Abstract
The ability to generate gaseous doubly charged cations of glycerophosphocholine (GPC) lipids via electrospray ionization has made possible the evaluation of electron transfer dissociation (ETD) for their structural characterization. Doubly sodiated GPC cations have been reacted with azobenzene radical anions in a linear ion trap mass spectrometer. The ion/ion reactions proceed through sodium transfer, electron transfer, and complex formation. Electron transfer reactions are shown to give rise to cleavage at each ester linkage with the subsequent loss of a neutral quaternary nitrogen moiety. Electron transfer without dissociation produces [M+2Na]+• radical cations, which undergo collision-induced dissociation (CID) to give products that arise from bond cleavage of each fatty acid chain. The CID of the complex ions yields products similar to those produced directly from the electron transfer reactions of doubly sodiated GPC, although with different relative abundances. These findings indicate that the analysis of GPC lipids by ETD in conjunction with CID can provide some structural information, such as the number of carbons, degree of unsaturation for each fatty acid substituent, and the positions of the fatty acid substituents; some information about the location of the double bonds may be present in low intensity CID product ions.
Keywords: Electron Transfer Dissociation, Ion/Ion Reaction, Glycerophosphocholine Lipid, Linear Ion Trap
INTRODUCTION
With the advent of electrospray ionization (ESI) [1, 2] and matrix-assisted laser desorption ionization (MALDI) [3, 4], the analysis of lipids using tandem mass spectrometry (MS) has been greatly facilitated since these techniques can generate the intact gaseous pseudo-molecular ions for most lipids ranging from simple fatty acids to complex lipids [5–10]. Among the various complex lipids, glycerophospholipids (GPLs) perform two important biological functions: one is making up most of the membranes of mammalian cells, and the other is acting as secondary messengers in metabolism [11, 12]. The structural determination (identities and positions of the fatty acid substituents) of GPLs for all five subclasses has been made via CID of negative ions formed by either ESI, fast atom bombardment (FAB), or MALDI [5, 13, 14]. Among the five main subclasses of GPLs, which are differentiated by the head-group (e.g., ethanolamine, choline, serine, glycerol, or inositol), glycerophosphocholine (GPC) species generally give stronger signals in positive ion mode than in negative mode due to the fixed charge on the quaternary nitrogen. The structural information mentioned above can be obtained by CID of alkali adducts of GPC species generated in positive ion ESI-MS [15]. Little or no information regarding the location of the double bonds in GPCs has been reported by these methods.
Both electron capture dissociation (ECD) [16–19] and electron transfer dissociation (ETD) [20–23] have been shown to be particularly useful in the structural determination of proteins and peptides [24–27]. This is mainly because more extensive sequence information can often be obtained via ECD or ETD than via CID and because, unlike CID, labile post-translational modifications are often retained, which is particularly valuable in establishing their location in the sequence [21, 28]. Thus, it is of interest to explore the ETD and ECD behavior of other ions derived from bio-molecules of interest.
In this communication, we report results from ion/ion electron transfer reactions of doubly sodiated GPC lipids. The major ETD channels show preferential cleavage at ester linkages. Thus the carbon numbers and degrees of unsaturation for each chain can be readily determined. Electron transfer without dissociation produces [M + 2Na]+• ions, which fragment to give products that arise from cleavage of the lipid backbone when subjected to collision-induced dissociation (CID). In addition, subsequent CID of the singly charged radical cations formed during the electron transfer reaction appears to be capable of yielding a limited degree of information regarding the positions of the double bonds.
EXPERIMENTAL SECTION
Materials
Chloroform and methanol were purchased from Mallinckrodt (Phillipsburg, NJ). Sodium salts and azobenzene were obtained from Sigma-Aldrich (St. Louis, MO). The 1-stearoyl-2-arachidonyl-sn-glycerol-3-phosphocholine ((18:0/20:4)-GPC) and 1-stearoyl-2-docosahexaenoyl-sn-glycerol-3-phosphocholine ((18:0/22:6)-GPC) lipids were obtained from Avanti Polar Lipids (Birmingham, AL) and used without further purification. These GPC lipids were dissolved to 0.5 mg/mL in a 50/50 (vol/vol) methanol/chloroform solution with 1mM (final concentration) NaCl added for generation of sodium adducts while 1% (v/v) acetic acid was added for singly protonated GPC formation by positive nano-ESI. It was noted that relatively low interface voltage gradients tended to maximize observation of the doubly charged ions.
Mass Spectrometry
All experiments were performed using a prototype version of a Q TRAP mass spectrometer [29] (Applied Biosystems/MDS SCIEX, Concord, Ontario, Canada) equipped with a home-made dual nano-ESI/APCI source, which was described in detail elsewhere [30]. All the electronics were controlled by Daetalyst 3.14 software, a research version of software provided by MDS SCIEX.
A typical scan function for an ion/ion electron transfer experiment consists of the following steps: positive ion injection into Q2 LIT (100 ms), anion injection into Q2 LIT (100 ms), mutual storage of cations and anions in Q2 LIT (400 ms) and transfer of ion/ion reaction product ions to mass analyzer Q3 (50 ms) for mass analysis. For an ion/ion electron-transfer reaction experiment, the positive high voltage power supply connected to the nano-ESI wire was pulsed on to generate analyte ions. Analyte ions were isolated by Q1 in RF/DC mode and injected axially into the Q2 LIT with nitrogen as the buffer gas at a pressure of ~ 5 mTorr. These ions were cooled in Q2 for 30 ms, during which time the high voltage on this emitter was turned off. After the cooling step, the power supply connected to the APCI wire, which was operated in the negative polarity, was triggered on to generate the electron transfer reagent anions. These reagent anions were isolated by Q1 in RF/DC mode before they entered Q2 LIT with relatively low kinetic energies (~5 eV). During this period, the DC potentials on the Q2 containment lenses, as well as on the Q2 rods themselves, were adjusted to a common value while an auxiliary RF voltage was applied to IQ3 lens. In the subsequent mutual ion polarity storage step, the potential applied to the APCI emitter was turned off and the auxiliary RF signals were applied to both containment lenses (i.e., IQ2 and IQ3) to store ions axially. After a defined period of mutual storage, the residual reagent ions were ejected from Q2 by applying attractive DC potentials to the Q2 containment lenses while the auxiliary RF signals were terminated. Then the positively charged product ions arising from ion/ion electron transfer reactions, as well as the residual precursor ions, were transferred from Q2 to Q3, and cooled for 50 ms before they were subjected to mass selective axial ejection (MSAE) [31] using a supplementary RF signal at a frequency of 380 kHz. When CID experiments on the product ions resulting from the ETD reactions were desired, the product ions were isolated in Q3 first in RF/DC mode and subsequently subjected to activation by applying an appropriate auxiliary AC on one pair of the Q3 rods.
RESULTS AND DISCUSSION
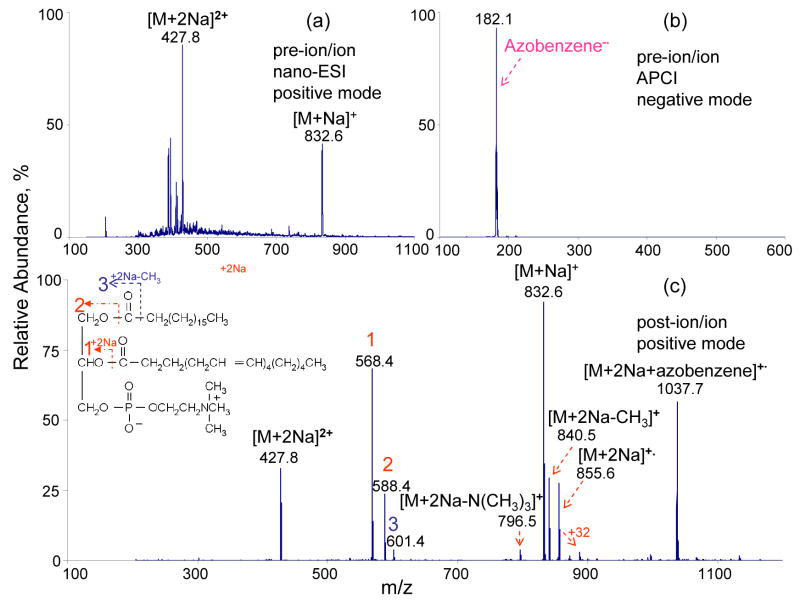
The ability to generate multiply charged positive ions of GPCs is a necessary condition for employing ETD in the analysis of GPCs. Figure 1a shows the nano-ESI spectrum of sodiated adducts of (18:0/20:4)-GPC. The relative abundances of doubly sodiated and singly sodiated adducts are largely determined by the solution and atmosphere/vacuum interface conditions. The ion/ion reactions between isolated doubly sodiated adducts of (18:0/20:4)-GPC and singly charged azobenzene radical anions (shown in Figure 1b) give rise to the mass spectrum of Figure 1c. The main reaction products can be grouped into 3 classes: (1) sodium transfer from the doubly sodiated (18:0/20:4)-GPC to the azobenzene anion to form [M+Na] +•; (2) complex formation to give [M+2Na+azobenzene]+•; (3) and electron transfer from the azobenzene anion to [M+2Na]2+ to generate [M+2Na]+• (i.e., electron transfer without dissociation) and [M+2Na-267]+, [M+2Na-287]+, [M+2Na-15]+, and [M+2Na-59] + ETD products. The losses of 15 Th and 59 Th corresponds to loss of a methyl radical and tri-methylamine, respectively, while the losses of 267 and 287 Th correspond to losses of each of the respective hydrocarbon chains in the form of a radical [RC=O•]. The former two products are reflective of the head group while the latter two products indicate that cleavage of the ester bonds is the dominant ETD process for doubly sodiated GPCs. From the masses of the latter two products, the carbon number and degree of unsaturation for each chain can be obtained.
Figure 1.
Mass spectra derived from (a) positive nano-ESI of (18:0/20:4)-GPC, (b) negative APCI of azobenzene, (c) ion/ion ETD reaction of isolated doubly sodiated (18:0/20:4)-GPC cations with singly charged azobenzene anions in Q2 LIT for 400 ms.
Moreover, the relative abundances of ions reflecting the losses of the fatty acid substituents from ETD of doubly sodiated GPC species might indicate the positions of these substituents, although a much larger data set is required to draw conclusions regarding this possibility. Comparison of the relative abundances of the ions at m/z 568 ([M+2Na-R2C=O]+) and m/z 588 ([M+2Na-R1C=O]+) in Figure 1c indicates that the abundance of the ion resulting from the loss of the sn-2 substituent exceeds that of the ion resulting from the loss of the sn-1 substituent. The same observation has been made in the ETD results of doubly sodiated (18:0/22:6)-GPC lipids (data not shown), although with a difference in the relative abundances of the relevant product ions. This possible tendency stands in contrast to that from the CID of (18:0/20:4)-GPC lipids in the form of [M+Li]+, [M-15]− or [M+acetate]−, which generally show the preferential loss of the sn-1 acyl group [10, 15, 32].
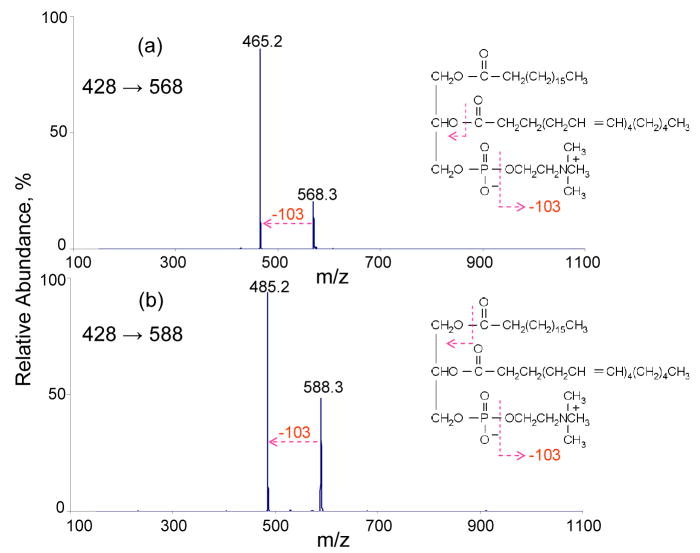
The fragmentation behavior of these ETD product ions were further explored with MS3 experiments using CID. The results of MS3 experiments on the m/z 568 and m/z 588 ions produced in the reaction discussed above are shown in Figure 2. Collisional activation in both cases leads exclusively to a loss of 103 Th, which corresponds to the neutral loss of [O(CH2)2N(CH3)3]. While this loss provides information about the head group, no new information about the fatty acid substituents is forthcoming from this reaction. Although it would be desirable to perform MS4 to examine whether dissociation of the CID product can provide further structural information, the absolute signal of the ion in the MS3 spectrum is low, which compromises the practical utility of such an experiment.
Figure 2.
(a) MS3 of m/z 568 [M+2Na-287]+. (b) MS3 of m/z 588 [M+2Na-267]+.
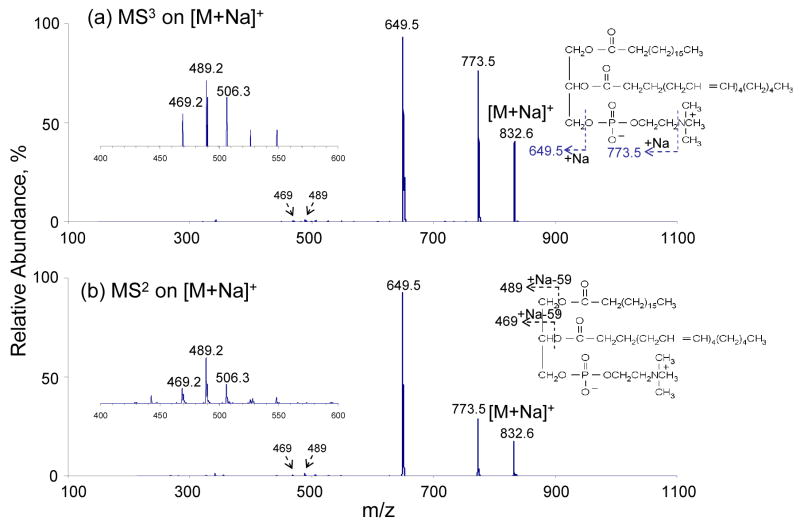
Figure 3a shows the MS3 data resulting from the CID of the [M+Na]+ ions generated from ETD of the doubly sodiated (18:0/20:4)-GPC. The major dissociation channels are the losses of the trimethylamine head-group and the cholinephosphate group. Low abundance peaks at m/z 489 and m/z 469 correspond to the loss of each fatty acid subsituent in the form of [M-RCOOH-59]+. Due to the LMCO of m/z 200 used for this in-trap ion activation, no ions below m/z 200 were recorded. For comparison, the CID spectrum of [M+Na]+ generated directly from solution-phase via nano-ESI is also included here. The results are shown in Figure 3b. The results are similar to the ions shown in Figure 3a in terms of ion identity and relative abundance. This result indicates that the singly sodiated (18:0/20:4)-GPC species formed via electron transfer dissociation and directly via nano-ESI can not be distinguished with ion-trap CID.
Figure 3.
(a) CID MS3 of m/z 833 generated from ETD of doubly sodiated (18:0/20:4)-GPCs. (b) CID MS2 of m/z 833 generated from nano-ESI of (18:0/20:4)-GPCs solution.
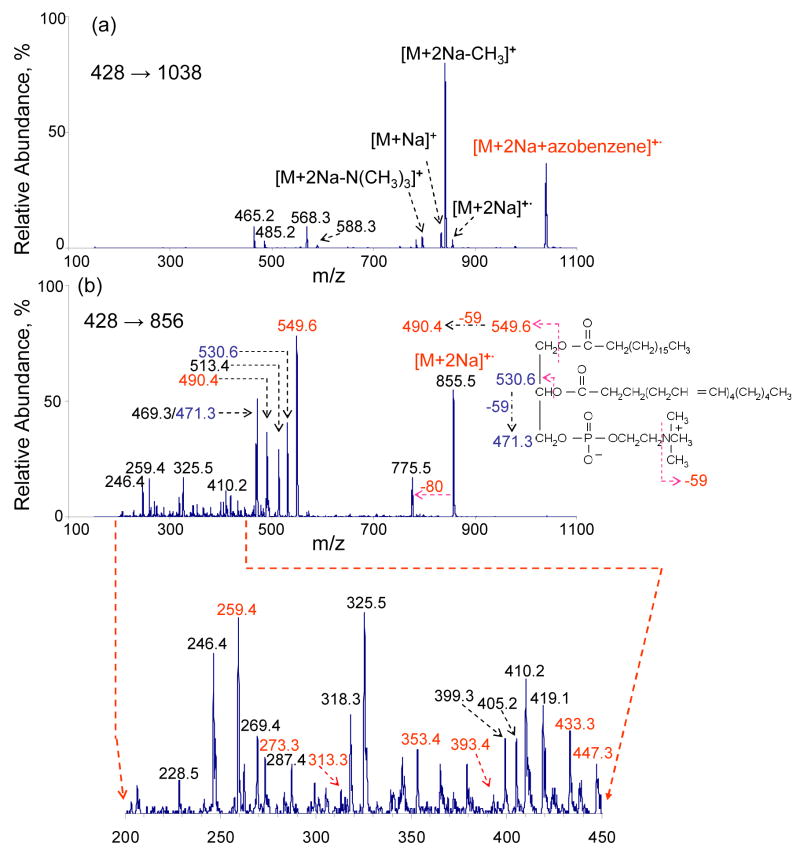
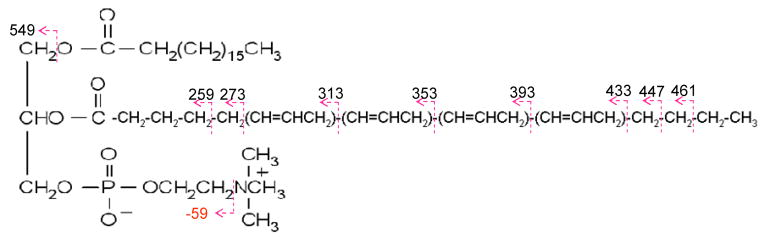
While CID of the major first generation ETD product ions does not provide additional information regarding the fatty acid chains, it is of interest to examine the ion/ion reaction products that do not lead to fragmentation. Figure 4a shows the CID product ion spectrum of the complex ions formed in the reaction of doubly sodiated (18:0/20:4)-GPC with the azobenzene anion. The product ion masses mirror those in the ETD spectrum of (18:0/20:4)-GPC and also include products ions at m/z 465 and m/z 485, which are the sequential fragmentation products of the ions at m/z 568 and m/z 588, respectively (see the MS3 data of Figure 2). Figure 4b shows the mass spectrum resulting from the collisional activation of the non-dissociative electron transfer product [M+2Na]+•. The CID product ion at m/z 549 likely results from cleavage of the C-O bond β to the carbonyl carbon of the ester linkage (see Figure 4b) of 18:0 fatty acid chain. The CID product ion at m/z 530 probably results from cleavage of the C-O bond β to the carbonyl carbon of the ester linkage of 20:4 fatty acid chain with, perhaps, a hydrogen transfer to the ion, but the mass is observed at 1 Da unit higher that expected. Along with the acyl chain losses, the concomitant losses of neutral trimethylamine head-group (59 Th) in both cases give rise to the ions of m/z 490 and m/z 471, respectively. Product ion at m/z 513 is tentatively assigned to [M+2Na-R1COO-N(CH3)3] (R1=18:0 fatty acid chain). The loss of 80 Th from CID of [M+2Na]+• may arise from the loss of HPO3, although it requires a rearrangement mechanism during the CID process. The product ions shown in the m/z range from 200 and 470 include some consecutive mass differences of 14 Th and 40 Th. This leads to some information about cleavage along the sn-2 fatty acid chain (20:4). For example, Scheme 1 shows plausible cleavages that give rise to product ions of the indicate mass-to-charge ratios.
Figure 4.
(a) MS3 of m/z 1038 [M+2Na+azobenzene] +•. (b) MS3 of m/z 856 [M+2Na] +•.
Scheme 1.
Proposed fragmentation pathway for the location of the double bond in (18:0/20:4)-GPC species.
CONCLUSIONS
Ion/ion reactions between doubly sodiated GPC species result in sodium transfer to the anion, adduct formation, electron transfer without dissociation, and ETD of the cation. ETD products directly provide carbon number and degree of unsaturation. The relative abundances of the ions may also yield fatty acid position but too little data are in hand to draw a firm conclusion on this point. Electron transfer without dissociation produces [M + 2Na]+• ions, which fragment upon collisional activation to give products that correspond to bond cleavage of each fatty acid chain. The CID of complex ions generates products that are similar to those produced directly from the electron transfer reactions of doubly sodiated GPC cations, in terms of ion identities, although with different relative abundances. These findings indicate that the ETD of doubly sodiated GPC lipids gives similar to that derived from CID of alkali adducts. CID of the singly charged radical cations formed via electron transfer may be able to yield some degree of information about double bond positions but further work is warranted to assess this potential. In any case, the signal levels associated with the potentially informative ions are relatively low, which affects the overall sensitivity of the approach.
Acknowledgments
This work was sponsored by MDS SCIEX, an Industrial Associate of the Department of Chemistry and the National Institutes of Health, National Institute of General Medical Sciences under Grants GM 45372 and GM 071534.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray Ionization Principles and Practice. Mass Spectrom Rev. 1990;9:37–70. [Google Scholar]
- 3.Karas M, Bachman D, Bahr U, Hillenkamp F. Matrix-Assisted Ultraviolet Laser Desorption of Non-Volatile Compounds. Int J Mass Spectrom Ion Proc. 1987;78:53–68. [Google Scholar]
- 4.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T. Protein and Polymer Analyses up to M/Z 100000 by Laser Ionization Time-of-Flight Mass Spectrometry. Rapid Commun Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 5.Murphy RC, Fiedler J, Hevko J. Analysis of Nonvolatile Lipids by Mass Spectrometry. Chem Rev. 2001;101:479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 6.Murphy RC, Raetz CR, Reynolds CM, Barkley RM. Mass Spectrometry Advances in Lipidomica: Collision-Induced Decomposition of Kdo2-Lipid A. Prostaglandins Other Lipid Mediat. 2005;77:131–140. doi: 10.1016/j.prostaglandins.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey DJ. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Phospholipids. J Mass Spectrom. 1995;30:1333–1346. [Google Scholar]
- 8.Kaufmann R, Wingerath T, Kirsch D, Stahl W, Sies H. Analysis of Carotenoids and Carotenol Fatty Acid Esters by Matrix-Assisted Laser Desorption Ionization (MALDI) and MALDI–Post-Source-Decay Mass Spectrometry. Anal Biochem. 1996;238:117–128. doi: 10.1006/abio.1996.0264. [DOI] [PubMed] [Google Scholar]
- 9.Asbury GR, Al-Saad K, Siems WF, Hannan RM, HillJr HH. Analysis of Triacylglycerols and Whole Oils by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry. J Am Soc Mass Spectrom. 1999;10:983–991. [Google Scholar]
- 10.Stubiger G, Belgacem O. Analysis of Lipids Using 2,4,6-Trihydroxyacetophenone as a Matrix for MALDI Mass Spectrometry. Anal Chem. 2007;79:3206 –3213. doi: 10.1021/ac062236c. [DOI] [PubMed] [Google Scholar]
- 11.Maxfield FR, Tabas I. Role of Cholesterol and Lipid Organization in Disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 12.Zeisel SH, Blusztajn JK. Choline and Human Nutrition. Annu Rev Nutr. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- 13.Hsu FF, Turk J, Owens RM, Rhoades ER, Russell DG. Structural Characterization of Phosphatidyl-Myo-Inositol Mannosides from Mycobacterium Bovis Bacillus Calmette Guerin by Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. I. Pims and Lyso-Pims. J Am Soc Mass Spectrom. 2007;18:466–478. doi: 10.1016/j.jasms.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu FF, Turk J, Owens RM, Rhoades ER, Russell DG. Structural Characterization of Phosphatidyl-Myo-Inositol Mannosides from Mycobacterium Bovis Bacillus Calmette Guerin by Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. Ii. Monoacyl- and Diacyl-Pims. J Am Soc Mass Spectrom. 2007;18:479–492. doi: 10.1016/j.jasms.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu FF, Bohrer A, Turk J. Formation of Lithiated Adducts of Glycerophosphocholine Lipids Facilitates Their Identification by Electrospray Ionization Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 1998;9:516–526. doi: 10.1016/S1044-0305(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 16.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 17.Zubarev RA, Kruger NA, Fridricksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron Capture Dissociation of Gaseous Multiply-Charged Proteins Is Favored at Disulfide Bonds and Other Sites of High Hydrogen Atom Affinity. J Am Chem Soc. 1999;121:2857–2862. [Google Scholar]
- 18.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron Capture Dissociation for Structural Characterization of Multiply Charged Protein Cations. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 19.Zubarev RA. Reactions of Polypeptide Ions with Electrons in the Gas Phase. Mass Spectrom Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 20.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc Natl Acad Sci US A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion Dependence in the Partitioning between Proton and Electron Transfer in Ion/Ion Reactions. Int J Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 22.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron Transfer Ion/Ion Reactions in a Three-Dimensional Quadrupole Ion Trap: Reactions of Doubly and Triply Protonated Peptides with SO2- Anal Chem. 2005;77:1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunawardena HP, He M, Chrisman PA, Pitteri SJ, Hogan JM, Hodges BD, McLuckey SA. Electron Transfer Versus Proton Transfer in Gas-Phase Ion/Ion Reactions of Polyprotonated Peptides. J Am Chem Soc. 2005;127:12627–12639. doi: 10.1021/ja0526057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top Down Characterization of Larger Proteins (45 Kda) by Electron Capture Dissociation Mass Spectrometry. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 25.Ge Y, El-Naggar M, Sze SK, Oh HB, Begley TP, McLafferty FW, Boshoff H, Barry CE., 3rd Top Down Characterization of Secreted Proteins from Mycobacterium Tuberculosis by Electron Capture Dissociation Mass Spectrometry. J Am Soc Mass Spectrom. 2003;14:253–261. doi: 10.1016/s1044-0305(02)00913-3. [DOI] [PubMed] [Google Scholar]
- 26.Sze SK, Ge Y, Oh H, McLafferty FW. Top-Down Mass Spectrometry of a 29-Kda Protein for Characterization of Any Posttranslational Modification to within One Residue. Proc Natl Acad Sci US A. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein Identification Using Sequential Ion/Ion Reactions and Tandem Mass Spectrometry. Proc Natl Acad Sci US A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone DH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Anal Chem. 2001;2001:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 29.Hager JW. A New Linear Ion Trap Mass Spectrometer. Rapid Commun Mass Spectrom. 2002;16:512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 30.Liang X, Xia Y, McLuckey SA. Alternately Pulsed Nanoelectrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Anal Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Londry FA, Hager JW. Mass Selective Axial Ion Ejection from a Linear Quadrupole Ion Trap. J Am Soc Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 32.Jackson SN, Wang HY, Woods AS. In Situ Structural Characterization of Phosphatidylcholines in Brain Tissue Using MALDI-MS/MS. J Am Soc Mass Spectrom. 2005;16:2052–2056. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]