Abstract
The low stability of natural proteins often limits their use in therapeutic, industrial and research applications. The scale and throughput of methods such as circular dichroism, fluorescence spectroscopy and calorimetry severely limit the number of variants that can be examined. Here we demonstrate a high-throughput thermal scanning (HTTS) method for determining the approximate stabilities of protein variants at high throughput and low cost. The method is based on binding to a hydrophobic dye akin to ANS, which fluoresces upon binding to molten globules and thermal denaturation intermediates. No inherent properties of the protein, such as enzymatic activity or presence of an intrinsic fluorophore, are required. Very small sample sizes are analyzed using a real-time PCR machine, enabling the use of high-throughput purification. We show that the apparent TM values obtained from HTTS are approximately linearly related to those from CD thermal denaturation for a series of four-helix bundle hydrophobic core variants. We demonstrate similar results for a small set of TIM barrel variants. This inexpensive, general and scaleable approach enables the search for conservative, stable mutants of biotechnologically-important proteins, and it provides a method for statistical correlation of sequence-stability relationships.
Natural proteins are often too unstable for therapeutic or industrial applications, or even for crystallography or directed evolution experiments.1 There is still no reliable way to predict stabilizing mutations, and biophysical characterization of proteins is generally large-scale and low-throughput.2 Except for enzymes, where enzymatic activity can be screened at elevated temperatures, high-throughput methods of screening for stability are lacking. Notably, the dominant classes of protein drugs—hormones, antibodies, cytokines, etc.—are binding proteins or ligands, not enzymes. Here we demonstrate that a dye-binding thermal shift screen, an extension of the ThermoFluor® method of screening for protein-ligand interactions,3 reports the relative thermal stabilities of libraries of protein variants. We call the method High-Throughput Thermal Scanning, or HTTS.
In ThermoFluor®, samples of a receptor protein are mixed with an analyte ligand and a fluorescent hydrophobic dye akin to 1-anilinonaphthalene-8-sulphonic acid (ANS). Folded proteins exclude these types of dyes, but molten globules and thermal denaturation intermediates bind them, resulting in a sharp increase in fluorescence. Binding of a ligand to the folded state of the receptor shifts the apparent melting temperature higher, which can be observed by heating the sample in a fluorimeter. Besides for drug discovery, this method has been applied to optimization of ligand and buffer conditions for crystallography.4
We wished to invert the format of the screen, instead using a library of protein variants under the same conditions of dye and buffer, to probe the approximate relative thermal stabilities of the mutants. Since dye binding is so physically different from circular dichroism (CD) spectroscopy or intrinsic fluorescence, and because many interesting mutants may have very different amino acid composition, it was not clear at the outset how the associated melting temperatures would correspond to standard large-scale approaches. Moreover, since a different protein is required in each well of the 96-well plate, compatible inexpensive and small-scale growth and purification had to be achieved.
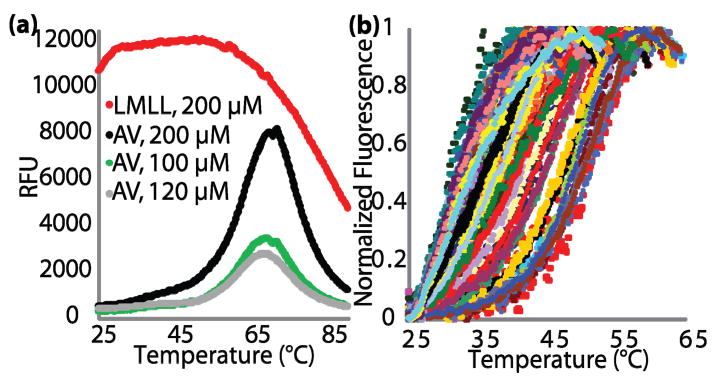
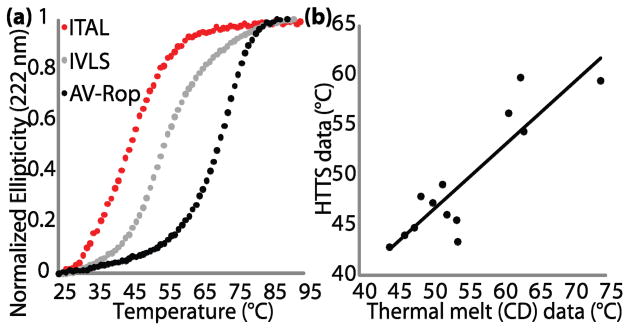
We first validated the method using a small set of variants of the simple, well-studied four-helix bundle protein, Rop.5 The Rop variants are from two combinatorial libraries in which the two central layers (residues 15, 19, 41 and 45) of the hydrophobic core were repacked. We name the library variants by the amino acids at the four varied positions; hence, wild-type Rop is ITLA. AV-Rop is an engineered cysteine-free variant (C38A C52V) with wild-type like thermal stability as measured by CD spectroscopy (S.B.H., Chang Byeon, J.J.L. & T.J.M., manuscript in preparation). When AV-Rop is mixed with dye, there is little fluorescence at room temperature, but there is a sharp increase in fluorescence between 52–63 °C (Fig. 1b). As expected from the use of an ANS-like hydrophobic dye, HTTS is readily able to distinguish molten globular proteins from native-like proteins. When a molten globular variant of Rop, LMLL, is mixed with dye, there is a large fluorescence signal at room temperature, and there is no increase in fluorescence upon heating (Fig. 3a).
Figure 1.
(a) Schematic of HTTS. Molten globular variants bind hydrophobic dyes such as ANS at room temperature. Native variants exclude the dye at low temperature but bind it strongly as the protein thermally denatures, resulting in a sharp fluorescence signal increase. (b) HTTS of three native-like variants of Rop which vary substantially in their thermal stabilities.
Figure 3.
(a) Full, un-normalized HTTS traces for a molten globular variant (LMLL) and AV-Rop at three concentrations in three independent experiments. (b) HTTS traces of native-like variants from a random selection of 96 core-repacked mutants of AV-Rop.
A real-time PCR machine was used for these experiments because it is compatible with very small volumes and because most multiwell plate readers cannot heat samples to 95 °C. Because the excitation and emission maxima of ANS are not accessible by commercial filter sets for RT-PCR machines, we use SYPRO® Orange, which has been used successfully for ThermoFluor®.6 By application of a variation on the Clarke & Fersht7 equation using data from 25 °C to the fluorescence maximum, we calculate an apparent TM value.
In general, the amount of protein required to obtain good signal-to-noise is about 100 μM with 5× dye (the absolute concentration of SYPRO® Orange is not disclosed by the manufacturer). For Rop and other proteins tested in the lab, sufficient protein can be obtained from 2 mL of culture grown in deep 96-well plates with T7 overexpression. Saturated overnight cultures are diluted and induced with IPTG, and cells are lysed with glass beads or detergent. Hexahistidine-tagged protein is captured on magnetic NiNTA beads, washed, and released by proteolysis using TEV protease. Curiously, the His6-TEV site tag must be removed from the sample, because it results in a dye-mediated fluorescence signal at about 45 °C. The NiNTA magnetic beads, the most expensive component of the method, can be regenerated for use many times.
To assess the validity of HTTS to measure the approximate stabilities of a series of protein variants, 13 Rop variants were also analyzed using CD spectroscopy (Fig. 2). The HTTS apparent TM values are roughly related linearly to the CD TM values, where the average offset between them depends upon buffer and dye conditions. Five variants bind large amounts of hydrophobic dye at room temperature, consistent with their non-cooperative CD thermal melts. CD urea denaturation data differ more substantially from either thermal method.
Figure 2.
(a) Thermal denaturation of the same three variants as in Fig. 1b, monitored by CD at 222 nm. (b) Plot of CD TMs versus HTTS apparent TMs for 13 native-like Rop variants.
We also subjected 7 variants of yeast triosephosphate isomerase (TIM) to both CD thermal melts and HTTS, with excellent correspondence (Supporting Information). TIM is a 240 a.a. (β/α)8 barrel protein, radically different in size and overallfold, supporting the generality of this method.
Randomly-selected active variants from an AV-Rop library were purified and analyzed in 96-well format. This library was constructed so that residues 15, 19, 41, and 45 were subrandomized to the hydrophobic residues (AVILMF) and alcohols (ST). The variants in this library range from slightly stabilized to molten globular, as might be expected from the radical repacking of the core of a small protein. Native-like variants (Fig. 3b) were separated from molten globules and variants that failed to express using empirical analysis of the fluorescence signals (i.e., identifying those with very high or very low fluorescence signals throughout the melt). A full sequence-stability analysis of this library will be presented separately.
Several of the variants from this library were identical in amino acid sequence. These repeats differ only in the amount of signal, but not in the midpoint of denaturation or overall shape of the curve. The differences in signal can be attributed to differences in protein expression and purification and random differences from the normalization procedure employed by the RT-PCR software used here. One normally expects concentration-dependent variation in TM values for multimeric proteins. However, Rop concentrations are quite uniform here, and Rop thermal stability is remarkably insensitive to concentration above 1 μM.
The CD thermal melts of all Cys-free Rop variants tested are reversible. In HTTS, after passing through the fluorescence maximum, there is always decreasing fluorescence observed upon continued heating. This may be caused by aggregation or temperature-dependent dye fluorescence or binding (Fig. 3a). Although we have called the midpoint values for the melting curves apparent TMs, the conditions are not fully reversible and the system may not be at equilibrium particularly after passing through the apparent melting points. Consequently, variants with very different unfolding rates may behave anomalously in HTTS.
Protein stability is typically characterized by calorimetry, or thermal or chemical denaturation observed by intrinsic fluorescence or CD spectroscopy. Automation increases the throughput, but not all proteins have an intrinsic fluorophore and specialized equipment is required.8 Indirect screens (binding or proteolysis resistance of phage-displayed variants, or protein solubility and expression) have also been applied.9 But here we present one of the first high-throughput collections of thermodynamic data on a single protein scaffold, using a method that is simple and inexpensive enough to be carried out in any lab with access to a real-time PCR machine.
Thermal shift assays have been used to optimize the thermal stability of a single protein under different conditions, either with a battery of ligands or with a battery of buffer conditions.3, 4, 6 Inverting the format of the screen to examine a library of protein variants, as we demonstrate here, opens the door to new kinds of discovery. Protein-based therapeutics can have remarkable specificity, but the marginal stability of natural proteins is problematic for storage and pharmacological properties.10 Protein engineering to increase solubility and expression,9 remove or introduce surface interactions,11 or generate stable subfragments12 has become a critical tool in structural biology in recent years. The HTTS method goes beyond these, enabling a search for conservative mutants of a protein with enhanced stability.
HTTS is a simple, low-cost method that uses commonly available instrumentation. It requires only microgram quantities of protein that can be purified using commercially-available reagents designed for high-throughput analysis. HTTS is able to provide an abundance of thermodynamic data on a large library of protein variants in a single one-hour experiment. The ability to acquire large sample sizes of thermodynamic and sequence data presents an obvious avenue to analyzing their relationship in a statistical manner.
Supplementary Material
Acknowledgments
JJL was an NIH CBIP fellow and is a pre-doctoral fellow of the AHA Great Rivers Affiliate. SBH was an OSU Biochemistry SURP fellow and Mayers Summer Research Intern. BJS is an NIH CBIP fellow. Thanks to Mike Zianni, PMGF, for assistance with real-time PCR. This work was supported by the NIH (R01GM083114 and U54NS058183) and The Ohio State University.
Footnotes
References
- 1.Arnold FH. Nature. 2001;409:253–7. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]; Bloom JD, et al. Proc Natl Acad Sci USA. 2006;103:5869–74. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Graddis TJ, Remmele RL, Jr, McGrew JT. Curr Pharm Biotechnol. 2002;3:285–297. doi: 10.2174/1389201023378148. [DOI] [PubMed] [Google Scholar]
- 2.Magliery TJ, Regan L. Eur J Biochem. 2004;271:1595–608. doi: 10.1111/j.1432-1033.2004.04075.x. [DOI] [PubMed] [Google Scholar]; Richards FM. Cell Mol Life Sci. 1997;53:790–802. doi: 10.1007/s000180050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings MD, Farnum MA, Nelen MI. J Biomol Screen. 2006;11:854–63. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]; Matulis D, et al. Biochemistry. 2005;44:5258–66. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]; Pantoliano MW, et al. J Biomol Screen. 2001;6:429–40. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 4.Ericsson UB, et al. Anal Biochem. 2006;357:289–98. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]; Mezzasalma TM, et al. J Biomol Screen. 2007;12:418–28. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]; Niesen FH, Berglund H, Vedadi M. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]; Vedadi M, et al. Proc Natl Acad Sci USA. 2006;103:15835–40. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yeh AP, McMillan A, Stowell MH. Acta Crystallogr D. 2006;62:451–7. doi: 10.1107/S0907444906005233. [DOI] [PubMed] [Google Scholar]
- 5.Munson M, et al. Protein Sci. 1996;5:1584–93. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo MC, et al. Anal Biochem. 2004;332:153–9. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 7.Clarke J, Fersht AR. Biochemistry. 1993;32:4322–9. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 8.Edgell MH, et al. Biochemistry. 2003;42:7587–93. doi: 10.1021/bi034063g. [DOI] [PubMed] [Google Scholar]
- 9.Roodveldt C, Aharoni A, Tawfik DS. Curr Opin Struct Biol. 2005;15:50–6. doi: 10.1016/j.sbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Frokjaer S, Otzen DE. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 11.Cooper DR, et al. Acta Crystallogr D. 2007;63:636–45. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 12.Hecky J, Muller KM. Biochemistry. 2005;44:12640–54. doi: 10.1021/bi0501885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.