Abstract
We recently described two opposing states of transcriptional competency. One is termed ‘competent’ whereby a gene is capable of responding to trans-acting transcription factors of the cell, such that it is active if appropriate transcriptional activators are present, though it can also be silent if activators are absent or repressors are present. The other is termed ‘occluded’ whereby a gene is silenced by cis-acting, chromatin-based mechanisms in a manner that blocks it from responding to trans-acting factors, such that it is silent even when activators are present in the cellular milieu. We proposed that gene occlusion is a mechanism by which differentiated cells stably maintain their phenotypic identities. Here, we describe chromatin analysis of occluded genes. We found that DNA methylation plays a causal role in maintaining occlusion for a subset of occluded genes. We further examined a variety of other chromatin marks typically associated with transcriptional silencing, including histone variants, covalent histone modifications and chromatin-associated proteins. Surprisingly, we found that although many of these marks are robustly linked to silent genes (which include both occluded genes and genes that are competent but silent), none is linked specifically to occluded genes. Although the observation does not rule out a possible causal role of these chromatin marks in occlusion, it does suggest that these marks might be secondary effect rather than primary cause of the silent state in many genes.
INTRODUCTION
A fundamental feature of multicellular organisms is the ability of differentiated cell types to maintain their phenotypic identities over time irrespective of fluctuations in the physiological environment of the cells (1). How, at the molecular level, the myriad cell types in an organism manage to stably preserve their distinct identities is an important but little understood question in biology. One attractive idea is that the identity of a given cell type is safeguarded by the irreversible silencing (or ‘occlusion’) of what can be referred to as lineage-inappropriate genes—i.e. genes whose aberrant expression would promote alternative lineages (2–6).
This idea received considerable support from our recent study, which identified a state of stable gene silencing that we termed the ‘occluded’ state (2). Specifically, we showed that the transcriptional competency of a gene can exist in one of two states: either competent or occluded. In the competent state, a gene is capable of responding to trans-acting transcription factors of the cell such that it is active if appropriate activators are present, though it can also be silent if activators are absent or repressors are present. In the occluded state, in contrast, a gene is no longer capable of responding to the cell's trans-acting milieu, presumably due to cis-acting chromatin marks, and remains silent even in the presence of activators. Experimentally, we used the ‘trans-complementation’ assay to identify occluded genes, which involves fusing two disparate cell types and searching in fused cell for genes silent in the genome of one fusion partner but active in the other. The active orthologs of these genes attest to the presence of a trans-acting cellular milieu that is conducive to the expression of the genes. Logically then, the silent orthologs, which are bathed in the same milieu, must have been blocked from the milieu's action by the cis effect of their chromatin state—i.e. they are occluded.
Using trans complementation, we uncovered occluded genes in a variety of mammalian cell types (2). Importantly, we found that occluded genes in a given cell type tend to include master regulators of alternative cell lineages. Furthermore, the occluded state is maintained during cell division and is extraordinarily stable under a wide range of physiological conditions. These results support the idea that the occlusion of lineage-inappropriate genes could be a key mechanism by which cell type identity is maintained.
An obvious next question to address is the biochemical basis of gene occlusion. Numerous studies have linked various chromatin marks—including DNA methylation, histone variants, covalent histone modifications and the binding of certain chromatin-associated proteins—to levels of gene expression (7–15). These chromatin marks are thus good starting points for investigating the biochemical basis of occlusion. Here, we analyze a wide variety of chromatin marks for their potential involvement in occlusion.
RESULTS
Analysis of DNA methylation using bisulfite sequencing
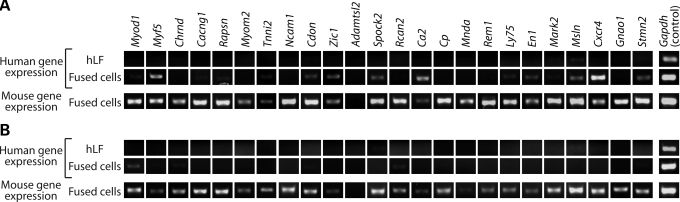
Our recent study identified occluded genes in a number of cell types (2), including the human lung fibroblasts MRC-5 (hereafter denoted hLF). In the study, we fused hLF with the mouse skeletal muscle myoblasts C2C12 (hereafter mSMM) to search for occluded genes in hLF. Given that this fusion was intended to interrogate the occlusion status of genes in the hLF genome, hLF was considered the responder in the fusion, whereas mSMM was considered the reprogrammer. We identified 24 occluded and 10 transactivated genes in hLF via fusion with mSMM (detailed descriptions of these genes are provided in Supplementary Material, Table S1). Experimentally, occluded and transactivated genes were defined by gene expression patterns in cells before and after fusion [see Table 1 and also refer Lee et al. (2), for detailed definition of occluded versus transactivated genes]. Basically, both occluded genes and transactivated genes are silent in the responder while active in the reprogrammer prior to fusion. For occluded genes, this expression pattern remains the same after fusion. For transactivated genes, in contrast, their expression status in the responder changes from silent to active upon fusion with the reprogrammer. Transactivated genes are thus competent but silent in the responder, and can turn on in response to the introduction of transcriptional activators upon fusion.
Table 1.
Expression patterns of occluded, transactivated and extinguished genes
| Expression pattern in reprogrammer |
Expression pattern in responder |
Conclusion | ||
|---|---|---|---|---|
| Before fusion | After fusion | Before fusion | After fusion | |
| Active | Active | Silent | Silent | Gene in responder occluded |
| Active | Active | Silent | Active | Gene in responder transactivated and hence competent |
| Active | Silent | Silent | Silent | Gene in reprogrammer extinguished |
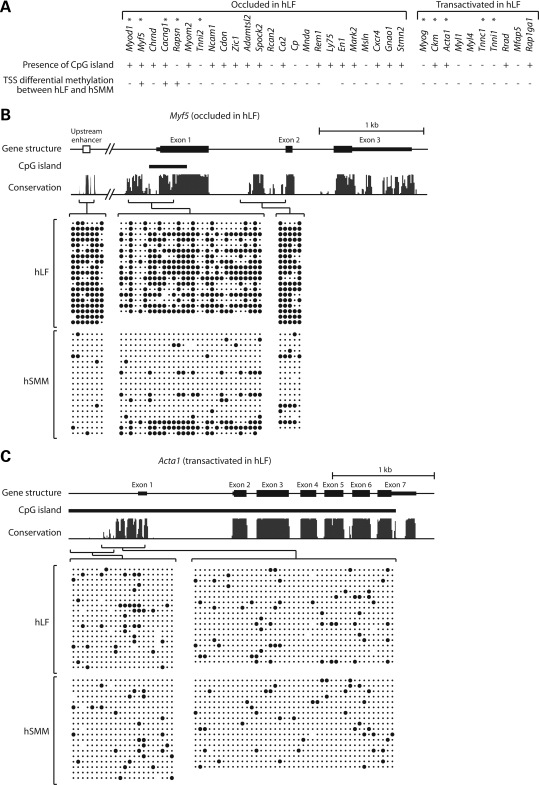
To examine the potential role of DNA methylation in occlusion, we first performed a bioinformatic survey that identified CpG islands in 18 of 24 occluded and 4 of 10 transactivated genes in hLF (Fig. 1A). Occluded genes thus appear somewhat enriched for CpG islands, though this is only marginally significant (P< 0.06 by Fisher's exact test). We then performed extensive bisulfite sequencing to analyze methylation patterns of a subset of 10 genes, five of which were chosen from the 24 occluded genes, whereas the other five were selected from the 10 transactivated genes. Analysis was carried out on two cell types: hLF and human skeletal muscle myoblasts (hSMM). RT–PCR confirmed that all 10 genes are expressed in hSMM (data not shown), indicating their competent state in these cells. Given that most of the genes are too big for bisulfite sequencing in their entirety, we focused on putative cis-regulatory regions identified by cross-species sequence conservation (see Materials and Methods). Also included in the analysis are regions surrounding transcription start sites (TSS) and experimentally validated enhancer elements irrespective of conservation.
Figure 1.
Bisulfite sequencing analysis of DNA methylation. (A) The 25 occluded and 10 transactivated genes in hLF that we uncovered previously (2). The presence or absence of CpG island in each gene is indicated, along with whether transcription start site (TSS) is differentially methylated between hLF and hSMM. (B) DNA methylation pattern of the occluded gene Myf5. (C) DNA methylation pattern of the transactivated gene Acta1. In the schema of gene structure, exons are shown in solid bars with thick bars indicating coding regions and thin bars indicating untranslated regions. Bioinformatically identified CpG islands are indicated. In the conservation graph, the height of peaks reflects the degree of cross-species conservation. Individual amplicons in bisulfite sequencing and their corresponding genomic regions are indicated by brackets. Within each block of bisulfite sequencing data, columns correspond to CpG sites while rows correspond to sequenced clones. Solid circles indicate methylated CpG; dots indicate unmethylated CpG.
Figure 1B and C shows representative results of the methylation analysis for one occluded gene (Myf5) and one transactivated gene (Acta1). Results for the remaining eight genes are presented in Supplementary Material, Fig. S1. In three of five occluded genes (Myf5, Cacng1 and Rapsn), strong differential methylation was observed between hLF and hSMM, with at least a subset of the regions sampled having much higher levels of methylation in hLF than hSMM. Of the remaining two occluded genes, Myod1 showed mild differential methylation in an enhancer far upstream of TSS, and Tnni2 did not show differential methylation between hLF and hSMM in any of the regions sampled. In contrast to the occluded genes, none of the transactivated genes showed discernable differential methylation between hLF and hSMM. We note that for Myf5 and Cacng1, at least some of the methylated regions in hLF fall within CpG islands. Although CpG islands are generally assumed to be unmethylated, there are clear exceptions such as many X-inactivated genes, some imprinted genes and genes abnormally silent in cancer cells (16–18). Furthermore, normal CpG methylation can occasionally be found in non-imprinted, non X-inactivated genes, often in a tissue-specific manner (19–21). The methylation within CpG islands of some occluded genes may therefore represent another example of such exceptions.
For the three occluded hLF genes that showed robust differential methylation between hLF and hSMM, the vicinity of TSS is invariably a part of the differentially methylated regions. We therefore used bisulfite sequencing to examine the methylation status of TSS for the remaining 23 genes (Ly75 is not included because it is technically refractory to bisulfite sequencing). These remaining genes showed little or no TSS differential methylation between hLF and hSMM, regardless of whether they are occluded in hLF or not (Supplementary Material, Fig. S2; data of TSS methylation analysis also summarized in Fig. 1A). (Primer sequences of all amplicons used in bisulfite sequencing are provided in Supplementary Material, Table S2).
The above results demonstrate that for a subset of occluded genes, the occluded state is characterized by increased methylation, especially around TSS. However, many occluded genes do not show appreciable differential methylation in TSS between occluded state in hLF and competent state in hSMM, suggesting that either methylation is not involved in conferring the occluded state to these genes or if it is involved, it does so by acting in regions other than TSS. Data below are in line with the latter possibility for at least some genes.
Effect of drug-induced demethylation on the occluded state
To further examine whether DNA methylation contributes causally to the occluded state, we treated hLF with the demethylating drug 5-aza-2′-deoxycytidine (AdC) prior to cell fusion. The treatment itself did not turn on the occluded hLF genes. Upon fusion with mSMM, however, about half of the occluded hLF genes showed variable levels of transactivation (Fig. 2A). Yet, for the majority of these, the expression levels of the hLF copies are notably lower than that of the mSMM copies. This could reflect either heterogeneous response of cells to drug treatment or the fact that for some of the occluded genes, demethylation only leads to partial erasure of the occluded state. It is noteworthy that AdC treatment can alter the occluded state of genes not showing appreciable differential TSS methylation between hLF and hSMM. It suggests that DNA methylation plays a role in maintaining the occluded state of these genes, but it does so by affecting regulatory regions outside of the immediate vicinity of TSS. These results, together with the bisulfite sequencing data, argue that DNA methylation is a causal factor contributing to the occlusion of at least some genes, whereas there are likely other mechanisms that also contribute to the occluded state.
Figure 2.
Effect of AdC (A) and TSA (B) treatment on occluded genes in hLF. RT–PCR analysis of gene expression is performed on drug-treated hLF without fusion and drug-treated hLF fused to mSMM. hLF, human lung fibroblasts; mSMM, mouse skeletal muscle myoblasts.
Analysis of 20 chromatin marks by chromatin immunoprecipitation
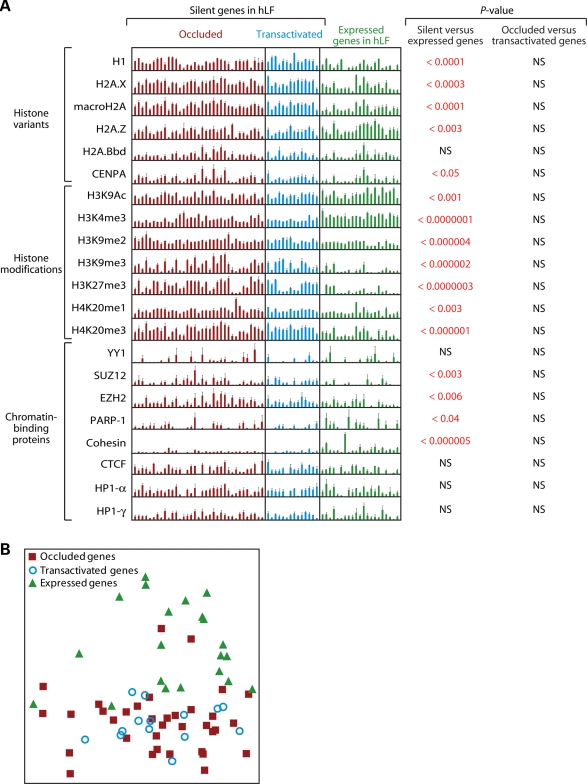
Besides DNA methylation, many chromatin marks have been found to be over- or underrepresented at either silent loci of the genome or regions believed to be heterochromatic (e.g. the inactive X and the centromere) (12,13). We examined 20 such marks by chromatin immunoprecipitation (ChIP) followed by PCR. These include seven histone modifications (H3K9Ac, H3K4me3, H3K9me2, H3K9me3, H3K27me3, H4K20me1 and H4K20me3), six histone variants (H1, H2A.X, macroH2A, H2A.Z, H2A.Bbd and CENPA) and seven chromatin-binding proteins (YY1 which is a key component of Polycomb repressive complex 1, SUZ12 and EZH2 which are key components of Polycomb repressive complex 2, PARP-1, cohesin and HP1-α and HP1-γ which are mammalian homologs of Drosophila heterochromatin protin-1). This list contains essentially all the major chromatin marks that have been implicated in the regulation of gene expression. We analyzed these marks for three classes of genes in hLF: the 24 occluded and 10 transactivated genes as depicted in Fig. 1A, and a set of 17 actively expressed genes randomly selected from the microarray data and validated by RT–PCR. For all these genes, we focused on the vicinity of TSS for chromatin analysis because it is the predominant site of differential chromatin modification in association with gene activity (12,13,22). For a few genes, validated enhancers were also included in the analysis. (primer sequences of all amplicons used in ChIP–PCR analysis are provided in Supplementary Material, Table S3).
For the great majority of these marks, there are significant differences between silent genes (including both occluded and transactivated genes) and expressed genes in a manner largely consistent with the literature (Fig. 3A). Specifically, five marks, H3K9Ac, H3K4me3, H2A.Z, PARP-1 and cohesin, are enriched in expressed genes relative to silent genes, with the enrichment being most notable for H3K4me3. In contrast, 11 marks, H3K9me2, H3K9me3, H3K27m3, H4K20me1, H4K20me3, H1, H2A.X, macroH2A, CENPA, SUZ12 and EZH2, show the opposite trend—i.e. they are enriched in silent relative to expressed genes, with the most notable enrichment seen in H3K9me2, H3K9me3, H3K27me3 and H4K20me3. Four marks, H2A.Bbd, YY1, HP1-α and HP1-γ, did not show any significant difference between silent and expressed genes. In the comparison between occluded and transactivated genes, however, none of the 20 marks showed a significant difference between the two categories of genes.
Figure 3.
ChIP analysis of 20 chromatin marks in hLF. Genes targeted by the analysis can be divided into silent and expressed categories, with the silent category further divided into occluded and transactivated groups. (A) PCR quantitation of ChIP for individual genes. Each bar represents a region interrogated by a PCR amplicon. The height of each bar represents fold-enrichment, relative to input, of each gene. P-values are calculated from these data using the t-test and indicate the statistical significance that two groups of genes are distinct for the chromatin mark surveyed. Error bars are based on multiple replicates of the experiment. NS, not significant; hLF, human lung fibroblasts. (B) Principal component analysis of ChIP data across the 20 chromatin marks.
To produce a visually more intuitive representation of the separation in chromatin signatures among the genes, we used principal component analysis to reduce the 20-dimensional data from the 20 marks to two dimensions. As expected, occluded genes and transactivated genes clustered closely with each other, whereas expressed genes clustered separately (Fig. 3B). These data indicate that, of the chromatin marks surveyed, silent genes (including both occluded and transactivated genes) and expressed genes are highly distinct from each other, whereas occluded genes and transactivated genes are rather similar.
Effect of histone deacetylase inhibitor on the occluded state
The above ChIP data failed to establish any difference in histone acetylation between occluded and transactivated states. To confirm that histone acetylation is not causally involved in occlusion, we treated hLF with the histone deacetylase inhibitor trichostatin A (TSA) prior to cell fusion. The treatment essentially did not alter the occluded state of hLF genes, with only Myod1 and Rcan2 showing very weak transactivation (Fig. 2B). This argues that, consistent with the ChIP data, histone hypoacetylation is not causally linked to the occluded state, even though histone hypoacetylation is robustly associated with the lack of expression as demonstrated by our data and many previous studies (9,12).
DISCUSSION
A variety of chromatin marks have been associated with the silent state of genes. These marks, often referred to as ‘epigenetic marks’ in the literature, include DNA methylation, histone variants, covalent histone modifications and the binding of chromatin-associated proteins, among others. When a gene's silent state is associated with certain chromatin marks, it is often said to be undergoing ‘epigenetic silencing’—a term that can sometimes invoke the sense that chromatin marks are the primary cause of silencing. In reality, it is rarely known whether chromatin marks at a silent locus are indeed the primary cause of silencing or just a secondary effect (23). Consider the following rather plausible scenario. When transcriptional activators of a gene are absent in the cell, that gene assumes a set of chromatin marks by default. But when transcriptional activators become available (say, as a result of some signaling event), the gene's silent state as well as the associated chromatin marks is readily reverted. In this case, chromatin marks associated with the silent state of this gene are not the primary cause of silencing. Rather, these chromatin marks are the secondary effect of the true primary cause (i.e. the lack of transcriptional activators), even if these makes play a role in the execution or enforcement of silencing.
The above example shows that it is important to differentiate between two situations of epigenetic silencing. In the first situation, cis-acting chromatin marks are the primary cause of silencing such that the affected gene remains silent irrespective of whether transcriptional activators for that gene are present in the cell or not. In the second situation, the trans-acting cellular milieu is the primary cause of silencing (i.e. either the absence of transcriptional activators or the presence of transcriptional repressors in the milieu), whereas chromatin marks are just a secondary effect even if they participate in executing or enforcing the silent state. The first situation is basically the occluded state, whereas the second situation is the competent but silent state. These two states can be distinguished by the trans-complementation assay (2). Thus, the trans-complementation assay further divides the silent portion of the genome into two classes: cis-silenced (or occluded) genes for which cis-acting chromatin marks are the primary cause of silencing and trans-silenced genes for which the trans-acting cellular milieu is the primary cause of silencing though the chromatin state of the genes could also be affected as a secondary effect of silencing.
In this study, we attempted to probe which specific chromatin marks contribute causally to occlusion. We uncovered evidence that DNA methylation plays a causal role in maintaining occlusion for a subset of occluded genes. However, it is much less clear whether a variety of other chromatin marks, including histone variants, covalent histone modifications and the binding of chromatin-associated proteins, contribute causally to occlusion. Indeed, although many of these marks are linked robustly to silent genes, none shows detectable differentiation between the two classes of silent genes: those that are occluded and those that are competent but silent (i.e. transactivated genes) (Fig. 3). As yet, it is too early to draw any firm conclusions from this study because it relies on a limited number of genes and focuses on the vicinity of TSS. Nevertheless, our data do raise the possibility that some of the repressive marks (i.e. chromatin marks traditionally associated with gene silencing such as H3K9 methylation) might only be a secondary effect rather than the primary cause of silencing in many cases. This is not to say that these repressive marks do not participate in the execution or enforcement of silencing. For example, a repressive mark can result in more closed (i.e. tightly packed) chromatin structure such that promiscuous binding by RNA polymerase is inhibited, thus reducing background expression. But such a repressive mark cannot be considered as the primary cause of silencing, if it can be readily erased by the introduction of transcriptional activators into the cellular milieu. In this case, trans-acting cellular milieu rather than the repressive mark is the true primary cause of silencing, with the existence of the repressive mark only secondary to the trans-acting milieu.
We thus suggest, as have others (23,24), that caution be taken when discussing the role of chromatin marks in gene silencing so as not to imply causality when that information is not available. We further suggest that the ability to identify occluded genes by the trans-complementation assay provides a hitherto unavailable functional readout of chromatin state. With this information, one can better investigate whether a chromatin mark (or combination of marks) plays a causal role or a secondary role in gene silencing.
MATERIALS AND METHODS
Cell culture
The hLF, MRC-5, were obtained from ATCC (CCL-171), and so were the mSMM C2C12 (CRL-1772). hSMM were obtained from Cambrex (CC-2580T25). These cells were cultured under standard conditions following vendors’ instructions or as described previously (2,25,26).
DNA methylation analysis
Genomic regions targeted for bisulfite sequencing were chosen on the basis of cross-species conservation as defined by the UCSC Genome Browser (Placental Mammal Conserved Elements by 28-way Multiz Alignment) (27). DNA methylation analysis was performed by bisulfite mutagenesis sequencing as described (28). Approximately 20 clones were sequenced for each region of interest and sequence files were analyzed using BiQ Analyzer software (29). Primer sequences of all amplicons used in bisulfite sequencing are provided in Supplementary Material, Table S2. The analysis of Ly75's TSS was abandoned after six primer sets failed to amplify the region.
Chromatin analysis using ChIP
ChIP was performed essentially as described previously (30), with the following modifications. Samples were sonicated in 7 ml aliquots for nine cycles of 20 s at 33% power using a Fisher Sonic Dismembrator Model 500 with a 0.5 in. flat tip horn. This amount of sonication yielded an average DNA fragment size of 300 – 700 bp (data not shown). For immunoprecipitation from 7 × 106 cells, 40 µl of a 1:1 mixture of protein A and protein G Dynabeads (Invitrogen) were coupled to 10 µg of antibody, and then incubated with sonicated chromatin samples overnight. Immunoprecipitated chromatin was eluted from beads in 150 µl elution buffer (50 mm Tris pH 8, 10 mm EDTA, 1% SDS), digested with proteinase K (Roche) and DNA was purified using the GenCatch PCR Cleanup Kit (Epoch Biolabs). Semi-quantitative PCR was performed with template concentration and PCR cycle tailored to each amplicon to obtain linear range amplification. PCR products were resolved on agarose gel and visualized by ethidium bromide staining. Densitometry analysis of background-subtracted images of PCR bands was performed using the Gel Analyzer module of ImageJ 1.37v (National Institutes of Health, http://rsb.info.nih.gov/ij). Measured PCR band intensities were normalized to IP input controls. The value of each data point was calculated as the average of at least three independent replicates. Primer sequences of all amplicons used in ChIP–PCR analysis are provided in Supplementary Material, Table S3.
Antibodies used for ChIP were as follows with catalog numbers in parentheses: H3K9Ac (ab4441), H3K4me3 (ab8580), H3K9me2 (ab1220), H3K9me3 (ab8898), H3K27me3 (ab6002), H4K20me1 (ab9051), H4K20me3 (ab9053), H2A.X (ab11175), macroH2A.1 (ab37264), H2A.Z (ab4174), H2A.Bbd (ab4175), cohesin (ab992) and HP1-γ (ab50365) were from Abcam; CENP-A (sc-22787), YY1 (sc-7341X) and PARP-1 (sc-53643) were from Santa Cruz Biotechnology; H1 (05-457) and SUZ12 (04-046) were from Millipore; HP1-α (2616) was from Cell Signaling Technology and EZH2 (36-6300) was from Invitrogen.
For drug inhibition of DNA methylation, cells were plated at 20–25% confluence and treated with 10 µm AdC until the cells had undergone two population doublings. Cell fusion was then carried out and fused cells were incubated for four more days without AdC. For drug inhibition of histone acetylation, cells were treated with 1 µm trichostatin A for 24 h until just before fusion. For both AdC and trichostatin A treatment, control cells not subject to fusion were exposed to the same temporal course of drug treatment.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
Funding to pay the Open Access charge was provided by Howard Hughes Medical Institute.
ACKNOWLEDGEMENTS
We thank Nitzan Mekel-Bobrov for assistance in statistical analysis of chromatin immunoprecipitation data. This work was partly supported by National Institutes of Health grants F32HL922792 (to J.G.), F32GM075503 (to G.E.S.) and HL07605 (to E.C.B.).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Waddington C.H. Principles of Development and Differentiation. New York: Macmillan; 1966. [Google Scholar]
- 2.Lee J.H., Bugarija B., Millan E.J., Walton N.M., Gaetz J., Fernandes C.J., Yu W.H., Mekel-Bobrov N., Vallender T.W., Snyder G.E., et al. Systematic identification of cis-silenced genes by trans complementation. Hum. Mol. Genet. 2009;18:835–846. doi: 10.1093/hmg/ddn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caplan A.I., Ordahl C.P. Irreversible gene repression model for control of development. Science. 1978;201:120–130. doi: 10.1126/science.351805. [DOI] [PubMed] [Google Scholar]
- 4.Fisher A.G., Merkenschlager M. Gene silencing, cell fate and nuclear organisation. Curr. Opin. Genet. Dev. 2002;12:193–197. doi: 10.1016/s0959-437x(02)00286-1. [DOI] [PubMed] [Google Scholar]
- 5.Macaluso M., Giordano A. How does DNA methylation mark the fate of cells? Tumori. 2004;90:367–372. doi: 10.1177/030089160409000401. [DOI] [PubMed] [Google Scholar]
- 6.Sparmann A., van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 8.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Vermaak D., Ahmad K., Henikoff S. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 2003;15:266–274. doi: 10.1016/s0955-0674(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg A.D., Allis C.D., Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Heard E., Clerc P., Avner P. X-chromosome inactivation in mammals. Annu. Rev. Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 17.Paulsen M., Ferguson-Smith A.C. DNA methylation in genomic imprinting, development, and disease. J. Pathol. 2001;195:97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 19.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L., Kondo Y., Guo Y., Zhang J., Zhang L., Ahmed S., Shu J., Chen X., Waterland R.A., Issa J.P. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki M., Sato S., Arai Y., Shinohara T., Tanaka S., Greally J.M., Hattori N., Shiota K. A new class of tissue-specifically methylated regions involving entire CpG islands in the mouse. Genes Cells. 2007;12:1305–1314. doi: 10.1111/j.1365-2443.2007.01136.x. [DOI] [PubMed] [Google Scholar]
- 22.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Ptashne M. On the use of the word ‘epigenetic. Curr. Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 24.Madhani H.D., Francis N.J., Kingston R.E., Kornberg R.D., Moazed D., Narlikar G.J., Panning B., Struhl K. Epigenomics: a roadmap, but to where? Science. 2008;322:43–44. doi: 10.1126/science.322.5898.43b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blau H.M., Pavlath G.K., Hardeman E.C., Chiu C.P., Silberstein L., Webster S.G., Miller S.C., Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 26.Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallender T.W., Lahn B.T. Localized methylation in the key regulator gene endothelin-1 is associated with cell type-specific transcriptional silencing. FEBS Lett. 2006;580:4560–4566. doi: 10.1016/j.febslet.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 30.Li Z., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl Acad. Sci. USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.