Abstract
The most consistent finding derived from the WTCCC GWAS for rheumatoid arthritis (RA) was association to a SNP at 6q23. We performed a fine-mapping of the region in order to search the 6q23 region for additional disease variants. 3962 RA patients and 3531 healthy controls were included in the study. We found 18 SNPs associated with RA. The SNP showing the strongest association was rs6920220 [P = 2.6 × 10−6, OR (95% CI) 1.22 (1.13–1.33)]. The next most strongly associated SNP was rs13207033 [P = 0.0001, OR (95% CI) 0.86 (0.8–0.93)] which was perfectly correlated with rs10499194, a SNP previously associated with RA in a US/European series. Additionally, we found a number of new potential RA markers, including rs5029937, located in the intron 2 of TNFAIP3. Of the 18 associated SNPs, three polymorphisms, rs6920220, rs13207033 and rs5029937, remained significant after conditional logistic regression analysis. The combination of the carriage of both risk alleles of rs6920220 and rs5029937 together with the absence of the protective allele of rs13207033 was strongly associated with RA when compared with carriage of none [OR of 1.86 (95% CI) (1.51–2.29)]. This equates to an effect size of 1.50 (95% CI 1.21–1.85) compared with controls and is higher than that obtained for any SNP individually. This is the first study to show that the confirmed loci from the GWA studies, that confer only a modest effect size, could harbour a significantly greater effect once the effect of additional risk variants are accounted for.
INTRODUCTION
Rheumatoid arthritis [RA, (MIM #180300)] is an autoimmune inflammatory disease characterized by inflammation and destruction of synovial joints leading to progressive joint damage and disability. It is the most common inflammatory arthritis, affecting up to 1% of the population worldwide (1). Although RA aetiology is not yet fully understood, it is known that a strong genetic component plays a major role in disease susceptibility, together with environmental factors (2).
The Wellcome Trust Case Control Consortium (WTCCC) identified, through a genome-wide association study (GWAS), nine new putative RA susceptibility loci, in addition to HLA and PTPN22 (3). One single nucleotide polymorphism (SNP) located in the chromosomal region 6q23, rs6920220, was unequivocally replicated in a validation study (4). Interestingly, association to the same SNP and a second, independently associated polymorphism in the region (rs10499194) was detected in a GWAS in a US population, confirming the locus as important in RA causation (5). Increased risk for RA is conferred by the minor allele of rs6920220 and protection by the minor allele of rs10499194. The effect size for both these alleles is relatively small (OR 1.22 and OR 0.75, respectively). Both associated variants map to an intergenic region of 6q23, between the OLIG3 and TNFAIP3 genes. OLIG3 is involved in the development and differentiation of neuronal cells and therefore not an obvious candidate gene for RA (6). Conversely, A20, the product of TNFAIP3, is a potent anti-inflammatory protein, since it is required for the termination of both tumor necrosis factor (TNF) and Toll-like receptor-induced NF-κB signals (7,8). In addition, A20 deficient mice develop severe inflammation, which includes inflammation of the joints (9). Therefore, TNFAIP3 seems a robust candidate gene for RA susceptibility. However, interrogation of data available from public databases shows no correlation between genotype at either SNP and TNFAIP3 expression in PBMCs (10). Identification of variants within the TNFAIP3 gene itself which were associated with RA susceptibility would lend support to the hypothesis that the intergenic region exerts its effect through regulation of this locus. The failure to identify correlation between genotype and expression may arise if the associated SNPs are in only modest LD with the true causal variants. Hence, the aim of the current study was to undertake fine-mapping of the 6q23 region in order to search for additional disease-associated variants both within the intergenic block encompassing the RA-associated SNPs and across the TNFAIP3 gene itself.
RESULTS
Three thousand nine hundred and sixty-two RA patients and 3531 healthy controls were genotyped for 70 SNPs in two regions within the 6q23 locus defined by recombination rate and LD. The first region spanned the LD block containing the rs6929220 SNP, association of which had previously been validated with RA susceptibility. This variant maps to an intergenic region but it was hypothesized that it may exert its functional effect via the TNFAIP3 gene. Hence, fine mapping of this gene was also undertaken.
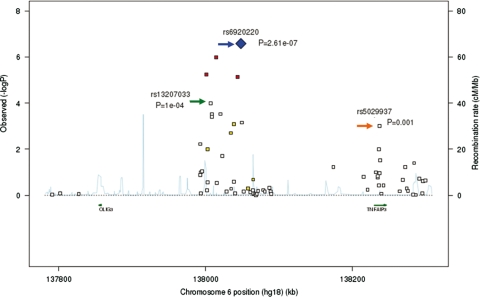
We found 18 SNPs associated with RA, trend P-value < 0.05 (Fig. 1 and Supplementary Material, Table S1). The SNPs showing the strongest association with RA remained rs6920220 [P = 2.61 × 10−7, OR (95% CI) 1.22 (1.13–1.32)] and the three SNPs (rs2327832, rs6933404 and rs6927172) in r2 = 1 with that variant. The next most strongly associated polymorphisms were the SNPs rs13207033 [P = 0.0001, OR (95% CI) 0.86 (0.80–0.93)], rs13192841 and rs1257282 all of which were perfectly correlated (r2 = 1) with rs10499194. As in the previous study, the minor allele was associated with protection against RA (5). This represents the first independent replication of the association of the rs10499194 polymorphism with RA susceptibility. Additionally, we found a number of new potential RA markers, including rs5029937, located in the intron 2 of TNFAIP3.
Figure 1.
Case–control association results of the OLIG3/TNFAIP3 region. The blue diamond indicates the strongest signal from the fine-mapping, i.e rs6920220. For the rest of the SNPs, the colour indicates the extent of LD with rs6920220 (red: r2 > 0.8, orange: 0.5≤ r2 <0.8, yellow: 0.5> r2 ≥ 0.2, white: r2 < 0.2).
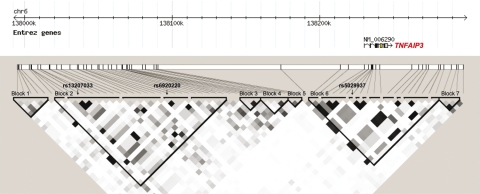
The LD pattern among the markers included in this study showed the presence of seven different blocks (Fig. 2). The most strongly associated SNPs are located in Block 2 which lies in the intergenic region between OLIG3 and TNFAIP3, spanning ∼63 kb and located between positions 138002057 and 138064935 of chromosome 6q. Haplotype analysis with Block 2 SNPs yielded six common haplotypes with frequency >5% (Table 1). Haplotype 1, tagged by the most strongly associated SNP in the single-point analysis, rs6920220 (highlighted in red), confers risk for RA [P = 1.8 × 10−6, OR (95% CI) 1.22 (1.13–1.33)], while Haplotype 2, tagged by rs13207033 and rs1878658 (highlighted in blue and green, respectively), is associated with protection [P = 6 × 10−5, OR (95% CI) 0.83 (0.76–0.91)]. The associated SNP in intron 2 of the TNFAIP3 gene, rs5029937, is located in a different LD block spanning 71 kb, but we found no evidence for a haplotypic effect of SNPs in this block.
Figure 2.
LD in the ∼500 kb region analysed, shown as r2 values, and location of the three independently associated SNPs.
Table 1.
Haplotype analysis for Block 2
| Haplotype | Effect | 10 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 20 | 21 | 22 | 23 | 24 | 26 | 27 | 28 | 30 | 31 | 32 | 33 | 34 | RA patients, n (%) 2n = 7096 | Controls, n (%) 2n = 5776 | P-value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Risk | C | G | G | G | C | G | G | A | G | A | G | T | A | G | A | A | C | A | C | G | G | 1766 (24.9) | 1231 (21.3) | 1.8 × 10−6 | 1.22 (1.13–1.33) |
| 2 | Protective | C | A | A | A | T | A | G | G | G | G | G | T | T | C | G | C | T | A | C | G | G | 1186 (16.7) | 1123 (19.4) | 6 × 10−5 | 0.83 (0.76–0.91) |
| 3 | Neutral | C | G | G | G | C | A | G | A | T | A | G | A | A | C | G | A | C | G | C | A | T | 1239 (17.5) | 1022 (17.7) | 0.73 | 0.98 (0.90–1.08) |
| 4 | Neutral | A | A | G | G | C | A | G | A | G | G | G | T | T | C | G | A | C | G | C | G | T | 1042 (14.7) | 802 (13.9) | 0.19 | 1.07 (0.97–1.18) |
| 5 | Neutral | C | A | G | G | C | A | G | A | G | G | T | T | T | C | G | C | T | A | A | G | T | 660 (9.3) | 561 (9.7) | 0.09 | 0.90 (0.80–1.02) |
| 6 | Neutral | C | A | A | A | T | A | A | A | G | G | G | T | T | C | G | C | T | A | C | G | G | 626 (8.8) | 527 (9.1) | 0.55 | 0.96 (0.85–1.09) |
| Others (MAF < 5%) | 557 (8.1) | 510 (8.8) |
Individuals with complete genotype data for all SNPs were included in the analysis. Haplotypes were estimated using Haploview.
10: rs566097, 12: rs9389526, 13: rs13207033, 14: rs13192841, 15: rs12527282, 16: rs2327832, 17: rs17066681, 18: rs1878658, 19: rs2069311, 20: rs12194935, 21: rs678385, 22: rs12206392, 23: rs9402914, 24: rs665668, 26: rs6927172, 27: rs6920220, 28: rs636393, 30: rs508214, 31: rs667520, 32: rs10499196, 33: rs9321627, 34: rs609438.
In order to determine whether the three effects observed were independent or not, conditional logistic regression analysis was undertaken. The 18 associated markers were conditioned on the effect of each of the other associated SNPs. Only three SNPs, rs6920220, rs13207033 and rs5029937, remained significant after conditioning on each associated marker (Table 2) and the results showed that, rs1878658, one of the SNPs tagging the protective haplotype, was not significantly associated with RA after conditioning on rs13207033. This suggests that each of the 3 SNPs is exerting an independent effect. As an additional test for independence, we conditioned the effect of the associated SNPs on both rs6920220 and rs13207033. Only the rs5029937 SNP showed significant association (P-value = 0.02, after conditioning on both rs6920220 and rs13207033). To further test the independent effect of the intron 2 SNP, rs5029937, we repeated the conditional logistic regression pooling the validation data from this study with that obtained from the previous WTCCC GWAS for RA (3) with the aim of increasing the statistical power, since allele frequencies were similar across both groups and there was no overlap in samples between these two cohorts. We used imputed data for rs5029937 in the WTCCC cohort, given that this SNP was not included in the initial GWAS (imputation confidence score 0.99). This polymorphism showed a highly significant association with RA after conditioning on rs6920220, rs13207033 and both (P-values 4.71 × 10−11, 6.95 × 10−6 and 7.59 × 10−6, respectively).
Table 2.
Conditional logistic regression analysis for all SNPs with single-point allelic P < 0.05
| SNP | Position | Unconditioned P-value | P-value conditional on rs6920220 | P-value conditional on rs13207033 | P-value conditional on rs6920220 and rs13207033 |
|---|---|---|---|---|---|
| rs590523 | 137992570 | 0.006 | 0.214 | 0.049 | 0.282 |
| rs6933404 | 138000928 | 5.71 × 10−06 | 0.957 | 0.0002 | 0.715 |
| rs9389526 | 138002555 | 0.01 | 0.996 | 0.435 | 0.164 |
| rs13207033 | 138007111 | 0.0001 | 0.011 | — | — |
| rs13192841 | 138008907 | 0.0003 | 0.014 | Collinear | — |
| rs12527282 | 138008945 | 0.0004 | 0.009 | Collinear | — |
| rs2327832 | 138014761 | 1.01 × 10−06 | 0.65 | 0.0003 | 0.506 |
| rs1878658 | 138020079 | 0.0003 | 0.021 | 0.495 | 0.283 |
| rs2069311 | 138022920 | 0.02 | 0.007 | 0.111 | 0.01 |
| rs678385 | 138034243 | 0.002 | 0.736 | 0.354 | 0.464 |
| rs665668 | 138038578 | 0.0008 | 0.546 | 0.202 | 0.593 |
| rs6927172 | 138043868 | 7.20 × 10−06 | Collinear | 0.0005 | — |
| rs6920220 | 138048197 | 2.61 × 10−07 | — | 8.77 × 10−5 | — |
| rs636393 | 138049223 | 0.0007 | 0.079 | 0.449 | 0.903 |
| rs508214 | 138055358 | 0.001 | 0.106 | 0.536 | 0.686 |
| rs5029937 | 138236844 | 0.001 | 0.041 | 0.019 | 0.02 |
| rs2230926 | 138237759 | 0.03 | 0.274 | 0.141 | 0.222 |
| rs6932056 | 138284130 | 0.04 | 0.442 | 0.223 | 0.336 |
Collinear, one allele determines the other in >99% of haplotypes.
For exploratory purposes, we performed a stepwise logistic regression analysis. The method begins with an empty model to which variables are added in an iterative process. This analysis showed that rs6920220 exhibits the strongest association with RA. Subsequent additions of rs5029937 and rs13207033 significantly improve the model, adding further evidence to support that multiple polymorphisms in the 6q23 region are independently associated with RA (Table 3).
Table 3.
Stepwise logistic regression of three SNPs model
| SNP added to the model | P-value | OR (95% CI) |
|---|---|---|
| rs6920220 | 0.001 | 1.04 (1.02–1.06) |
| rs5029937 | 0.042 | 1.11 (1.01–1.23) |
| rs13207033 | 0.049 | 0.98 (0.96–0.99) |
Stratification analysis of the three independently associated RA SNPs showed that all three SNPs were associated with both male and female RA cases (Supplementary Material, Table S3). rs6920220 and the intron 2 SNP, rs5029937, appeared to be associated with auto-antibody positive cases, with strong association in both RF positive and anti-CCP positive cases. In contrast, rs13207033 was associated with both RF positive and negative RA cases, but only anti-CCP negative cases. Further studies in larger cohorts will be required in order to clarify the role of these polymorphisms with auto-antibody status, as only a subset of individuals with available serological data was included in the present analysis.
In order to determine the combined contribution to the risk of developing RA, logistic regression analysis was undertaken (Table 4). Loci were defined as carriage of the minor risk alleles A and T at rs6920220 and rs5029937, respectively, combined with the different genotypes of the protective SNP rs13207033. In order to avoid a loss of power due to the high degree of stratification, we pooled the validation data from this study with that obtained from the previous WTCCC GWAS for RA (3). For OR calculations, the reference was the lowest risk combination. The combination of the carriage of both risk alleles of rs6920220 and rs5029937 together with the absence of the protective allele of rs13207033 was strongly associated with RA when compared with carriage of none [OR of 1.86 (95% CI) (1.51–2.29)]. This equates to an effect size of 1.50 (95% CI 1.21–1.85) compared with controls and is higher than that obtained for any SNP individually (4.4% of the RA patients versus 2.3% of the controls carried this high-risk combination).
Table 4.
Association study of all possible gene combinations of the three independent RA associated SNPs
| Carriage of rs6920220 risk allele | Carriage of rs5029937 risk allele | rs13207033 | RA patients, n = 5126 | Controls, n = 5510 | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| GG | GG | AA | 280 (5.5) | 392 (7.1) | Ref | — |
| GG | GG | AG | 1210 (23.6) | 1475 (26.8) | 1.09 (1.03–1.17) | 0.006 |
| GG | GG | GG | 1164 (22.7) | 1322 (24) | 1.20 (1.05–1.36) | 0.006 |
| GG | TT+GT | AA | 21 (0.4) | 12 (0.2) | 1.45 (1.17–1.79) | 0.001 |
| GG | TT+GT | AG | 98 (1.9) | 77 (1.4) | 1.58 (1.27–1.98) | 5.1 × 10−5 |
| GG | TT+GT | GG | 79 (1.5) | 75 (1.4) | 1.73 (1.35–2.22) | 1.5 × 10−5 |
| AA+GA | GG | AA | 4 (0.1) | 5 (0.1) | 1.19 (1.09–1.29) | 7.8 × 10−5 |
| AA+GA | GG | AG | 575 (11.2) | 618 (11.2) | 1.30 (1.19–1.42) | 1.8 × 10−8 |
| AA+GA | GG | GG | 1396 (27.2) | 1311 (23.8) | 1.42 (1.24–1.62) | 2.3 × 10−7 |
| AA+GA | TT+GT | AA | 1 (0.02) | 0 | 1.55 (1.29–1.86) | 2.6 × 10−6 |
| AA+GA | TT+GT | AG | 74 (1.4) | 60 (1.1) | 1.70 (1.41–2.04) | 2.02 × 10−8 |
| AA+GA | TT+GT | GG | 224 (4.4) | 163 (2.3) | 1.86 (1.51–2.29) | 5.2 × 10−9 |
DISCUSSION
We present evidence that the recently described RA susceptibility locus at 6q23 contains at least three independent effects, two conferring susceptibility (rs6920220 and rs5029937) and one protection (rs13207033). Carriage of both susceptibility alleles and absence of protective alleles confers a far greater risk for RA compared with the effect size initially assigned to this locus.
The P-value for rs5029937 in the single point analysis (P = 0.001), although statistically significant, is not as impressive as those found for rs6920220 and rs13207033 and would not have remained significant had a very conservative Bonferroni correction to account for 70 independent tests been applied. However, a number of statistical analyses each support an independent effect of this SNP. First, P-values obtained from the conditional analyses are statistically significant. Secondly, the stepwise logistic regression also suggested an independent effect at this SNP (P = 0.04). Finally, the combination analysis (Table 4) showed a substantial increase in the OR (from 1.42 to 1.86) when adding the rs5029937 susceptibility T allele to the carriage of the rs6920220 risk allele and the absence of the rs13207033 protective allele. The lack of LD with the previously described RA susceptibility markers in the 6q23 region adds further evidence to support the independent effect of rs5029937. The results provide convincing evidence to support a third allele in the 6q23 region but replication in an independent cohort is required to confirm this unequivocally.
The present work constitutes only the first stage of the complete study of the 6q23 chromosomal region. We restricted our fine-mapping to the region surrounding the rs6920220 SNP that was detected in the second tier of significance of our previous GWAS (P = 10−5−5 × 10−7) and to fine-mapping the TNFAIP3 gene, since it was the most suggestive candidate gene in the neighbouring region. Although we found three independent variants associated with RA, it is possible that the locus harbours additional independent RA markers, as we did not explore the entire region. In fact, the haplotype data obtained in the present study support this hypothesis. For example, the haplotype analysis showed that haplotypes 1 and 2 were associated with susceptibility to and protection against RA, respectively. This was in accordance with the results found by Plenge et al. (5). However, haplotype 6, which was associated with protection in the study by Plenge et al. (5), was neutral in our study. rs13207033 tags haplotypes 2 and 6 and, therefore, it seems that the presence of the protective A allele of rs13207033 is not sufficient to provide protection for RA. The SNPs included in this study were able to capture 89% of the variation within this LD block. Therefore, additional SNPs not tagged in the current study might be involved in the protective effect together with rs13207033.
Interestingly, several markers in the TNFAIP3 region have recently been shown to be associated with the related auto-immune disease, systemic lupus erythematosus (SLE), in two independent studies (11,12). However, the results were conflicting. The SNP showing the strongest association with RA in the present study, rs6920220, was also associated with SLE in the study by Graham et al. (11), but it failed to show independent association with SLE in the study by Musone et al. (12) after conditional analysis. Conversely, Graham et al. (11) did not find evidence for association for the rs10499194 RA protective SNP, whereas Musone et al. (12) found that rs13192841 (a perfect proxy of rs10499194) was the most strongly associated variant in their cohort. Despite this disagreement, it is clear that the TNFAIP3 region plays an important role in SLE, as both studies found additional markers significantly associated with the disease including the functional exon 3 rs2230926 polymorphism. Interestingly, this variant was not independently associated with RA in our study. A recent robust study has shown that rs6920220 and rs10499194 are also independently associated with T1D (13). Moreover, a GWAS has demonstrated strong association of the TNFAIP3 with psoriasis (14). These findings confirm the importance of the 6q23 locus. Future studies will be required to ascertain whether RA, SLE and T1D are associated with the same or different causative variants within the 6q23 region, and whether the locus harbours risk alleles for other auto-immune or inflammatory diseases.
A consistent observation from successful GWAS is that the effect sizes of new loci are small (OR < 1.2) and that the combined effect of all known loci can account for only a fraction of the genetic risk of disease. This may be because very large numbers of as yet undefined loci explain complex traits, but may also be explained by a significant underestimation of risk for new loci. Re-sequencing and fine mapping studies may be essential in order to identify all risk/protective alleles at a locus and only once the effect of combinations of these are assessed can the true risk for a given locus be ascribed. Here we demonstrate that this is indeed the case for the recently described RA locus at 6q23, for which we have characterized three independent effects that can increase the risk from 1.22 to 1.50, establishing 6q23 as the next most important locus for RA in a UK population alongside HLA-DRB1 and PTPN22.
MATERIALS AND METHODS
In order to search the 6q23 region for additional disease variants, we analysed a panel of 70 SNPs across chromosome 6q23 encompassing two regions defined by recombination rate and LD blocks. The first region contained the strongest signal (rs6929220) and spanned the LD block around it from position 137990000 to 138090000. The second encompassed the TNFAIP3 gene from position 138215000 to 138300000, where we had suggestive evidence of independent effects (Fig. 2). Tagging SNPs across the blocks were selected with a minor allele frequency (MAF) > 0% and r2 = 0.8 and were enriched with proxies for known susceptibility SNPs (rs6920220 and rs10499194). Statistical power to detect effect sizes between 1.15 and 2.00 was >80% at a 5% significance level.
Patients with RA (n = 3962) were recruited from six centres (Manchester, Sheffield, Leeds, Aberdeen, Oxford and London) across the UK. All cases were Caucasian of Northern European descent and all fulfilled the 1987 American College of Rheumatology classification criteria modified for genetic studies (15,16). Clinical and demographic characteristics of the cohort are detailed in Supplementary Material, Table S1. Briefly, 72% subjects were female, 72% were rheumatoid factor positive and 67% carried ACPA antibodies as recognized by the anti-CCP antibody test. Therefore, this cohort is representative of a hospital-based series of RA subjects. Healthy controls (n = 3531) were recruited from five of the six centres (cases only recruited from London). All participants were recruited after providing informed consent and the study was approved by the North West Research Ethics Committee (MREC 99/8/84).
Serum RF and anti-CCP antibody titre were measured using commercially available kits [RF-PAIA Immunoturbidimetric Assay for rheumatoid factor, Diastat™ Anti-CCP Kit (Axis-Shield Diagnostics Limited, UK)]. Patients with titres ≥40 units/ml and ≥5 units/ml were defined as positive for RF and anti-CCP antibodies, respectively.
Genotyping was performed using the Sequenom iPlex platform (www.sequenom.com) according to the manufacturer's instructions. All genotyping was performed at the arc Epidemiology Unit, Manchester. Only samples and SNPs exceeding a 90% success rate were used in the analysis.
Genotype counts in cases and controls were analysed using PLINK software using the χ2 test for trend (17). Stratification analysis by gender, RF status and anti-CCP antibody status was performed. Haplotype analysis was undertaken using the software programs UNPHASED (18) and Haploview (19). Conditional and stepwise logistic regression was used to determine whether independent effects existed. These analyses were carried out utilizing STATA version 9.2.
SUPPLEMENTARY MATERIAL
Supplementary Material is available atHMG online.
FUNDING
We acknowledge the use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. This work was supported by the Arthritis Research Campaign [arc grant reference number 17552]. We would also like to acknowledge the National Health Service (NHS) Research and Development Support Fund for Guy's and St Thomas' and Lewisham NHS Trusts. We are grateful to the Manchester Biomedical Research Centre and the EU FP6 programme AutoCure for their support. Funding to pay the Open Access charge was provided by the arc grant ref 17552.
ACKNOWLEDGEMENTS
IT support was provided by Mark Lay, statistical advice from Mark Lunt and technical support by Paul Gilbert, Bhaneeta Lad and Stephen G. Martin. Nursing support was provided by Janet Grumley, Julie Shotton and Claire Farrar.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Firestein G.S. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.MacGregor A.J., Snieder H., Rigby A.S., Koskenvuo M., Kaprio J., Aho K., Silman A.J. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson W., Barton A., Ke X., Eyre S., Hinks A., Bowes J., Donn R., Symmons D., Hider S., Bruce I.N., et al. Rheumatoid arthritis association at 6q23. Nat. Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plenge R.M., Cotsapas C., Davies L., Price A.L., de Bakker P.I., Maller J., Pe'er I., Burtt N.P., Blumenstiel B., DeFelice M., et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filippi A., Tiso N., Deflorian G., Zecchin E., Bortolussi M., Argenton F. The basic helix-loop-helix olig3 establishes the neural plate boundary of the trunk and is necessary for development of the dorsal spinal cord. Proc. Natl. Acad. Sci. USA. 2005;102:4377–4382. doi: 10.1073/pnas.0407284102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertz I.E., O'Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 8.Boone D.L., Turer E.E., Lee E.G., Ahmad R.C., Wheeler M.T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 9.Lee E.G., Boone D.L., Chai S., Libby S.L., Chien M., Lodolce J.P., Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham R.R., Cotsapas C., Davies L., Hackett R., Lessard C.J., Leon J.M., Burtt N.P., Guiducci C., Parkin M., Gates C., et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musone S.L., Taylor K.E., Lu T.T., Nititham J., Ferreira R.C., Ortmann W., Shifrin N., Petri M.A., Ilyas K.M., Manzi S., et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung E., Smyth D.J., Howson J.M., Cooper J.D., Walker N.M., Stevens H., Wicker L.S., Todd J.A. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2008;10:188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 14.Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnett F.C., Edworthy S.M., Bloch D.A., McShane D.J., Fries J.F., Cooper N.S., Healey L.A., Kaplan S.R., Liang M.H., Luthra H.S. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 16.MacGregor A.J., Bamder S., Silman A.J. A comparison of the performance of different methods of disease classification for rheumatoid arthritis. Results from an analysis from a nationwide twin study. J. Rheumatol. 1994;21:1420–1426. [PubMed] [Google Scholar]
- 17.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., De Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet. Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 19.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.