis. These observations suggest that observed the linkage at 14q32 was unlikely to be related to the Gly206Ala PSEN1 mutation.
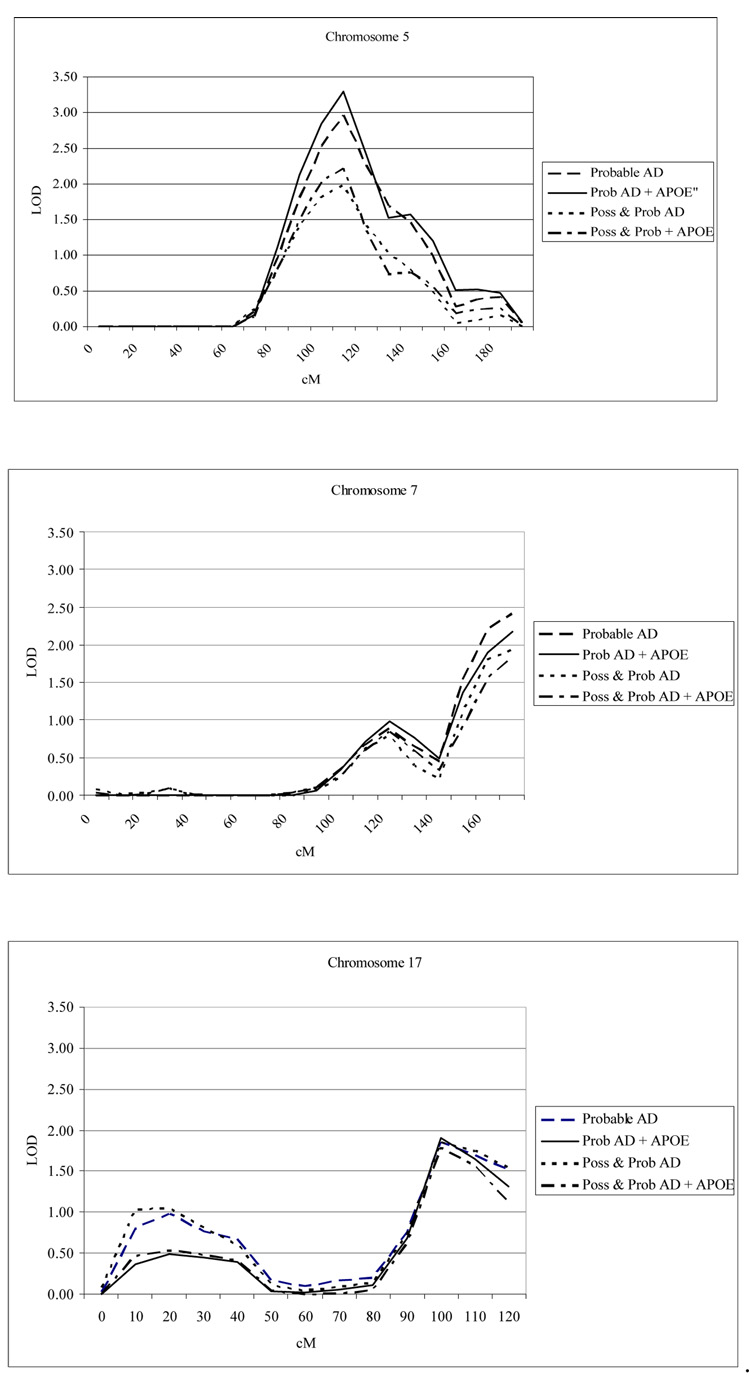
The multipoint analysis in general yielded stronger evidence for linkage using probable AD than probable and possible LOAD as the definition of “affected” (Figures 1a, b and c). For probable LOAD, the strongest evidence for linkage was observed at 5q15 (LOD=2.96 for unadjusted and 3.29 for APOE adjusted), followed by 7q36 (LOD 2.41 for unadjusted and 2.17 for APOE adjusted) and 17q25 (1.86 for unadjusted and 1.9 for APOE adjusted). Comparable, but somewhat weaker, LOD scores were observed for probable and possible AD. The highest LOD score was 1.97 (P=0.0013) at chromosome 5q15, and when adjusted for the APOE-ε4 allele the LOD score increased to 2.22 (P=0.0007). The multipoint LOD scores for 7q36 and 17q25 were suggestive (1.75 to 1.92) and declined somewhat when adjusted for the effect of APOE-ε4
Figure 1.
a–c. Illustrative multipoint variance component linkage analysis for chromosomes 5, 7, and 17. For Model 2 (using affected members) under two different definitions of affection, the results are presented for the multipoint linkage analysis with and without APOE adjustment.
Lastly, our family-based association analysis showed that marker g10693 at 17p13 was significant in both linkage analysis (p=0.009) and association analysis (p=0.002). In addition, several markers showed significant evidence for association with age at onset (Table 3). The most significant results were obtained at markers AGAT113Z (13q, p<0.001), D12S297 (12q13, p<0.001) and D14S592 (14q23, p<0.001). Microsatellite marker D19S245, located close to APOE, showed a significant association (p=0.001) with age at onset.
Table 3.
Summary of results of family-based test of association*
| chr | Marker | Cytogenetic Location |
cM | p-value |
|---|---|---|---|---|
| 1 | D1S1594 | 1q43 | 265.5 | 0.004 |
| 2 | D2S1334 | 2q21 | 145.1 | 0.006 |
| 2 | D2S2968 | 2q37 | 251.9 | 0.010 |
| 3 | D3S3038 | 3p24 | 44.8 | 0.009 |
| 4 | D4S2397 | 4p15 | 43 | 0.004 |
| 4 | TAGA006 | 4q27 | 124.6 | 0.003 |
| 4 | ATT077P | 4q35 | 195.4 | 0.001 |
| 6 | D6S1056 | 6q16 | 103 | 0.004 |
| 6 | D6S1277 | 6q26 | 173 | 0.007 |
| 7 | D7S2477 | 7p22 | 0 | 0.006 |
| 7 | D7S2204 | 7q21 | 91 | 0.009 |
| 7 | D7S1799 | 7q22 | 114 | 0.001 |
| 7 | D7S1804 | 7q32 | 137 | 0.006 |
| 8 | ATT070 | 8p22 | 30 | 0.004 |
| 10 | D10S1222 | 10q26 | 156 | 0.004 |
| 11 | ATA34E08N | 11p14 | 33 | 0.007 |
| 12 | D12S297 | 12q13 | 65 | 0.00002 |
| 12 | D12S2078 | 12q24 | 150 | 0.004 |
| 13 | AGAT113Z | 13q33.3 | 107.9 | 0.00043 |
| 14 | D14S750 | 14q23 | 60 | 0.003 |
| 14 | D14S592 | 14q23 | 67 | 0.00046 |
| 16 | AAT107 | 16q21 | 81.1 | 0.003 |
| 17 | G10693 | 17p13 | 14.7 | 0.002 |
| 17 | D17S2193 | 17q24 | 89 | 0.003 |
| 19 | D19S245 | 19q13 | 78 | 0.001 |
| 22 | D22S1045 | 22q12 | 43 | 0.002 |
Probable or possible AD was considered affected.
DISCUSSION
We report five candidate susceptibility loci for age at onset of AD phenotype based on a genome wide search of Caribbean Hispanic families with multiple members affected with AD. The five candidate loci include 5q15, 7q36.3, 14q32.12, 17q25.1, and 17p13, and two of these loci (7q36.3 and 14q32.12) were previously reported.
With few exceptions [13, 16], reports of genome-wide scans of familial AD have relied on the clinical diagnosis as the trait of interest [5]. However, Li et al. [14, 40] argued that the age-at-onset of disease is equally important because the “regulation of onset could make it possible to delay onset beyond an individual’s normal life span.” Indeed, allelic variation in APOE profoundly affects risk by lowering the age-at-onset of AD between three to five years with each copy of the ε4 variant [6, 8, 9]. The likelihood that variants in other genes could have similar effects on the age-at-onset was strengthened by a study finding that as many as four additional loci contribute to the variance in age-at-onset of LOAD [12]. The ability to identify these genes depends not only on the number of such genes but also the magnitude of their contribution to the age-at-onset. Naturally, gene identification would become increasingly difficult as the number of genes grows larger and their effect size smaller. The past few years of research would support this view, because a large number of loci have been implicated and are currently the subject of intensive investigation.
No single method is superior in identifying genetic factors underlying complex diseases; thus, we performed linkage and family based associated analysis using the same set of 9.3 cM spaced microsatellite markers covering the entire genome. In our previous study of LOAD [20], seven loci were identified with LOD scores greater than 2.0, but the highest LOD score was 3.1 at chromosome 3q28. In the current study of age-at-onset, we observed weak to modest linkage or association for several markers that were also significant in our previous study of AD linkage genome scan. These include 3p22, 4p15, 7p22, 14q23, and 17q25. More importantly, we identified several markers that were not previously observed in the LOAD scan, suggesting that the results may are related to the different phenotypes.
Avramopoulos and colleagues [41] reported significant evidence for linkage near PSEN1 at 14q24 related to a phenotype that included psychosis. Although 14q32.12 is some distance away (~33 cM) from 14q24 and the PSEN1 gene, we as others [16] observed significant linkage at this locus. Further, we found a significant association with marker locus D14S592 at 14q23, located 5 cM away from the PSEN1 gene. The linkage signal remained strong after excluding the families carrying the Gly206Ala mutation in the PSEN1 gene, suggesting presence of additional genetic locus in this region.
We found suggestive evidence of linkage at 7q36.3 that deserves further investigation. Previously, Rademakers et al [17] reported that LOAD phenotype was linked to 7q36. In one family, they identified a mutation in PAX transcription activation domain interacting protein, but it is unclear as to the role of this gene in LOAD. Since the highest LOD score on 7q36 was obtained from the most distal marker, additional markers will need to be examined to exclude the possibility of a false positive result.
The family based association tests revealed two chromosomal regions, 12q13 and 14q23, that have been reported in literature as the chromosomal regions that may harbor genes contributing to AD. In addition, marker g10693 at 17p13 was found to be significant under both linkage and association. The convergence of the two complementary strategies provides further support for considering this locus as a candidate region.
We observed only modest overlap between linkage and association results. This is not a contradiction in that linkage and association are techniques that examine two different but related phenomena. Linkage analysis tests the co-segregation of the disease and marker loci within families without reference to any specific allele. Association investigates the co-occurrence of alleles with disease. It is possible to observe an allelic association without evidence of linkage, especially when allele frequency is high. Moreover, association between marker alleles and disease locus can be maintained by factors other than linkage such as demographic history, population admixture, recent mutation or inbreeding. When a disease-associated allele explains only a small portion of the variance even though it occurs more often in affected individuals, power for linkage analysis can be limited [4]. In these situations, family-based association analyses can provide addition statistical power [43]. Therefore, linkage and association tests may be used as complementary approaches when identifying susceptibility loci underlying complex traits.
Several reasons may explain the lack of signal near the APOE locus in our linkage analysis. It has been reported that APOE explains only 4% of the age at onset variability in familiar AD [44] making it difficult to detect. Second, microsatellite markers, spaced at an average distance of 9 cM in a genome wide search, may not have insufficient marker information content for mapping, thereby further reducing statistical power. In addition, the frequency of the ε4 allele in this set of FAD was approximately 30%, indicating that approximately 50% of the individuals will be homozygous. As a result, power for linkage analysis at this locus will be lowered, since co-segregation of a putative allele with disease will be difficult to detect. When using family-based association tests however, we observed modest support for association with D19S245, located approximately 10 cM away from the APOE locus. In a recent paper, Coon et al [45] demonstrated that the APOE locus as a susceptibly locus for late onset AD was identified when they saturate the genome with over 300, 000 SNP markers.
The findings in this analysis are distinct from those in the previous investigations of age-at-onset. One of the main reasons for differences is the fact that the current study examines familial AD based on Caribbean Hispanic ethnicity, while others primarily used Caucasians of European descents. Moreover, each study highlighted different loci for regions harboring susceptibility loci. Olson et al [16] observed evidence for an interaction between 20p12.2 and the APP region (21q21), limited to the oldest age group and to those lacking APOE-ε4 alleles. This finding has been supported in follow-up analyses [46], but we found no evidence for linkage in that region. The closest marker showing any relation to age-at-onset in our dataset was at 20q13.2 (LOD ranging from 1.06 to 1.15; P=0.015). Li et al [14, 40] combined individuals with AD and Parkinson’s disease to identify loci related to the age-at-onset. Loci near chromosome 10q24.32 provided the maximum LOD score, and in a later study [40], these investigators found that single nucleotide polymorphisms in glutathione S-transferase omega-1, GSTO1, were associated with this phenotype. However, other studies have not confirmed this association thus far [47–49]. We did not find evidence for linkage for GSTO1 at 10q26.12 for the age-at-onset phenotype, but found weak evidence for linkage with the AD phenotype [21]. Holmans et al [13] performed an affected sib-pair linkage analysis with age-at-onset, and observed the strongest support for linkage at chromosome 21q21, which was previously reported by Olson and colleagues [16]. In our dataset, locus 21q22 yielded a modest LOD score of 1.08.
No previous study has reported 5q15, 17q25, 13q or 17p13 as loci harboring susceptibility genes influencing age at onset of familiar late onset AD. Among the most interesting candidate genes underlying the linkage peak at 5q15 is calpastatin (CAST, 5q15-q21). Calcium-dependent proteases, calpains, may be involved in the pathogenesis of neurodegenerative diseases such as AD by participating in excitotoxic stress-induced [50, 51]. CAST is a specific natural inhibitor of calpain and can alter neuritic degeneration in rodent models of AD. At 17q25, in addition to two members of the G protein coupled receptor family, galanin receptor 2 (GALR2) is located. GALR2 is expressed in the hypothalamus and hippocampus, and is involved primarily in signaling through the phospholipase C/protein kinase C pathway. Fathi et al [52] demonstrated that GALR2 coupled efficiently to both the Gq and the Gi proteins to simultaneously activate two independent signal transduction pathways. At 17p13 region, MYH8 and TM4SF5 genes could be considered as possible candidate genes because their expression is significantly down regulated in hippocampus in AD brain [53].
Age-at-onset analysis using a genome-wide scan of polymorphic microsatellites suggests that five candidate loci: 5q15, 7q36.3, 14q32.12, 17q25.1 and 17p13, deserve further investigation. Two of these loci have been identified in previous linkage analyses: 7q36.3 and 14q32.12. Given the support for linkage or association or both at 5q15, 7q36.3, 13q, 17p13 and 17q25, fine mapping of these regions is imperative.
ACKNOWLEDGEMENTS
Funding for this project was provided by the National Institutes on Aging, National Institutes of Health R37 AG15473, National Heart, Lung and Blood Institute HV48141 and the Canadian Institutes of Health Research. In addition, support was provided by the Charles S. Robertson Gift for Research on Alzheimer’s Disease from the Banbury fund. We also thank the members of Estudio Familiar de Influencia Genetica en Alzheimer, The Sociedad Dominicana de Geriatria y Gerontologia, The Sociedad Dominicana de Neurologia y Neurocirugia, The Sociedad Dominicana de Psiquiatria and the Associacion Dominicana Alzheimer y Similares, Inc, and The Bioethics National Committee for Research in the Dominican Republic.
REFERENCES
- 1.Nussbaum RL, Ellis CE. Alzheimer's disease and Parkinson's disease. N Engl J Med. 2003;348(14):1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 2.St George-Hyslop PH, Petit A. Molecular biology and genetics of Alzheimer's disease. C R Biol. 2005;328(2):119–130. doi: 10.1016/j.crvi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120(4):545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):20373–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 5.Kamboh MI. Molecular genetics of late-onset Alzheimer's disease. Ann Hum Genet. 2004;68(Pt 4):381–404. doi: 10.1046/j.1529-8817.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 6.Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D, Albert MS, Tanzi R. ApoE-4 and age at onset of Alzheimer's disease: the NIMH genetics initiative. Neurology. 1997;48(1):139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 7.Breitner JC, Jarvik GP, Plassman BL, Saunders AM, Welsh KA. Risk of Alzheimer disease with the epsilon4 allele for apolipoprotein E in a population-based study of men aged 62–73 years. Alzheimer Dis Assoc Disord. 1998;12(1):40–44. doi: 10.1097/00002093-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 9.Harwood DG, Barker WW, Ownby RL, St George-Hyslop P, Mullan M, Duara R. Apolipoprotein E polymorphism and age of onset for Alzheimer's disease in a bi-ethnic sample. Int Psychogeriatr. 2004;16(3):317–326. doi: 10.1017/s104161020400033x. [DOI] [PubMed] [Google Scholar]
- 10.Nicodemus KK, Stenger JE, Schmechel DE, Welsh-Bohmer KA, Saunders AM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA, Martin ER. Comprehensive association analysis of APOE regulatory region polymorphisms in Alzheimer disease. Neurogenetics. 2004;5(4):201–208. doi: 10.1007/s10048-004-0189-9. [DOI] [PubMed] [Google Scholar]
- 11.Wijsman EM, Daw EW, Yu X, Steinbart EJ, Nochlin D, Bird TD, Schellenberg GD. APOE and other loci affect age-at-onset in Alzheimer's disease families with PS2 mutation. Am J Med Genet B Neuropsychiatr Genet. 2005;132(1):14–20. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- 12.Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66(1):196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DeVrieze F, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O'Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135(1):24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 14.Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70(4):985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nacmias B, Latorraca S, Piersanti P, Forleo P, Piacentini S, Bracco L, Amaducci L, Sorbi S. ApoE genotype and familial Alzheimer's disease: a possible influence on age of onset in APP717 Val-->Ile mutated families. Neurosci Lett. 1995;183(1–2):1–3. doi: 10.1016/0304-3940(94)11100-w. [DOI] [PubMed] [Google Scholar]
- 16.Olson JM, Goddard KA, Dudek DM. A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet. 2002;71(1):154–161. doi: 10.1086/341034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, Aulchenko Y, Weckx S, De Pooter T, Van den Broeck M, Corsmit E, De Rijk P, Del-Favero J, van Swieten J, van Duijn CM, Van Broeckhoven C. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77(4):643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wavrant-De Vrieze F, Crook R, Holmans P, Kehoe P, Owen MJ, Williams J, Roehl K, Laliiri DK, Shears S, Booth J, Wu W, Goate A, Chartier-Harlin MC, Hardy J, Perez-Tur J. Genetic variability at the amyloid-beta precursor protein locus may contribute to the risk of late-onset Alzheimer's disease. Neurosci Lett. 1999;269(2):67–70. doi: 10.1016/s0304-3940(99)00417-6. [DOI] [PubMed] [Google Scholar]
- 19.Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75(3):398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, Medrano M, Arriaga A, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Expanded Genome-Wide Scan Implicates a Novel Locus at 3q28 Among Caribbean Hispanics with Familial Alzheimer's Disease. Archives of Neurology. 2006;63(11):1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Mayeux R, Mayo D, Mo J, Santana V, Williamson J, Flaquer A, Ciappa A, Rondon H, Estevez P, Lantigua R, Kawarai T, Toulina A, Medrano M, Torres M, Stern Y, Tycko B, Rogaeva E, St George-Hyslop P, Knowles JA. Fine mapping of 10q and 18q for familial Alzheimer's disease in Caribbean Hispanics. Mol Psychiatry. 2004;9(11):1042–1051. doi: 10.1038/sj.mp.4001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romas SN, Santana V, Williamson J, Ciappa A, Lee JH, Rondon HZ, Estevez P, Lantigua R, Medrano M, Torres M, Stern Y, Tycko B, Mayeux R. Familial Alzheimer disease among Caribbean Hispanics: a reexamination of its association with APOE. Arch Neurol. 2002;59(1):87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 23.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 24.Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 25.Pittman J, Andrews H, Tatemichi T, Link B, Struening E, Stern Y, Mayeux R. Diagnosis of dementia in a heterogeneous population. A comparison of paradigm-based diagnosis and physician's diagnosis. Arch Neurol. 1992;49(5):461–467. doi: 10.1001/archneur.1992.00530290043010. [DOI] [PubMed] [Google Scholar]
- 26.Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, Mayeux R. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49(5):453–460. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- 27.Stricks L, Pittman J, Jacobs DM, Sano M, Stern Y. Normative data for a brief neuropsychological battery administered to English- and Spanish-speaking community-dwelling elders. J Int Neuropsychol Soc. 1998;4(4):311–318. [PubMed] [Google Scholar]
- 28.Ganguli M, Dodge HH, Chen P, Belle S, DeKosky ST. Ten-year incidence of dementia in a rural elderly US community population: the MoVIES Project. Neurology. 2000;54(5):1109–1116. doi: 10.1212/wnl.54.5.1109. [DOI] [PubMed] [Google Scholar]
- 29.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–548. [PubMed] [Google Scholar]
- 30.Maestre G, Ottman R, Stern Y, Gurland B, Chun M, Tang MX, Shelanski M, Tycko B, Mayeux R. Apolipoprotein E and Alzheimer's disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37(2):254–259. doi: 10.1002/ana.410370217. [DOI] [PubMed] [Google Scholar]
- 31.Athan ES, Williamson J, Ciappa A, Santana V, Romas SN, Lee JH, Rondon H, Lantigua RA, Medrano M, Torres M, Arawaka S, Rogaeva E, Song YQ, Sato C, Kawarai T, Fafel KC, Boss MA, Seltzer WK, Stern Y, St George-Hyslop P, Tycko B, Mayeux R. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. Jama. 2001;286(18):2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 32.Preisser JS, Koch GG. Categorical data analysis in public health. Annu Rev Public Health. 1997;18:51–82. doi: 10.1146/annurev.publhealth.18.1.51. [DOI] [PubMed] [Google Scholar]
- 33.Zeger SL, Liang KY. An overview of methods for the analysis of longitudinal data. Stat Med. 1992;11(14–15):1825–1839. doi: 10.1002/sim.4780111406. [DOI] [PubMed] [Google Scholar]
- 34.Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, Medrano M, Arriaga A, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean hispanics with familial Alzheimer disease. Arch Neurol. 2006;63(11):1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- 35.Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54(3):535–543. [PMC free article] [PubMed] [Google Scholar]
- 36.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comuzzie AG, Williams JT. Correcting for ascertainment bias in the COGA data set. Genet Epidemiol. 1999;(17 Suppl 1):S109–S114. doi: 10.1002/gepi.1370170719. [DOI] [PubMed] [Google Scholar]
- 38.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9(4):301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 39.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50(4):211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 40.Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, Hauser MA, Scott WK, Small GW, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Goetz CG, Mastaglia F, Middleton LT, Roses AD, Saunders AM, Schmechel DE, Gullans SR, Haines JL, Gilbert JR, Vance JM, Pericak-Vance MA, Hulette C, Welsh-Bohmer KA. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12(24):3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 41.Avramopoulos D, Fallin MD, Bassett SS. Linkage to chromosome 14q in Alzheimer's disease (AD) patients without psychotic symptoms. Am J Med Genet B Neuropsychiatr Genet. 2005;132(1):9–13. doi: 10.1002/ajmg.b.30074. [DOI] [PubMed] [Google Scholar]
- 42.Greenberg DA, Doneshka P. Partitioned association-linkage test: distinguishing "necessary" from "susceptibility" loci. Genet Epidemiol. 1996;13(3):243–252. doi: 10.1002/(SICI)1098-2272(1996)13:3<243::AID-GEPI2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405(6788):847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 44.Tunstall N, Owen MJ, Williams J, Rice F, Carty S, Lillystone S, Fraser L, Kehoe P, Neill D, Rudrasingham V, Sham P, Lovestone S. Familial influence on variation in age of onset and behavioural phenotype in Alzheimer's disease. Br J Psychiatry. 2000;176:156–159. doi: 10.1192/bjp.176.2.156. [DOI] [PubMed] [Google Scholar]
- 45.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68(4):613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 46.Goddard KA, Olson JM, Payami H, van der Voet M, Kuivaniemi H, Tromp G. Evidence of linkage and association on chromosome 20 for late-onset Alzheimer disease. Neurogenetics. 2004;5(2):121–128. doi: 10.1007/s10048-004-0174-3. [DOI] [PubMed] [Google Scholar]
- 47.Kolsch H, Linnebank M, Lutjohann D, Jessen F, Wullner U, Harbrecht U, Thelen KM, Kreis M, Hentschel F, Schulz A, von Bergmann K, Maier W, Heun R. Polymorphisms in glutathione S-transferase omega-1 and AD, vascular dementia, and stroke. Neurology. 2004;63(12):2255–2260. doi: 10.1212/01.wnl.0000147294.29309.47. [DOI] [PubMed] [Google Scholar]
- 48.Nishimura M, Sakamoto T, Kaji R, Kawakami H. Influence of polymorphisms in the genes for cytokines and glutathione S-transferase omega on sporadic Alzheimer's disease. Neurosci Lett. 2004;368(2):140–143. doi: 10.1016/j.neulet.2004.06.076. [DOI] [PubMed] [Google Scholar]
- 49.Ozturk A, Desai PP, Minster RL, Dekosky ST, Kamboh MI. Three SNPs in the GSTO1, GSTO2 and PRSS11 genes on chromosome 10 are not associated with age-at-onset of Alzheimer's disease. Neurobiol Aging. 2005;26(8):1161–1165. doi: 10.1016/j.neurobiolaging.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Higuchi M, Iwata N, Saido TC. Understanding molecular mechanisms of proteolysis in Alzheimer's disease: progress toward therapeutic interventions. Biochim Biophys Acta. 2005;1751(1):60–67. doi: 10.1016/j.bbapap.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Higuchi M, Tomioka M, Takano J, Shirotani K, Iwata N, Masumoto H, Maki M, Itohara S, Saido TC. Distinct mechanistic roles of calpain and caspase activation in neurodegeneration as revealed in mice overexpressing their specific inhibitors. J Biol Chem. 2005;280(15):15229–15237. doi: 10.1074/jbc.M500939200. [DOI] [PubMed] [Google Scholar]
- 52.Fathi Z, Battaglino PM, Iben LG, Li H, Baker E, Zhang D, McGovern R, Mahle CD, Sutherland GR, Iismaa TP, Dickinson KE, Zimanyi IA. Molecular characterization, pharmacological properties and chromosomal localization of the human GALR2 galanin receptor. Brain Res Mol Brain Res. 1998;58(1–2):156–169. doi: 10.1016/s0169-328x(98)00116-8. [DOI] [PubMed] [Google Scholar]
- 53.Taguchi K, Yamagata HD, Zhong W, Kamino K, Akatsu H, Hata R, Yamamoto T, Kosaka K, Takeda M, Kondo I, Miki T. Identification of hippocampus-related candidate genes for Alzheimer's disease. Ann Neurol. 2005;57(4):585–588. doi: 10.1002/ana.20433. [DOI] [PubMed] [Google Scholar]