Abstract
The Drosophila eggshell provides an in vivo model system for extracellular matrix assembly, in which programmed gene expression, cell migrations, extracellular protein trafficking, proteolytic processing, and cross-linking are all required to generate a multi-layered and regionally complex architecture. While abundant structural components of the eggshell are known and are being characterized, less is known about non-abundant structural, regulatory, and enzymatic components that are likely to play critical roles in eggshell assembly. We have used sensitive mass spectrometry-based analyses of fractionated eggshell matrices to validate six previously predicted eggshell proteins and to identify eleven novel components, and have characterized the expression patterns of many of their mRNAs. Among these are several putative structural or regulatory (non-enzymatic) proteins, most larger in mass than the major eggshell proteins and often showing preferential expression in follicle cells overlying specific structural features of the eggshell. Of particular note are the putative enzymes, some likely to be involved in matrix cross-linking (two yellow family members previously implicated in eggshell integrity, a heme peroxidase, and a small-molecule oxidoreductase) and others possibly involved in matrix proteolysis or adhesion (proteins related to cathepsins B and D). This work provides a framework for future molecular studies of eggshell assembly.
Keywords: Drosophila, eggshell assembly, mass spectrometry, gene expression, vitelline membrane, chorion
Introduction
The Drosophila eggshell provides a remarkable in vivo model for processes involved in assembly of complex extracellular matrix architectures (reviewed by Waring, 2000). Over the last 30 hours of oogenesis, somatic follicle cells overlying the oocyte secrete a sequence of eggshell components that together form five eggshell layers, including the oocyte-proximal vitelline membrane, the lipid wax layer, the crystalline inner chorionic layer (ICL), the tripartite endochorion containing a floor, pillars, and roof, and the non-proteinaceous exochorion (Margaritis et al., 1980). This sequential secretion involves precise regulation of gene expression and, in the case of the major chorion genes, gene amplification to enable synthesis of required quantities of chorion proteins in less than 5 hours (Petri et al., 1976; Spradling and Mahowald, 1980). Eggshell morphogenesis also involves follicle cell migrations, of main body follicle cells to form a continuous columnar epithelium over the oocyte (Schulz et al., 1993; Zarnescu and Thomas, 1999), of border cells that participate in forming an anterior sperm-entry structure, the micropyle (Montell et al., 1992), and of dorsal anterior follicle cells that migrate out from the oocyte while synthesizing the paired respiratory appendages (Dorman et al., 2004). Cell signaling between the follicle cells and oocyte is important for these migration events, as well as for establishing and maintaining polarity of the oocyte and eggshell (for reviews, see Dobens and Raftery, 2000; Montell, 2003; van Eeden and St. Johnston, 1999). A role for the eggshell in embryonic patterning has also been shown by the anchoring in the vitelline membrane of Torsolike, a spatial cue involved in patterning the embryo termini (Stevens et al., 2003); similar anchoring has been suggested, but not shown, for an as-yet-unknown spatial cue involved in establishing the embryonic dorsoventral axis (Anderson et al., 1992).
Despite the orderly secretion first of vitelline membrane proteins (sV23, sV17, VM32E, VM34C) in the mid-oogenesis Stages 8−10, and then chorion proteins – these divided into “early” (s36, s38 in Stages 11−12), “middle” (s16, s19 in Stage 13), and “late” (s15, s18 in Stage 14) classes – immunolocalization and Western blot studies have revealed a surprising complexity in what happens to these proteins after their secretion. These events include temporally regulated trafficking of proteins between layers and their proteolytic processing after deposition. For example, a large proportion of the secreted s36 chorion protein initially localizes to the vitelline membrane, with only a small fraction present over the forming chorion, then late in oogenesis it cannot be detected in the vitelline membrane and instead is distributed throughout the endochorion (Pascucci et al., 1996); similar behavior is seen for the s36 and s38 homologues in D. virilis (Trougakos and Margaritis, 1998). Conversely, while the vitelline membrane proteins sV23 and sV17 appear to localize exclusively to the vitelline membrane (Pascucci et al., 1996), VM32E shows partial relocalization to the inner chorionic layer and endochorion late in oogenesis (Andrenacci et al., 2001). The major chorion proteins do not undergo proteolytic processing, while the sV23 and sV17 vitelline membrane proteins do appear to undergo sequential N- and C-terminal processing after their secretion (Manogaran and Waring, 2004; Pascucci et al., 1996). Products of the complex dec-1 (defective chorion-1) locus, required for formation of an organized endochorion, show the most dramatic range of complex extracellular regulation (Nogueron et al., 2000). Following sequential processing of the major fc106 isoform within the vitelline membrane, some products re-localize to the chorion while others remain in the vitelline membrane. The late-expressed fc177 isoform also appears to be processed in the vitelline membrane, and its cleavage products re-localize to the chorion or to the oocyte cytoplasm. The enzymes that carry out all of these proteolytic processing events are not known, nor is it known what protein-protein interactions are responsible for the dynamic trafficking of eggshell proteins or their cleavage products between eggshell layers.
The chorion and vitelline membrane are additionally stabilized by covalent cross-linking, which renders the major eggshell proteins completely insoluble in laid eggs. The vitelline membrane proteins are completely cross-linked by disulfide bonds by Stage 12 (sV17, sV23) or by Stage 14 (VM32E), then are believed to be further dityrosine cross-linked by a peroxidase-type enzyme following ovulation (Andrenacci et al., 2001; Heifetz et al., 2001; LeMosy and Hashimoto, 2000; Pascucci et al., 1996; Petri et al., 1979). The chorion proteins remain soluble until Stage 14 and then undergo both disulfide- and dityrosine-based cross-linking rendering these proteins insoluble prior to ovulation (Mindrinos et al., 1980; Pascucci et al., 1996). Histochemical and antibody staining for peroxidases has been demonstrated in the endochorion but not the vitelline membrane (Keramaris et al., 1991; Konstandi et al., 2005; Mindrinos et al., 1980), and follicle cells have been shown to secrete the required substrate H2O2 in concert with chorion hardening (Margaritis, 1985). The Drosophila Pxd (Peroxidase) gene has recently been shown to be expressed in follicle cells and is proposed to encode the chorion peroxidase (Konstandi et al., 2005), yet it is unclear whether it is the only such enzyme involved in eggshell hardening or how the vitelline membrane undergoes dityrosine cross-linking at a distinct time and without apparent histochemical or antibody evidence for a peroxidase in this layer.
There is clearly much to learn about how the major eggshell proteins function in the assembly of the eggshell layers, as well as about regulation of their localization, interactions, processing, and cross-linking. In this work, we have utilized sensitive mass spectrometry methods, in combination with the completed Drosophila genome sequence (Adams et al., 2000), to identify minor proteins associated with the eggshell matrix that may play important structural or regulatory roles in eggshell biogenesis. These methods validate some previous assignments of predicted eggshell proteins based upon the presence of their genes in proximity to known eggshell genes, but also identify several additional structural proteins, enzymes, and enzyme-related proteins. RNA expression analysis provides support for these new species being authentic eggshell components, and reveals an even greater complexity of temporal and spatial regulation of gene expression than previously described, likely reflecting subtleties of function within this group of non-abundant eggshell proteins.
Methods
Flies
Fly strains used for preparation of eggshell matrices for 2-D gel analysis included Oregon R and w1118, which gave nearly identical spot patterns on 2-D gels. For LC-MS/MS and RNA in situ hybridization studies, the w1118 strain was used. Typically, young females (1 − 4 days after eclosion) were incubated in heavily yeasted vials with males present for 2 − 3 days at room temperature to stimulate development of mid- to late-stage egg chambers in which eggshell synthesis is occurring.
Eggshell matrix preparations
Ovaries, rather than embryos, were used as source material in this study because tyrosine-based cross-linking of the eggshell at the end of oogenesis and upon ovulation renders eggshell proteins insoluble, making 2-D gel analysis impossible, and introduces inter-peptide bonds that would complicate identification of unknown proteins by the trypsinization and LC-MS/MS approach. Ovaries were hand-dissected away from other fly tissues in cold Drosophila Ringer's buffer (Verheyen and Cooley, 1994), and transferred to microfuge tubes on ice. Only 50 ovary pairs were placed in each tube to allow thorough washing, and 150−300 ovary pairs were harvested and processed for each 2-D gel sample, using a modification of an eggshell preparation protocol described by Fargnoli and Waring (1982). Ovaries were homogenized in lysis buffer (50 mM NaPO4, pH 6.9, 2% Triton-X 100, 0.4 M NaCl, plus protease inhibitor cocktail), pelleted at 100 g, and supernatant discarded. The low-speed pellet was washed 3 times with 1 ml lysis buffer (twice re-homogenizing with Kontes mini-pestle to ensure thorough breakage of all egg chambers) and twice with low-salt wash buffer (10 mM NaPO4, pH 6.9, 50 mM NaCl, protease inhibitors), each time pelleted at 100 g except for the last spin, which was at 9300 g. The low speed spins enable pelleting of eggshell fragments while most other cellular structures remain in the supernatant and are removed. For 2-D gel analysis, the washed eggshell pellet was extracted by heating to 80° C for 10 minutes in 20 μl of either standard loading buffer (8 M urea, 5% CHAPS, 40 mM Tris base, protease inhibitors) or reducing loading buffer (7.2 M urea, 4.5% CHAPS, 100 mM DTT, 36 mM Tris base, protease inhibitors), followed by recovery and pooling of supernatant, which was stored at −70° C; spot patterns on 2-D gels were identical with extraction at 37° C (not shown). Yield was estimated by microBCA assay (Pierce). Samples prepared for LC-MS/MS analysis were similarly treated but with several modifications. Protease inhibitors were not included in the low-salt wash buffer and additional washes in this buffer were used to remove the TX-100 and protease inhibitors; protease inhibitors would interfere with trypsinization, and detergents are detrimental to peptide separation by liquid chromatography. The washed, damp pellets for LC-MS/MS analysis were stored at −70° C without extraction. After an initial LC-MS/MS analysis revealed significant nuclear contamination of the “standard” eggshell matrix preparation, we introduced a 30-minute incubation with RNase A and DNase I (200 μg/ml each in lysis buffer + 10 mM MgCl2) after the homogenization steps and prior to extensive washing in low-salt wash buffer, this time with intervening spins at 9300 g as we found the eggshell fragments pelleted poorly at 100 g following nuclease treatment. Propidium iodide staining was used to monitor nuclear contamination of pilot preparations. The final preparation obtained in this manner was devoid of yolk contamination as judged by LC-MS/MS, and contained only trace amounts of 10 nuclear protein contaminants; by contrast, the original preparations lacking the nuclease treatment contained at least 46 nuclear protein contaminants and all three major yolk proteins.
2-D gel electrophoresis, HPLC and mass spectrometry
Two-dimensional gels included a pI 3−10 isoelectric focusing dimension and 8.5% SDS-PAGE second dimension. In some cases, samples were labeled with Cy5 fluorescent dye prior to isoelectric focusing; however, this was limited to samples extracted from eggshell preparations without DTT as the presence of DTT is incompatible with the dye-labeling reaction. Typically, 200 − 600 μg total protein was loaded on gels. After electrophoresis, the gels were fixed and stained with Sypro Ruby. Using automated equipment (GE Healthcare), spots were picked, subjected to in-gel digestion with trypsin, and loaded onto an ABI 4700 Proteomics Analyzer for MALDI-ToF MS/MS analysis of peptides. Peptide masses and MS/MS ions were compared to the NCBI non-redundant Drosophila database using Mascot and ProteinProspector algorithms.
For LC-MS/MS analysis, eggshell preparations were denatured in urea, reduced, alkylated, and digested with sequencing grade trypsin (Promega) following standard procedures (Wells et al., 2002). The resulting peptides were desalted using reverse-phase C18 spin columns (The Nest Group) and Nitrogen bomb-loaded onto 0.075 mm × 10 cm in-house packed C18 column/emitters (New Objective). Peptides were eluted for two hours with a linear gradient of acetonitrile in 0.1% formic acid at a flow rate of approximately 250 nl/min into a linear ion trap (LTQ, ThermoElectron). One full ms scan followed by 8 CID MS/MS scans of the most abundant ions were collected. Dynamic exclusion was set at 2. All data analysis was performed using TurboSequest (ThermoFinnigan) against the non-redundant Drosophila database, as well as an inverted database to calculate false discovery rate (less than 1% in each run), and using stringent filtering of data. In the “standard” matrix preparation described above, 679 peptides were identified, leading to the identification of approximately 100 proteins with 2 or more fragmented peptides; of these 100 proteins, 27 had predicted signal sequences suggesting they were genuine secreted proteins, while 46 were nuclear proteins and 11 were cytoskeletal proteins. Following optimization of nuclease treatment as described above, a smaller number of peptides (395) were identified in a new preparation, leading to the identification of approximately 41 proteins with 2 or more fragmented peptides; of these, 32 had predicted signal sequences, 4 were nuclear proteins, and 4 were cytoskeletal proteins.
Protein sequence analysis and source of DNAs
Protein sequences derived from FlyBase entries were examined using SignalP, and BLAST programs on the ExPASy Proteomics and FlyBase servers. The following BDGP EST clones were obtained from the Drosophila Genome Resource Center: GM32356 (CG9050), RE74364 (CG11381), GM06507 (CG3074), SD01663 (CG9280), LD15803 (CG3322), and GM06086 (CG7981). For CG12398, CG13083, CG13084, CG4009, CG14796, CG15570, and CG31928, no cDNA clones were available, so primers to the 5’ and 3’ ends of the predicted transcripts, or to selected regions of the transcripts in the case of CG14796 and CG15570, were used to amplify the genes from genomic DNA by PCR, and products inserted into pGEM11Zf(+) (Promega) or pBS-SK(−) (Stratagene).
RNA in situ hybridization
Template DNAs were linearized with appropriate enzymes, and single-stranded digoxigenin-labeled sense and antisense RNA probes were synthesized using T3, T7, or SP6 versions of the MegaScript kit (Ambion), substituting digoxigenin-11-UTP (Roche) at a 1:2 ratio relative to UTP. RNA in situ hybridization on fixed w1118 egg chambers was performed at 56° C using the method described by Lehmann and Tautz (1994), with detection involving AP-conjugated anti-digoxigenin antibody (Roche) preabsorbed against fixed ovaries, and NBT/BCIP (Roche) color development for 5−25 minutes depending on signal strength and background in the sense control samples. Samples were examined using DIC optics at 100× and 200× magnification at a Zeiss AxioSkop 2, photographed using a MRC5 camera and AxioVision software, and re-sized using Adobe PhotoShop. Sense probes usually gave no staining at all or very weak staining of all cell types, or just nurse cells, in previtellogenic egg chambers. An exception was the CG9050 sense probe, which gave strong staining of nurse cells that was not seen with antisense; there is no predicted gene on the opposite strand so this may represent a technical artifact or cross-reactivity with another mRNA expressed by nurse cells.
Results
Proteomics approaches to identify components of the eggshell matrix
The starting point for this study was an interest in using proteomics methods to identify glycoproteins that might, similarly to many mammalian matrix proteins, play roles in follicle cell migrations and communications with the oocyte, and to identify enzymes that might play roles in matrix remodeling within the eggshell. Members of these protein classes are likely to have larger masses than those of the major structural proteins of the eggshell (see Table 1) and may or may not be tightly bound within the eggshell matrix. Extensive eggshell purification, which we will show is essential for detection of minor eggshell proteins, was based on a protocol described by Fargnoli and Waring (1982) involving several washes and low-speed spins for isolation of vitelline membranes from Stage 10 egg chambers, but instead we used whole ovaries as starting material in order to capture all stages of eggshell development. We found that both 2% Triton X-100 and 0.4 M NaCl were required during ovary homogenization to prevent the appearance of a large smear of yolk spots in the central region of 2-D gels, together with many associated contaminants (defined as known cytoplasmic components not having signal sequences; data not shown). These stringent extraction conditions may result in the loss of some important eggshell components, and provide a limitation to this study. However, we were encouraged by data showing that a variety of mammalian matrix glycoproteins are retained in insoluble matrices under similar extraction conditions (e.g., Carter and Hakomori, 1981, and references therein); also, the disulfide-based cross-linking of the vitelline membrane, present from Stage 12 of oogenesis onward, may serve to covalently anchor some minor components of the eggshell.
Table 1.
Proteins identified in eggshell matrix preparations
| Spot # | Gene Identity | Protein Name | Theoretical Mr (Da) | Cytology | Description/Function |
|---|---|---|---|---|---|
| Gene Products Having Signal Sequences | |||||
| Putative structural/regulatory (non-enzymatic) components of eggshell | |||||
| 1 | CG9050 | 40,531 | 26A4 | internal APY-rich repeats | |
| 2 | CG11381 | 49,913 | 1E4 | novel, glutamine-rich | |
| 3 | CG13083 | 31,776 | 37E4 | novel | |
| 4 | CG13084 | 51,299 | 37E3−4 | rel. to CG13083, proline-rich extension | |
| CG13114 | 44,779 | 30B10 | proline-rich | ||
| CG13992 | 70,749 | 26A4 | novel; weak Ig domain homology | ||
| CG13997 | 18,540 | 26A3 | conserved VM (vitelline membrane) motif | ||
| CG14796 | 194,464 | 2B2 | chitin-binding motif, mucin-like domain | ||
| CG15350 | Cp7Fb | 56,094 | 7F1 | predicted chorion protein | |
| CG15570 | 136,890 | 4B3 | mucin-like domain | ||
| CG33962 | Cp7Fa | 32,270 | 7F1 | predicted chorion protein | |
| Putative enzymes and enzyme-related proteins of eggshell | |||||
| 5 | CG5717 | yellow-g | 43,026 | 62D5 | eggshell integrity; dopa metabolism? |
| 6 | CG12398* | 71,415 | 13A1 | GMC oxidoreductase | |
| CG3074 | 48,786 | 58C7-D1 | cathepsin B-like (inactive?) | ||
| CG4009 | 70,756 | 89E10 | heme peroxidase | ||
| CG13804 | yellow-g2 | 42,526 | 62D5 | dopa metabolism? | |
| CG31928 | 45,878 | 22A2 | cathepsin D-like (inactive?) | ||
| Previously known eggshell proteins | |||||
| 7 | CG2175 | dec-1 | 108,371 | 7C1 | chorion; multiple isoforms and processing |
| 8 | CG6524 | chorion protein 19 | 18,525 | 66D12 | structural protein of chorion |
| 9 | CG1478 | chorion protein 36 | 29,916 | 7F1 | structural protein of chorion |
| 10 | CG9048 | VM26Aa (sV17) | 14,321 | 26A3 | structural protein of vitelline membrane |
| CG11213 | chorion protein 38 | 30,448 | 7F1 | structural protein of chorion | |
| CG6517 | chorion protein 18 | 17,231 | 66D12 | structural protein of chorion | |
| CG6533 | chorion protein 16 | 14,265 | 66D12 | structural protein of chorion | |
| CG6519 | chorion protein 15 | 11,981 | 66D12 | structural protein of chorion | |
| CG16874 | VM32E | 12,100 | 32E1 | structural protein of vitelline membrane | |
| CG9271 | VM34C | 11,954 | 34B11 | structural protein of vitelline membrane | |
| Other known secreted proteins | |||||
| 11 | CG2979 | yolk protein 2 | 49,660 | 9A5 | nutrient source for egg |
| 12 | CG9280 | glutactin | 118,749 | 29E3 | basement membrane, esterase-like domain |
| 13 | CG3322 | laminin B2 | 182,339 | 67C2 | basement membrane |
| 14 | CG7981 | perlecan | 461,787 | 3A3−4 | basement membrane; multiple isoforms |
| 15 | CG11411* | Nasrat | 238,761 | 1F3 | oocyte surface; patterning; eggshell |
| CG10129 | Nudel | 292,492 | 65B5 | oocyte surface; patterning; eggshell | |
| CG10236 | laminin A | 411,157 | 65A8−9 | basement membrane | |
| CG7123 | laminin B1 | 198,673 | 28C4-D1 | basement membrane | |
| CG33103 | papilin | 299,154 | 98D3−4 | basement membrane; multiple isoforms | |
| CG12908 | nidogen/entactin | 149,080 | 47A1 | basement membrane | |
| CG4145 | collagen IV | 174,119 | 25C1 | basement membrane | |
| CG16858 | viking (coll. IV) | 193,777 | 25C1 | basement membrane | |
| Gene Products Lacking Signal Sequences | |||||
| 16 | CG10067 | Actin 57B | 40,258 | 57B5 | cytoskeleton |
| 17 | CG2184 | myosin light chain 2 | 21,542 | 99E1 | cytoskeleton |
| 18 | CG4466* | hsp27 | 23,617 | 67B3 | molecular chaperone in nucleus |
| 19 | CG4264 | hsp cognate 4 | 71,104 | 88E4 | required for endocytosis, exocytosis |
| 20 | CG10527 | farnesoic acid O-methyltransferase | 31,556 | 57B16 | hormonal control of ovary maturation |
| 21 | CG30366* | 33,235 | 44C4 | novel | |
| 22 | CG8409 | Su(var)205 | 23,185 | 28F2−3 | heterochromatin assembly |
Proteins lacking spot numbers were identified only by LC-MS/MS, while those marked with an * were identified only by 2-D DIGE-MALDI-MS/MS. Additional contaminants identified by LC-MS/MS are noted in the Materials and Methods, but not shown here. Bolded genes were analyzed by RNA in situ hybridization in this work.
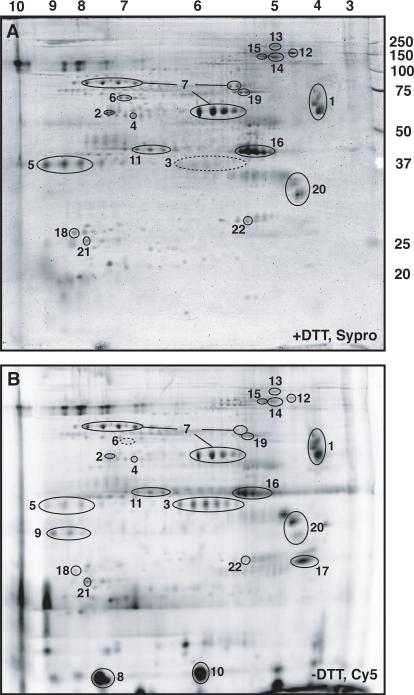
The first proteomics approach used was 2-D gel electrophoresis of proteins extracted from the purified eggshells with urea/CHAPS, followed by spot-picking, in-gel trypsin digestion, and MALDI MS/MS analysis. Two-dimensional electrophoresis was performed using a low-percentage (8.5%) SDS-PAGE dimension that allowed clear separation of several previously uncharacterized proteins in the 30 − 150 kDa range, larger than most of the major eggshell proteins and consistent with our emphasis on glycoproteins and enzymes (Figure 1). One application of two-dimensional electrophoresis is 2-D difference gel electrophoresis (2-D DIGE) in which two – three samples are separately labeled with Cy dyes and then run together on a single 2-D gel to identify differences in amount or migration of spots between the samples (Unlu et al., 1997). This method could in the future be used to compare eggshells from different genotypes, but has a potential limitation for analysis of the disulfide cross-linked eggshell in that the dye labeling reactions are strongly inhibited by the presence of reducing agents such as DTT. For this reason, we were interested in comparing the 2-D spot patterns of proteins extracted from the purified eggshells in either the presence or the absence of DTT, followed in both cases by DTT reduction of the extracted supernatants prior to loading the first dimension of the 2-D gel. Grossly similar amounts of the disulfide cross-linked vitelline membrane proteins sV17 and sV23 could be released from purified eggshells by urea/CHAPS extraction with or without DTT present, as judged by Western blotting with antibodies to these proteins (not shown), suggesting that urea/CHAPS extraction is sufficient to solubilize most of the partially-formed eggshells independent of their disulfide cross-linking status. We found that the overall pattern of spots obtained on 2-D gels was also nearly identical in samples extracted without DTT (Figure 1B) or with DTT (Figure 1A). Inconsistent and often poor staining of some major eggshell proteins with Sypro Ruby (e.g., spots 8, 9 and 10 in Figure 1B are barely detectable in Figure 1A) was also seen on gels containing only DTT-extracted proteins (not shown), so does not represent a DTT-dependent difference in the spot patterns. There were two authentic and reproducible differences based on the presence or absence of DTT in the samples. One was spot cluster 6, which could only be detected if proteins were extracted under reducing conditions (Figure 1A), and might represent a protein that is held in an insoluble disulfide-bonded complex. Conversely, spot cluster 3 gave a much weaker and indistinct spot pattern in DTT-extracted samples (Figure 1A) as compared to samples extracted without DTT present (Figure 1B), and might represent a protein that is unstable under reducing conditions.
Figure 1.
Two-dimensional gel electrophoresis of enriched eggshell preparations. Panels A and B represent Sypro Ruby staining and Cy5 fluorescent dye labeling, respectively, of a single gel on which was loaded 300 μg unlabeled proteins extracted in DTT-containing sample buffer and 60 μg proteins extracted in non-reducing sample buffer and labeled with Cy5. Thus, most of the protein spot intensity in panel A of this gel is derived from the DTT-extracted sample. Identified spots are numbered, and described in Table 1. Spot cluster 6 is only observed in DTT-extracted samples, while spot cluster 3 is weak or absent in these samples (indicated by dashed circles). Numbers above panel A indicate pI values in the isoelectric focusing dimension, while numbers at right indicate molecular masses (in kDa) of marker proteins in the SDS-PAGE dimension.
We were able to reproducibly identify 22 distinct proteins on 2-D gels with high confidence. Table 1 provides details about these proteins, which are indicated by spot cluster numbers in the first column corresponding to clusters shown on Figure 1. In order to provide a conceptual framework for classifying these proteins, we first determined whether or not they had predicted signal sequences, and therefore whether they were likely to represent secreted proteins that could be genuine eggshell matrix components, or were intracellular proteins most likely to be contaminants in the preparations. Next, we divided the signal sequence-containing proteins into four groups. Two of these groups represented well-characterized secreted proteins, either eggshell structural proteins (“previously known eggshell proteins” in Table 1), or others including yolk, basement membrane, and oocyte surface proteins (“other known secreted proteins” in Table 1). The other two groups represented less well-characterized or novel proteins, either enzyme-related proteins that had not previously been shown to be physically associated with the eggshell matrix (“putative enzymes and enzyme-related proteins of eggshell” in Table 1), or non-enzymatic proteins that had not previously been shown to be physically associated with the eggshell matrix and might have structural or regulatory roles in this matrix (“putative structural/regulatory proteins of eggshell” in Table 1). The assignment to this last group was in some cases supported by structural features similar to known eggshell structural proteins, e.g., presence of a vitelline membrane motif or enrichment in amino acids such as alanine and proline, or to other matrix motifs, e.g., chitin-binding and mucin-like motifs, but should not be construed as a statement of known function in the eggshell.
Because of the limited sensitivity of the 2-D gel approach, we turned to an LC-MS/MS approach. In this approach (see Methods), trypsinized peptides are generated directly from the whole eggshell preparation, so the outcome does not rely on the size of proteins or on their solubility in buffers used in 2-D gels, which can be a difficult problem for hydrophobic and extracellular matrix proteins (Wu and Yates, 2003). Also, LC-MS/MS peptide analysis has greater dynamic range and sensitivity than in-gel digestion followed by MALDI MS/MS (Wu and Yates, 2003). As demonstration of these advantages, Table 1 shows that between 4 and 7 additional proteins were identified in each of the categories of secreted proteins when compared to 2-D gel results, and these proteins ranged in mass from under 12 kDa to over 410 kDa. Two secreted proteins, CG12398 and Nasrat, were only detected by the 2-D gel method, which might reflect benefits of concentration of some minor constituents into spots on 2-D gels and/or differences in extractability of certain proteins under the somewhat different protocols used for these two methods. Several of the minor proteins, including CG3074, CG13114, CG14796, CG15570, and CG31928, were only detected by LC-MS/MS after a nuclease treatment was incorporated into the eggshell enrichment protocol that substantially reduced contamination by nuclear, yolk, and cytoskeletal proteins (see Methods), indicating the importance of extensive purification of the matrix fraction to success of proteomics studies.
These studies do not provide a comprehensive analysis of the eggshell matrix, as made clear by the absence of three known eggshell components in Table 1. The first of these is sV23, a major vitelline membrane protein that we know is present in the preparations by Western blotting (not shown). Because of its unusual amino acid composition, sV23 lacks predicted tryptic peptides in the size range (1−3 Da) that can be reliably analyzed by mass spectrometry, emphasizing a weakness of these approaches in analyzing the eggshell. The minor chorion component, Femcoat (Kim et al., 2002), and Torsolike, a spatial determinant embedded in the vitelline membrane (Stevens et al., 2003), were also not detected. These proteins each have multiple tryptic peptides that could have provided identification, so their absence may reflect insufficient sensitivity of our current protocols for detection of some minor species or loss of these proteins due to the stringency of the wash steps used in eggshell purification.
Validation of several previously predicted eggshell genes within vitelline membrane and chorion gene clusters
Six of the putative structural/regulatory and enzyme-related proteins correspond to known genes that were previously predicted to encode molecules important for eggshell biogenesis based upon their expression in follicle cells during late oogenesis and their cytological localization in close physical proximity to known eggshell genes. The major chorion genes, in particular, are located in two gene clusters, at region 7F on the X chromosome (including s36 and s38; Parks et al., 1986) and at region 66D on the third chromosome (including s15, s16, s18, and s19; Spradling, 1981, Griffin-Shea et al., 1982). These chorion gene clusters, as well as recently described regions at 30B and 62D (Claycomb et al., 2004), undergo gene amplification in order to allow rapid synthesis of large amounts of chorion proteins over the span of a few hours when the chorion is assembled. The vitelline membrane genes do not undergo amplification and while two of these genes are in isolated locations at 32E and 34C on the second chromosome, there has been reported an apparent cluster of vitelline membrane genes at 26A that includes the sV17 and sV23 genes (Popodi et al., 1988). Coordinated expression of closely linked genes at specific developmental times contributes to the organized synthesis of the eggshell layers, i.e., 26A cluster in Stages 9−10, 7F cluster primarily in Stages 11−12, and 66D cluster in Stages 13−14.
Three of the putative structural genes, CG13997, CG9050, and CG13992, are found immediately adjacent to the vitelline membrane genes encoding sV17 and sV23 in the 26A cluster (Figure 2A). The CG13997 gene product contains a region of homology (60% identical over 25 amino acids) to a 38-amino acid motif shared among the known vitelline membrane proteins and containing three cysteines shown to be important for disulfide cross-linking (Scherer et al., 1988; Waring, 2000). Also, the CG13997 gene corresponds to a previously described transcription unit, TU-3, expressed in concert with sV17 and sV23 (Popodi et al., 1988). While CG9050 lacks a vitelline membrane motif, it encodes 7 copies of an APY-rich motif similar to a repeat motif found in sV23, and corresponds to a previously described transcription unit, TU-1, expressed at 40−80 fold lower levels than sV23 and sV17, respectively (Popodi et al., 1988; Waring, 2000). CG13992 was not previously included in this putative vitelline membrane gene cluster and lacks all known eggshell gene motifs, but has been reported to contain a weak Ig domain homology (Vogel et al., 2003).
Figure 2.
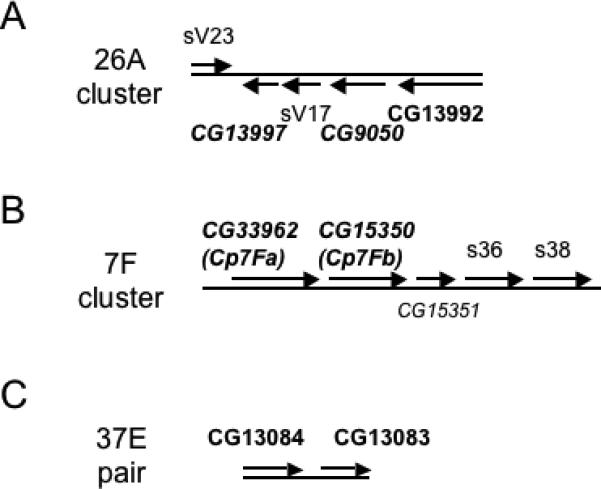
Six genes are located in clusters adjacent to major eggshell genes. The well-characterized genes in the clusters in A and B are indicated by normal type. Genes predicted by previous studies are italicized and those identified by this study are bolded, so predicted genes that are validated by the proteomics approaches are bolded and italicized. (A) Vitelline membrane gene cluster at position 26A on chromosome 2L (Popodi et al., 1988). (B) Chorion gene cluster at position 7F1 on the X chromosome (Parks et al., 1986). (C) Two of the new genes are adjacent to each other at position 37E on chromosome 2L.
The putative structural genes, CG33962 and CG15350, correspond respectively to the predicted chorion protein genes, Cp7Fa and Cp7Fb, in the 7F early chorion gene cluster containing the major s36 and s38 chorion genes (Figure 2B; Parks et al., 1986). Also included in this cluster is CG15351, or Cp7Fc, whose product is predicted to be secreted but which was not identified in our studies. The yellow-g and yellow-g2 genes, encoding putative dopachrome-conversion enzymes, have previously been shown to lie near the site of peak amplification in the 62D amplicon region (Claycomb et al., 2004). These genes are expressed in concert with the early chorion genes and yellow-g has been shown to be essential for vitelline membrane integrity (Claycomb et al., 2004).
In addition to the validation of the above six genes as encoding components of the eggshell matrix, the proteomics studies identified three putative structural components whose cytogenetic localization is in regions previously implicated in eggshell biogenesis and, in the first two cases, where no specific candidate genes had been identified in the region. The CG13114 gene, encoding a small, proline-rich protein, lies within the 30B amplicon, supporting the idea that this amplicon, like those at 7F, 62D, and 66D, contains genes important for eggshell formation (Claycomb et al., 2004). The CG11381 gene, at 1E4 on the tip of the X chromosome, encodes an ∼50 kDa glutamine-rich protein that might correspond to a minor chorion protein, s70, which has not been cloned but was mapped to a region including 1E4 by recombination analysis of an electrophoretic variant (Yannoni and Petri, 1984). The CG15570 gene lies adjacent to the Femcoat chorion gene, although at a distance of almost 19 kb, and encodes an ∼137 kDa protein predicted to have extensive mucin-like O-linked glycosylation (Julenius et al., 2005).
Seven genes showed no relationship to chromosomal regions previously implicated in eggshell biogenesis. Among these are three encoding putative structural or regulatory proteins, including CG14796 encoding an ∼195 kDa protein containing a single chitin-binding motif (Shen and Jacobs-Lorena, 1998) and predicted to have extensive mucin-like O-linked glycosylation, and the CG13083 and CG13084 genes residing next to each other at 37E4 (Figure 2C), where the ∼51 kDa CG13084 protein weakly resembles the ∼32 kDa CG13083 protein in an N-terminal region and has a proline-rich C-terminal extension absent in CG13083. Four of the putative enzyme or enzyme-related genes are in this category. These include CG12398, a member of the glucose-methanol-choline (GMC) family of small-molecule oxidoreductases (Cavener, 1992); CG4009, a heme peroxidase family member (Kimura and Ikeda-Saito, 1988); CG3074, a cathepsin B family member most closely related to the tubulointerstitial nephritis antigen related protein and predicted to be catalytically inactive (Wex et al., 2001); and CG31928, an aspartyl protease family member having potentially inactivating mutations in active site residues close to both catalytic aspartic acids (Dunn, 2002).
Expression analysis of putative eggshell genes
To gain insight into how the putative structural proteins and enzymes identified by proteomics could contribute to eggshell layers or regions, we performed RNA in situ hybridization studies of 10 genes, focusing mainly on those genes not associated with known eggshell gene clusters (see bolded genes in Table 1). Based on a comprehensive in situ hybridization study of the genes in the 7F and 66D chorion gene clusters (Parks and Spradling, 1987), we expected that authentic eggshell genes would be expressed in follicle cells at some but not all time points during the developmental stages when the eggshell is synthesized, and possibly in spatial patterns suggesting functions in formation of particular regions of the eggshell, e.g., dorsal appendages. Changes in spatial pattern of expression may occur secondarily to migration of expressing cells or as a consequence of gene activation under control of multiple regulatory elements, rather than from a specific local requirement for function (Parks and Spradling, 1987). Therefore, patterns of expression that are clearly strongest over a particular eggshell structure or are distinctly different than those of other eggshell genes would be stronger evidence in support of a localized requirement.
The simplest expression pattern was seen for two genes, CG9050 and CG12398. CG9050, one of the genes in the 26A vitelline membrane cluster, showed expression in all follicle cells overlying the oocyte only in Stages 9 and 10, with peak accumulation in Stage 10A (Figure 3A), correlating well to the previous Northern blot analysis of this transcript (Popodi et al., 1988) and indicating its presence during the peak expression of the major vitelline membrane genes. CG12398, the GMC oxidoreductase family member, showed an identical expression pattern (Figure 3B), suggesting it might play a role in vitelline membrane formation.
Figure 3.
Genes expressed uniformly in all follicle cells overlying the oocyte. RNA in situ hybridization with antisense probes against CG9050 (A) and CG12398 (B) shows expression exclusively in follicle cells in the mid-oogenesis stages 9 and 10A. Panels C-F. CG11381 expression in Stage 10B (C), Stage 11 (D, left), early Stage 12 (D, right), early Stage 13 (E), and late Stage 13 (F). Arrows in D-F point to strongest CG11381 expression in cells surrounding the developing micropyle.
The CG11381 gene, encoding a predicted ∼50 kDa glutamine-rich protein, showed an expression pattern temporally bridging that of the major vitelline membrane and early-intermediate chorion proteins. It was first expressed uniformly but relatively weakly in all follicle cells overlying the oocyte (main body follicle cells) in Stage 10B (Figure 3C), then showed intense staining in Stage 11 and early Stage 12 (Figure 3D). Its staining in these cells decreased in late Stage 12 (Figure 3E), and could no longer be detected by Stage 14 (not shown). In Stages 12 and 13, CG11381 was most strongly expressed in cells surrounding the developing micropyle, a specialized structure for sperm entry at the anterior end of the egg that consists of both vitelline membrane and chorion layers, and especially in the cells at the tip of this structure (arrows in Figure 3, panels D, E, and F). This expression pattern is reminiscent of that described for yellow-g and yellow-g2, which are similarly strongly expressed in main body follicle cells in Stage 12 but also preferentially expressed in cells surrounding the micropyle; however, the yellow-g and yellow-g2 genes were first expressed later than CG11381, in Stage 11, and initially just in follicle cells at the dorsal anterior end of the oocyte (Claycomb et al., 2004). Interestingly, the genes in the 7F and 66D chorion clusters are excluded from expression in the cells at the tip of the micropyle (Parks and Spradling, 1987), suggesting that this structure may be formed using just a subset of chorion genes.
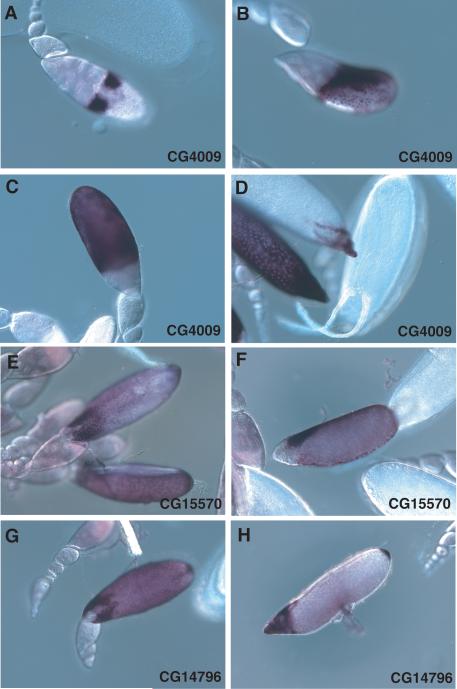
The CG4009 gene, encoding a heme peroxidase family member, is first expressed in two large patches adjoining the dorsal anterior surface of the Stage 10B egg chamber (Figure 4A), presumably corresponding to the positions being fated for the future dorsal appendages. Its expression then quickly extends from the dorsal side (Figure 4B) to cover the entire main body follicle cell epithelium by Stage 11 (Figure 4C). In Stage 12, all follicle cells, including those at the anterior end covering the nurse cells, strongly express CG4009, while in Stage 13, expression is quite weak except for staining of the forming dorsal appendages along their entire lengths, and staining is completely absent by Stage 14 (Figure 4D). The expression pattern of CG4009 in Stages 10B and 11 shares features with the patterns described for the genes in the 7F chorion cluster including the early expression in subsets of dorsal anterior follicle cells, seen for all 7F transcripts, and the transient appearance of nuclear transcripts (Figure 4B), described for Cp7Fb (Parks and Spradling, 1987).
Figure 4.
Genes whose expression initiates in dorsal anterior patches of follicle cells. Panels A-D. CG4009 expression in early Stage 10B (A), later Stage 10B (B), Stage 11 (C), Stage 12 (D, left center), Stage 13 (D, left top), and Stage 14 (D, right). Panels E and F. CG15570 expression in Stage 12, a dorsal view (E) and a lateral view (F). Panels G and H. CG14796 expression in Stage 12 (G) and in Stage 13 (H).
The CG15570 and CG14796 genes encode large proteins predicted to be highly O-glycosylated but, except for a chitin-binding motif in CG14796, having no structural relationship to known proteins or to each other. CG15570 is preferentially expressed in dorsal appendage-forming follicle cells, and is first seen in Stage 11 (or rarely, Stage 10B) in dorsal anterior patches as seen for CG4009 (not shown). It is most strongly expressed in Stage 12 in the follicle cells migrating anteriorly over the nurse cells (Figure 4E, F), with weaker staining of the main body follicle cells, and by Stage 13 only weak staining of the dorsal appendages persists (not shown). Similarly, CG14796 is first expressed in the 2 dorsal anterior patches in Stage 11 (not shown), then shows intense staining of follicle cells migrating anteriorly over the nurse cells in Stage 12 (Figure 4G), with moderate to strong staining of main body follicle cells. There appears to be a wider gap in staining at the dorsal midline in CG14796 than in CG15570, where in some cases there appears to be a single row of unstained cells at the midline (cf. Figure 4G and 4E). Unique to CG14796 among this collection of minor eggshell genes is consistent and strong preferential staining of a patch of posterior follicle cells overlying the aeropyle, visible in Stage 12 (Figure 4G) but most obvious in Stage 13 after staining is lost in the main body follicle cells (Figure 4H). Also seen at Stage 13 is persistent strong staining of the dorsal appendage-forming cells and transient staining of other anterior cells, apparently including nurse cells and a ring of staining just at the anterior margin of the oocyte.
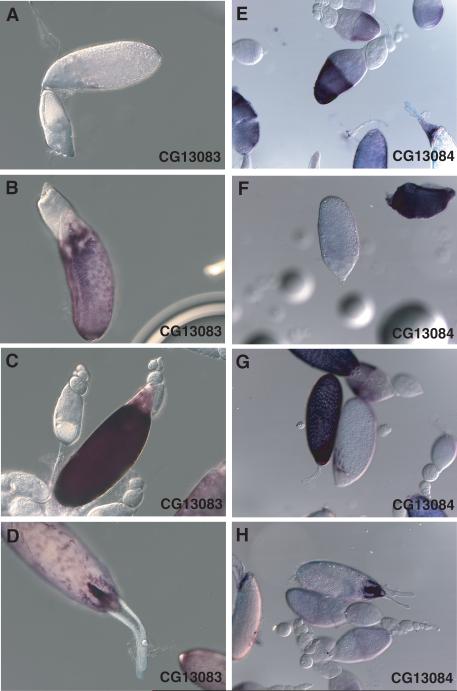
The clustered genes, CG13083 and CG13084, show a remarkably similar expression pattern in late oogenesis, Stages 12−14 (Figure 5). They are first detected in main body follicle cells in early Stage 12 (Figure 5B, G), reach peak expression during Stage 12 (Figure 5C, G), show diminished expression in Stage 13 (not shown), and are largely absent from these cells in Stage 14 (Figure 5D, H). Additional strong, and striking, expression is seen in Stages 13 and 14 in the midline of the anterior operculum, a specialized region of the eggshell involved in larval hatching, and in the proximal portion of the paired dorsal respiratory appendages to either side of the operculum (Figure 5D, H). In contrast to this common late expression pattern shared by CG13083 and CG13084, the CG13084 gene shows an additional early expression exclusively in Stage 10A egg chambers. In these egg chambers, CG13084 expression is usually moderate to strong in intensity and is often uniformly expressed by follicle cells overlying the oocyte, but can also show quite variable spatial distribution, e.g., ventral-only or strongest in posterior half of follicle cells overlying the oocyte (latter shown in Figure 5E). This early CG13084 expression is turned off by Stage 10B, and expression is not resumed until Stage 12, in concert with CG13083 expression (see Figure 5A, F for negative Stage 11 egg chambers). A similar transient early expression of the major late chorion gene, s15, has been reported by Northern blot analysis (Thireos et al., 1980), but could not be detected by ovary RNA in situ hybridization (Parks and Spradling, 1987).
Figure 5.
RNA in situ hybridization with antisense probes against CG13083 (A-D) and CG13084 (E-H). (A) Stage 10A, left, and Stage 11, top. (B) Early Stage 12. (C) Stage 12. (D) Early Stage 14. (E) Labeled egg chambers at top and center are Stage 10A. (F) Stage 11 egg chambers, center, are always negative for staining. (G) Early stage 12 (right), and later Stage 12 (left) showing return of strong staining. (H) Stage 14. Arrows in D and H point to strong staining of the midline region of the operculum, with lighter staining in the proximal third of the paired dorsal appendages. Unlike staining for CG4009 (Figure 4D), these probes never show staining of more distal regions of the dorsal appendages.
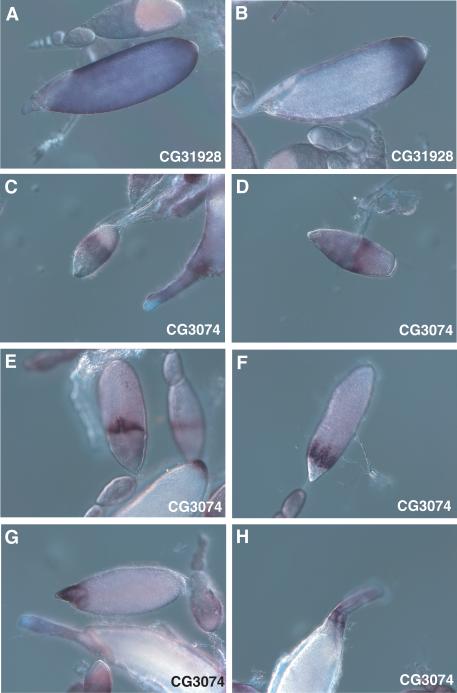
The two protease-related genes show some of the most unusual expression patterns. First, CG31928, which encodes an aspartyl protease-like protein, is initially expressed in Stage 12 in most follicle cells overlying the oocyte but is completely excluded from those that will contribute to anterior specializations and almost always excluded from the posterior follicle cells overlying the aeropyle (Figure 6A). During Stage 13, there is increased expression in a ring of follicle cells covering the posterior quarter of the egg chamber, except for the most posterior follicle cells, and decreased expression in more central regions, giving rise to a quite striking pattern seen in all Stage 14 egg chambers (Figure 6B).
Figure 6.
RNA in situ hybridization with antisense probes against the protease-related genes CG31928 (A-B) and CG3074 (C-H). (A) Stage 12. (B) Stage 14. (C) Stage 9, center. (D) Stage 10A. (E) Stage 11, center. (F) Stage 12. (G) Stage 13, above, and late Stage 14, below. (H) early Stage 14.
Finally, CG3074, encoding a cathepsin B-related protein containing a serine instead of a cysteine in its potential active site, shows a dynamic expression pattern across oogenesis. It is first detected in Stage 9 in a wide circumferential band centered at the nurse cell-oocyte boundary, within follicle cells that are migrating posteriorly over the oocyte, but is not expressed by the border cells migrating posteriorly between the nurse cells (Figure 6C). In Stage 10, expression strengthens but is usually limited to a narrow band of follicle cells adjacent to the anterior margin of the oocyte (Figure 6D). In Stage 11 and 12 (Figure 6E, F), staining becomes limited to cells in the region that will form the dorsal appendages, and is initially seen just in cells at the medial and anterior border of the follicle cell patches that will migrate forward over the nurse cells (may correspond to the floor cells of the dorsal appendages; Ward and Berg, 2005). In Stage 13 (Figure 6G), only the dorsal appendages are stained, and staining is stronger at more distal regions of these structures; this is also seen in Stage 14 (Figure 6H).
These results, summarized in Table 2, support the identification of these genes as authentic eggshell genes expressed by follicle cells during the time when the eggshell is being synthesized. In contrast, RNA in situ analysis of the basement membrane genes encoding glutactin, laminin B2, and perlecan showed only weak and variable expression with no linkage to specific events in eggshell formation (not shown). We suggest that the suite of basement membrane proteins identified in the eggshell matrix fraction (Table 1) is present because the basement membrane on the basal side of the follicle cells co-purifies with eggshell in this simple purification scheme based on detergent extraction and centrifugation.
Table 2.
Summary of follicle cell expression patterns for minor eggshell genes
| Gene | Stage 8 | Stage 9 | Stage 10A | Stage 10B | Stage 11 | Stage 12 | Stage 13 | Stage 14 |
|---|---|---|---|---|---|---|---|---|
| CG9050 | − | ++ | ++++ | + | − | − | − | − |
| CG12398 | − | ++ | ++++ | +/− | − | − | − | − |
| CG11381 | − | − | − | ++ | +++ | ++++/++ | +/− MB | − |
| ++ MP | ||||||||
| CG4009 | − | − | − | ++/+++ DP | ++++ | ++++ | +/− MB | − |
| +++ DA | ||||||||
| CG15570 | − | − | − | +/− DP | ++ DP | ++ MB | − MB | − |
| ++++ DA | + DA | |||||||
| CG14796 | − | − | − | − | +++ DP | ++++/++ MB | +/− MB | − |
| ++++ DA, PFC | ++ DA, PFC | |||||||
| CG13083 | − | − | − | − | − | +/++++ | ++ MB | − MB |
| +++ O/DA | ++/+ O/DA | |||||||
| CG13084 | − | − | +/+++ | − | − | +/++++ | ++ MB | − MB |
| +++ O/DA | ++/+ O/DA | |||||||
| CG31928 | − | − | − | − | − | ++ | +++ PR | +++ PR |
| CG3074 | − | ++ AR | +++ AR | ++ AR | +++ DAM | +++ DAM | ++ DA | ++ DA |
MB, main body, MP, micropyle, DP, dorsal anterior patches, DA, dorsal appendages, PFC, posterior follicle cells, O, operculum, PR, posterior ring, AR, anterior ring, DAM, dorsal appendage margin.
Discussion
Sensitive mass spectrometry analyses of purified eggshells effectively identified minor eggshell proteins, confirming the presence in the eggshell of 6 previously predicted proteins and demonstrating the presence of 11 novel components. Extensive purification of the eggshells was essential for the success of these approaches, in order to maximize loadings of minor proteins in the 2-D gel approach, and for detection via LC-MS/MS, where our samples were relatively complex and demanding for the LTQ to sequence all peptides as they eluted from the column. In the future, nuclease treatment to remove the major contaminants of eggshell preparations may make it feasible to use milder wash conditions less likely to strip the matrix of weakly bound proteins; these modifications, together with use of two-dimensional LC-MS/MS, may allow fuller characterization of the eggshell proteome. Gene microarrays involving purification of follicle cell-specific mRNAs (Bryant et al., 1999) offer a complementary approach to identifying secreted proteins expressed by these cells during development.
Temporal control of eggshell gene expression is important for the ordered synthesis of the eggshell layers (reviewed by Waring, 2000), and this timing provides one predictor of function for novel components. The time course of expression of eight of the novel genes described in this study (Table 2) shows that CG12398 expression overlaps with that of the major vitelline membrane genes, while CG11381, CG4009, CG15570, and CG14796 are expressed in concert with the early chorion genes of the 7F cluster, and CG13083, CG13084 and CG31928 are expressed similarly to the intermediate and late chorion genes of the 66D cluster. Changing spatial expression patterns across time may reflect gene induction in different follicle cell populations and/or migration of expressing cells, rather than indicating distinct functional requirements in different regions of the eggshell (Parks and Spradling, 1987). Indeed, this is likely to be the case for most of the newly isolated eggshell proteins, where expression occurs, across time, in all follicle cells contributing to the eggshell. However, preferential expression in follicle cells overlying certain specialized regions may result in differing ratios of eggshell components that could contribute to the different structural properties of the chorion in these regions (Margaritis et al., 1980). For example, CG15570 and, to a lesser extent, CG14796 are likely to be enriched in the highly fenestrated dorsal appendages compared to other regions, while CG13083 and CG13084 may, together with the co-expressed 66D cluster genes, contribute to the structure of the central operculum. A more exclusive expression is seen for CG11381 in the cells surrounding the tip of the forming micropyle, which appear to lack expression of all other eggshell genes except for yellow-g and yellow-g2 (Parks and Spradling, 1987; Claycomb et al., 2004). Conversely, CG31928 is specifically excluded from the cells overlying the anterior and posterior specializations of the chorion. Determining the significance of these differences in expression, and overall function in eggshell, will require knockdown or mutagenesis of these genes, as their sequences provide few clues to function or interactions.
The CG3074 gene is unique among the new genes in being expressed across all stages of eggshell development and in several distinct patterns restricted to the anterior end of the oocyte, suggesting that its product plays a different sort of role in the eggshell matrix than the other new eggshell genes that are more broadly expressed. CG3074 shares sequence homology and its overall domain structure with members of the tubulo-interstitial nephritis antigen family of inactive cathepsin-B-like proteins (Ikeda et al., 2000; Mukai et al., 2003; Nelson et al., 1995; Wex et al., 2001). These mammalian proteins have been shown to enhance integrin-mediated cell adhesion (Kalfa et al., 1994) and to promote tubulogenesis in the developing kidney (Kanwar et al., 1999). In the fly ovary, CG3074 is well-positioned to have a role in promoting cell shape changes and/or migration of centripetal follicle cells and dorsal appendage-forming cells (Horne-Badovinac and Bilder, 2005), and we are in the process of exploring this possibility.
The four potentially active enzymes identified in this study are globally expressed and have potential functions in tyrosine-based cross-linking of the eggshell. The yellow-g and yellow-g2 proteins have previously been shown to be critical for eggshell integrity and proposed to act in the tyrosine-based cross-linking of the eggshell (Claycomb et al., 2004), based upon their similarity to two dopachrome-conversion enzymes, yellow-f and yellow-f2, that appear to act in the melanization of cuticle (Han et al., 2002). The heme peroxidase family member, CG4009, may directly function, together with the Pxd peroxidase (Konstandi et al., 2005), in catalyzing the formation of di- and tri-tyrosine cross-links. The CG12398 oxidoreductase is more enigmatic; while the substrates of the GMC oxidoreductases are typically small molecules, especially sugars, hydrogen peroxide is noted as a byproduct of such reactions, e.g., the oxidation of glucose by pyranose oxidase (Giffhorn, 2000). Thus it is possible that CG12398 could produce the hydrogen peroxide necessary for tyrosine cross-linking, especially of the vitelline membrane where no source of hydrogen peroxide has been found. A similar mechanism of hydrogen peroxide production has been postulated for hardening of the Aedes aegypti chorion, involving a malate dehydrogenase activity that has not been cloned (Han et al., 2000).
Overall, this proteomics study of the eggshell extracellular matrix has revealed a richly diverse collection of new eggshell components, most of which were not previously implicated by genetic studies or proximity to known eggshell genes. Some of the enzymatic components, such as a GMC oxidoreductase and a heme peroxidase, may ultimately have significant explanatory power regarding the cross-linking of the eggshell, while the identification of novel structural components provides new information towards the long-term goal of understanding how the distinct layers and regions of the eggshell are constructed.
Acknowledgments
We are grateful for the resources of FlyBase, the Berkeley Drosophila Genome Project, the Drosophila Genome Resource Center, ExPASy and the Swiss Institute for Bioinformatics. EKL also thanks the two anonymous reviewers for their substantial improvement of the manuscript, and Michael Dinkins for helpful comments about protease-related genes. This work was supported in part by grants from March of Dimes and NIH to EKL, and by a grant from the Georgia Cancer Coalition to LW.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra L, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Schneider DS, Morisato D, Jin Y, Ferguson EL. Extracellular morphogens in Drosophila embryonic dorsal-ventral patterning. Cold Spring Harbor Symp. Quant. Biol. 1992;57:409–417. doi: 10.1101/sqb.1992.057.01.046. [DOI] [PubMed] [Google Scholar]
- Andrenacci D, Cernilogar FM, Taddei C, Rotoli D, Cavaliere V, Graziani F, Gargiulo G. Specific domains drive VM32E protein distribution and integration in Drosophila eggshell layers. J. Cell Sci. 2001;114:2819–2829. doi: 10.1242/jcs.114.15.2819. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu CR, Li MJ, van den Engh G, Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl. Acad. Sci. USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter WG, Hakomori S. A new cell surface, detergent-insoluble glycoprotein matrix of human and hamster fibroblasts. The role of disulfide bonds in stabilization of the matrix. J. Biol. Chem. 1981;256:6953–6960. [PubMed] [Google Scholar]
- Cavener DR. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev. Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Dobens LL, Raftery LA. Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev. Dyn. 2000;218:80–93. doi: 10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev. Biol. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Dunn BM. Structure and mechanism of the pepsin-like family of aspartyl peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- Fargnoli J, Waring GL. Identification of vitelline membrane proteins in Drosophila melanogaster. Dev. Biol. 1982;92:306–314. doi: 10.1016/0012-1606(82)90177-4. [DOI] [PubMed] [Google Scholar]
- Giffhorn F. Fungal pyranose oxidase: occurrence, properties and biotechnological applications in carbohydrate chemistry. Appl. Microbiol. Biotechnol. 2000;54:727–740. doi: 10.1007/s002530000446. [DOI] [PubMed] [Google Scholar]
- Griffin-Shea R, Thireos G, Kafatos FC. Organization of a cluster of four chorion genes in Drosophila and its relationship to developmental expression and amplification. Dev. Biol. 1982;91:325–336. doi: 10.1016/0012-1606(82)90039-2. [DOI] [PubMed] [Google Scholar]
- Han Q, Fang J, Ding H, Johnson JK, Christensen BM, Li J. Identification of Drosophila yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 2002;368:333–340. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Li G, Li J. Chorion peroxidase-mediated NADH/O2 oxidoreduction cooperated by chorion malate dehydrogenase-catalyzed NADH production: a feasible pathway leading to H2O2 formation during chorion hardening in Aedes aegypti mosquitoes. Biochim. Biophys. Acta. 2000;1523:246–253. doi: 10.1016/s0304-4165(00)00129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev. Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Takemura T, Hino S, Yoshioka K. Molecular cloning, expression, and chromosomal localization of a human tubulointerstitial nephritis antigen. Biochem. Biophys. Res. Comm. 2000;268:225–230. doi: 10.1006/bbrc.2000.2103. [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kalfa TA, Thull JD, Butkowski RJ, Charonis AS. Tubulointerstitial nephritis antigen interacts with laminin and type IV collagen and promotes cell adhesion. J. Biol. Chem. 1994;269:1654–1659. [PubMed] [Google Scholar]
- Kanwar YS, Kumar A, Yang Q, Tian Y, Wada J, Kashihara N, Wallner EI. Tubulointerstitial nephritis antigen: an extracellular matrix protein that selectively regulates tubulogenesis vs. glomerulogenesis during mammalian renal development. Proc. Natl. Acad. Sci. USA. 1999;96:11323–11328. doi: 10.1073/pnas.96.20.11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramaris KE, Stravopodis D, Margaritis LH. A structural protein that plays an enzymatic role in the eggshell of Drosophila melanogaster. Cell Biol. Int. Rep. 1991;15:151–159. doi: 10.1016/0309-1651(91)90106-s. [DOI] [PubMed] [Google Scholar]
- Kim C, Han K, Kim J, Yi JS, Kim C, Yim J, Kim YJ, Kim-Ha J. Femcoat, a novel eggshell protein in Drosophila: functional analysis by double stranded RNA interference. Mech. Dev. 2002;110:61–70. doi: 10.1016/s0925-4773(01)00559-7. [DOI] [PubMed] [Google Scholar]
- Kimura S, Ikeda-Saito M. Human myeloid peroxidase and thyroid peroxidase, two enzymes with separate and distinct physiological functions, are evolutionarily related members of the same gene family. Proteins. 1988;3:113–120. doi: 10.1002/prot.340030206. [DOI] [PubMed] [Google Scholar]
- Konstandi OA, Papassideri IS, Stravopodis DJ, Kenoutis CA, Hasan Z, Katsorchis T, Wever R, Margaritis LH. The enzymatic component of Drosophila melanogaster chorion is the Pxd peroxidase. Insect Biochem. Mol. Biol. 2005;35:1043–1057. doi: 10.1016/j.ibmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: practical uses in cell and molecular biology. Academic Press; New York: 1994. pp. 575–598. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Hashimoto C. The Nudel protease of Drosophila is required for eggshell biogenesis in addition to embryonic patterning. Dev. Biol. 2000;217:352–361. doi: 10.1006/dbio.1999.9562. [DOI] [PubMed] [Google Scholar]
- Manogaran A, Waring GL. The N-terminal prodomain of sV23 is essential for the assembly of a functional vitelline membrane network in Drosophila. Dev. Biol. 2004;270:261–271. doi: 10.1016/j.ydbio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Margaritis LH. The eggshell of Drosophila melanogaster II. Covalent crosslinking of the chorion proteins involves endogenous hydrogen peroxide. Tissue Cell. 1985;17:553–559. doi: 10.1016/0040-8166(85)90031-x. [DOI] [PubMed] [Google Scholar]
- Margaritis LH, Kafatos FC, Petri WH. The eggshell of Drosophila melanogaster. I. Fine structure of the layers and regions of the wild-type eggshell. J. Cell Sci. 1980;43:1–35. doi: 10.1242/jcs.43.1.1. [DOI] [PubMed] [Google Scholar]
- Mindrinos MN, Petri WH, Galanopolous VK, Lombard MF, Margaritis LH. Crosslinking of the Drosophila chorion involves a peroxidase. Wilhelm Roux Arch. Dev. Biol. 1980;189:187–196. doi: 10.1007/BF00868677. [DOI] [PubMed] [Google Scholar]
- Montell DJ. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Mukai K, Mitani F, Nagasawa H, Suzuki R, Suzuki T, Suematsu M, Ishimura Y. An inverse correlation between expression of a preprocathepsin B-related protein with cysteine-rich sequences and steroid 11beta-hydroxylase in adrenocortical cells. J. Biol. Chem. 2003;278:17084–17092. doi: 10.1074/jbc.M301477200. [DOI] [PubMed] [Google Scholar]
- Nelson TR, Charonis AS, McIvor RS, Butkowski RJ. Identification of a cDNA encoding tubulointerstitial nephritis antigen. J. Biol. Chem. 1995;270:16265–16270. doi: 10.1074/jbc.270.27.16265. [DOI] [PubMed] [Google Scholar]
- Nogueron MI, Mauzy-Melitz D, Waring GL. Drosophila dec-1 eggshell proteins are differentially distributed via a multistep extracellular processing and localization pathway. Dev. Biol. 2000;225:459–470. doi: 10.1006/dbio.2000.9805. [DOI] [PubMed] [Google Scholar]
- Parks S, Spradling A. Spatially regulated expression of chorion genes during Drosophila oogenesis. Genes Dev. 1987;1:497–509. [Google Scholar]
- Parks S, Wakimoto B, Spradling A. Replication and expression of an X-linked cluster of Drosophila chorion genes. Dev. Biol. 1986;117:294–305. doi: 10.1016/0012-1606(86)90372-6. [DOI] [PubMed] [Google Scholar]
- Pascucci T, Perrino J, Mahowald AP, Waring GL. Eggshell assembly in Drosophila: processing and localization of vitelline membrane and chorion proteins. Dev. Biol. 1996;177:590–598. doi: 10.1006/dbio.1996.0188. [DOI] [PubMed] [Google Scholar]
- Petri WH, Mindrinos MN, Lombard MF. Independence of vitelline membrane and chorion cross-linking in the Drosophila melanogaster eggshell. Dev. Biol. 1979;83:23a. [Google Scholar]
- Petri WH, Wyman AR, Kafatos FC. Specific protein synthesis in cellular differentiation. II. The eggshell proteins of Drosophila melanogaster and their program of synthesis. Dev. Biol. 1976;49:185–199. doi: 10.1016/0012-1606(76)90266-9. [DOI] [PubMed] [Google Scholar]
- Popodi E, Minoo P, Burke T, Waring GL. Organization and expression of a second chromosome follicle cell gene cluster in Drosophila. Dev. Biol. 1988;127:248–256. doi: 10.1016/0012-1606(88)90312-0. [DOI] [PubMed] [Google Scholar]
- Scherer LJ, Harris DH, Petri WH. Drosophila vitelline membrane genes contain a 114 base pair region of highly conserved coding sequence. Dev. Biol. 1988;130:786–788. doi: 10.1016/0012-1606(88)90367-3. [DOI] [PubMed] [Google Scholar]
- Schulz RA, The SM, Hogue DA, Galewsky S, Guo Q. Ets oncogene-related gene Elg functions in Drosophila oogenesis. Proc. Natl. Acad. Sci. USA. 1993;90:10076–10080. doi: 10.1073/pnas.90.21.10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Jacobs-Lorena M. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. Cloning, expression, and characterization. J. Biol. Chem. 1998;273:17665–17770. doi: 10.1074/jbc.273.28.17665. [DOI] [PubMed] [Google Scholar]
- Spradling A. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27:193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- Spradling A, Mahowald AP. Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1980;77:1096–1100. doi: 10.1073/pnas.77.2.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. The Drosophila embryonic patterning determinant torsolike is a component of the eggshell. Curr. Biol. 2003;13:1058–1063. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- Thireos G, Griffin-Shea R, Kafatos FC. Untranslated mRNA for a chorion protein of Drosophila melanogaster accumulates transiently at the onset of specific gene amplification. Proc. Natl. Acad. Sci. USA. 1980;77:5789–5793. doi: 10.1073/pnas.77.10.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trougakos IP, Margaritis LH. Immunolocalization of the temporally ”early” secreted major structural chorion proteins, Dvs38 and Dvs36, in the eggshell layers and regions of Drosophila virilis. J. Struct. Biol. 1998;123:111–123. doi: 10.1006/jsbi.1998.4028. [DOI] [PubMed] [Google Scholar]
- Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- van Eeden F, St. Johnston D. The polarisation of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr. Opin. Genet. Dev. 1999;9:396–404. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- Verheyen E, Cooley L. Looking at oogenesis. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: practical uses in cell and molecular biology. Academic Press; New York: 1994. pp. 545–561. [Google Scholar]
- Vogel C, Teichmann SA, Chothia C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech. Dev. 2005;122:241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. Int. Rev. Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Wex T, Lipyansky A, Bromme NC, Wex H, Guan XQ, Bromme D. TIN-ag-RP, a novel catalytically inactive cathepsin B-related protein with EGF domains, is predominantly expressed in vascular smooth muscle cells. Biochemistry. 2001;40:1350–1357. doi: 10.1021/bi002266o. [DOI] [PubMed] [Google Scholar]
- Wu CC, Yates JR., 3rd The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- Yannoni CZ, Petri WH. Localization of a gene for a minor chorion protein in Drosophila melanogaster: a new chorion structural locus. Dev. Biol. 1984;102:504–508. doi: 10.1016/0012-1606(84)90217-3. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Thomas GH. Apical spectrin is essential for epithelial morphogenesis but not apicobasal polarity in Drosophila. J. Cell Biol. 1999;146:1075–1086. doi: 10.1083/jcb.146.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]