Abstract
In mammals, adhesion and fusion of the palatal shelves are essential mechanisms during the development of the secondary palate; failure of these processes leads to the congenital anomaly, cleft palate. The mechanisms that prevent pathological adhesion between the oral and palatal epithelia while permitting adhesion and subsequent fusion of the palatal shelves via their medial edge epithelia remain obscure. In humans, mutations in the transcription factor interferon regulatory factor 6 (IRF6) underlie Van der Woude syndrome and popliteal pterygium syndrome. Recently, we have demonstrated that mice homozygous for a mutation in Irf6 exhibit abnormalities of epithelial differentiation that results in cleft palate as a consequence of adhesion between the palatal shelves and the tongue. In the current paper, we demonstrate that Irf6 is essential for oral epithelial differentiation and that IRF6 and the Notch ligand Jagged2 function in convergent molecular pathways during this process. We further demonstrate that IRF6 plays a key role in the formation and maintenance of the oral periderm, spatio-temporal regulation of which is essential for ensuring appropriate palatal adhesion.
INTRODUCTION
Development of the secondary palate involves a complex series of highly integrated events that are frequently disturbed resulting in the congenital malformation, cleft palate. Cleft palate, which has an estimated incidence of 1 in 2500 live births depending on geographic origin, racial and ethnic variation and socio-economic status (1,2), results in considerable morbidity to affected families as individuals who exhibit this condition may experience problems with eating, speaking and hearing which can be corrected to varying degrees by surgery, dental treatment, speech therapy and psychosocial intervention (3,4). The frequent occurrence and significant healthcare burden imposed by cleft palate highlight the need to dissect the aetiology and molecular pathogenesis of this distressing condition.
In mice, the developmental events underlying palatogenesis closely mirror those occurring in man; consequently, the mouse is the pre-eminent model organism for the study of mammalian palatogenesis (5). The first sign of overt development of the murine secondary palate occurs on embryonic day 11 (E11) with the outgrowth from the maxillary processes of paired palatal shelves that initially grow vertically down the sides of the developing tongue (E12–E13). At this stage, each palatal shelf consists of a central core of neural crest cell-derived mesenchyme surrounded by an epithelium composed of a basal layer of cuboidal ectodermal cells and a surface layer of flattened periderm cells similar to the remainder of the oral epithelia (6–8). During E14, the palatal shelves rapidly re-orientate to a horizontal position above the dorsum of the tongue and contact. The medial edge epithelia (MEE) of the apposed shelves adhere to form a midline epithelial seam (MES) which subsequently degenerates to allow mesenchymal continuity across the palate (E15) (9–16). Transforming growth factor β3 (TGFβ3) has been shown to play a key role in this process via multiple synergistic effects including stimulation of initial adhesion of the palatal shelves, increasing the surface area of the MEE by the induction of cellular bulges and filopodia and by promoting MEE degeneration (13,14,17–22). Although the epithelia of the vertical palatal shelves are in intimate contact with the mandibular and lingual epithelia, pathological fusions between the palate and the mandible and/or the tongue are rare (23–26); nevertheless, the MEE must rapidly acquire the capability to fuse if the palatal shelves are not to remain cleft. These observations suggest that competence for oral and palatal shelf adhesion is precisely spatio-temporally regulated; however, the mechanisms that control this process remain uncharacterized.
Mutations in the transcription factor interferon regulatory factor 6 (IRF6) underlie Van der Woude syndrome (VWS) and popliteal pterygium syndrome (PPS) (27), which are characterized by varying degrees of cleft lip, cleft palate, lip pits, skin-folds, syndactyly and intra-oral adhesions (28). Irf6 is expressed in the developing ectoderm with high levels observed in the MEE prior to and during palatal fusion (29,30). Intriguingly, mice that are homozygous for the mutation Arg84Cys (designated Irf6R84C/R84C), which is the most common mutation observed in PPS, or for a null allele of Irf6 (designated Irf6gt1/gt1), exhibit a hyper-proliferative epidermis that fails to undergo terminal differentiation leading to multiple soft tissue fusions which result in the hind-limbs, tail and body wall being fused together and the lumen of the oesophagus being obliterated (31,32). Severe intra-oral epithelial adhesions which occlude the oral cavity and result in cleft palate as the consequence of abnormal fusion between the epithelia covering the palatal shelves and the tongue are also observed in Irf6R84C/R84C and Irf6gt1/gt1 embryos (31,32). In contrast, although Irf6+/R84C embryos exhibited mild inter-epithelial adhesions between the maxilla and mandible in the region of the molar tooth germs, these did not interfere with development of the secondary palate which elevated and fused according to the normal in vivo schedule observed in wild-type embryos (31). Interestingly, the oral adhesions displayed by Irf6 mutant mice partially phenocopy those observed in mice carrying homozygous mutations in Jagged2 which is one of the five related cell surface ligands for the Notch family receptors in mice (Jag2ΔDSL/ΔDSL and Jag2sm/sm). Both strains of Jag2 mutant mice exhibit cleft palate as the result of fusion between the palatal shelves and the tongue, and syndactyly involving all four limbs (26,33).
In the current paper, we demonstrate that IRF6 and Jagged2 function in convergent molecular pathways during development of the oral ectoderm. Moreover, we show that IRF6 plays a key role in the formation of the oral periderm, spatio-temporal regulation of which is essential in ensuring appropriate palatal adhesion.
RESULTS
Irf6+/R84C;Jag2+/ΔDSL mice exhibit cleft palate
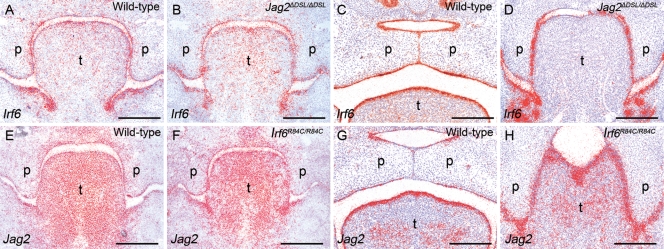
On the basis of the phenotypic overlap between the Irf6 and Jag2 mutant mice (33), we hypothesized that IRF6 and Jagged2 may function in the same or a convergent molecular pathway. Initially, we investigated whether Irf6 was down-regulated in Jag2ΔDSL/ΔDSL homozygous mice using section in situ hybridization (Fig. 1A–D). Irf6 expression was observed throughout the oral epithelia in E12.5 and E14.5 wild-type mice with no variation along the anterior–posterior axis of the secondary palate (Fig. 1A and C). There was no apparent decrease in the expression levels of Irf6 in Jag2ΔDSL/ΔDSL mice compared with their wild-type littermates (Fig. 1B and D). Similarly, we obtained no evidence that Jag2 was down-regulated in Irf6R84C/R84C mice (Fig. 1E–H). In vitro co-immunoprecipitation experiments also failed to reveal any evidence of a direct protein–protein interaction between Jagged2 and IRF6 (data not shown).
Figure 1.
Irf6 and Jag2 are expressed normally in the oral epithelia of Jag2ΔDSL/ΔDSL and Irf6R84C/R84C homozygous mice, respectively. Section in situ hybridization of wild-type (A, C, E, G), Jag2ΔDSL/ΔDSL (B, D) and Irf6R84C/R84C (F, H) mice. At E12.5 (A, B) and E14.5 (C, D), Irf6 is expressed throughout the oral epithelia of both wild-type and Jag2ΔDSL/ΔDSL littermate embryos. Similarly, at E12.5 (E, F) and E14.5 (G, H), Jag2 is expressed with a comparable pattern in wild-type and Irf6R84C/R84C littermate embryos. p, palate; t, tongue. Scale bars: 300 μm.
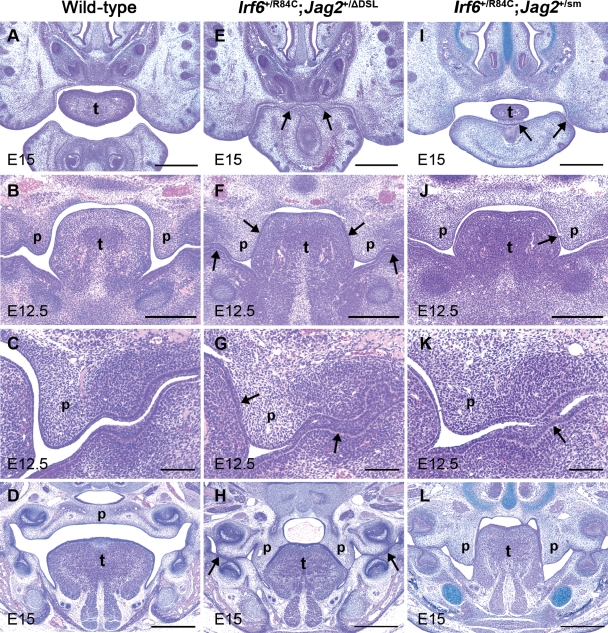
Subsequently, we inter-crossed Irf6+/R84C with either Jag2+/ΔDSL or Jag2+/sm mice. Compound heterozygotes (Irf6+/R84C;Jag2+/ΔDSL and Irf6+/R84C;Jag2+/sm), but not wild-type embryos or embryos carrying a mutant Irf6 or Jag2 allele alone, displayed fully penetrant intra-oral epithelial adhesions which resulted in cleft palate (Fig. 2). Anteriorly, in Irf6+/R84C;Jag2+/ΔDSL compound heterozygous mice, the oral cavity was completely closed and the epithelia covering the maxilla and mandible adhered to one another (Fig. 2A and E). Similarly, throughout E12—E15, multiple epithelial adhesions were observed between the palatal shelves and the tongue and between the epithelia covering the maxillary and mandibular surfaces of the oral cavity in Irf6+/R84C;Jag2+/ΔDSL embryos (Fig. 2F and G); consequently, by E15, the compound heterozygotes exhibited a cleft of the secondary palate with multiple epithelial adhesions within the oral cavity, highly similar to those observed in Jag2ΔDSL/ΔDSL and Irf6R84C/R84C mice (Fig. 2H) (26,33). In general, the intra-oral adhesions observed in Irf6+/R84C;Jag2+/sm embryos were less severe than those observed in Irf6+/R84C;Jag2+/ΔDSL embryos; however, the phenotype was fully penetrant, mild adhesions between the epithelia covering the ventral surface of the anterior tongue and the floor of the mouth, and cleft palate being observed in all embryos examined (Fig. 2I–L).
Figure 2.
Irf6+/R84C;Jag2+/ΔDSL and Irf6+/R84C;Jag2+/sm compound heterozygous embryos exhibit intra-oral epithelial adhesions and cleft palate. (A–L) Histological analysis of wild-type (A–D), Irf6+/R84C;Jag2+/ΔDSL (E–H) and Irf6+/R84C;Jag2+/sm (I–L) mice. (A–D) In wild-type mice, no adhesion between the oral epithelia is observed until the palatal shelves fuse to one another above the dorsum of the tongue (D). (E–H) In contrast, severe inter-epithelial adhesions are observed in Irf6+/R84C;Jag2+/ΔDSL mice which result in occlusion of the anterior region of the oral cavity (E). In addition, the palatal epithelia adhere to those covering the tongue and the mandible (arrows in F and G) such that the palatal shelves fail to elevate and remain cleft (H). Intra-oral adhesions are also observed between the maxilla and mandible in the regions of the molar tooth germs (arrows in H). (I–L) The oral adhesions observed in Irf6+/R84C;Jag2+/sm embryos are less severe than those observed in their Irf6+/R84C;Jag2+/ΔDSL counterparts at all developmental stages (arrows in I, J, K); nevertheless, cleft palate is observed in all Irf6+/R84C;Jag2+/sm embryos (L). p, palatal shelf; t, tongue. Scale bars: A, D, E, H, I, L, 500 μm; B, F, J, 300 μm; C, G, K, 100 μm.
Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice exhibit abnormal oral periderm development
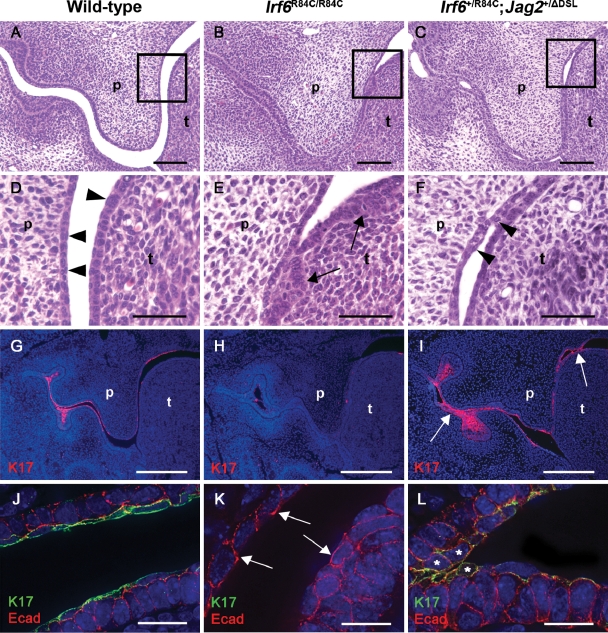
In light of the genetic interaction between Irf6 and Jag2, and the observation that the oral adhesions observed in Jag2ΔDSL/ΔDSL mice result from abnormal development of the epithelium covering the tongue (33), we analysed oral periderm formation in Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice (Fig. 3). Histological analysis at E13.5, when the periderm has formed over the oral epithelium in wild-type embryos, revealed clearly separated, bi-layered epithelia covering the tongue and future nasal surface of the palate (Fig. 3A, D and J). At this stage, both the lingual and palatal epithelia consisted of a regimented series of cuboidal basal cells overlaid with highly flattened, elongated periderm cells (arrowheads in Fig. 3D). In contrast, Irf6R84C/R84C homozygous mice only exhibited a layer of disorganized basal cells which appeared thicker in some regions of the tongue and lacked any obvious formation of periderm cells (Fig. 3B and E). In E13.5 Irf6+/R84C;Jag2+/ΔDSL mice, the epithelia covering the tongue and the future nasal surface of the palate were bi-layered in patches but the basal layer appeared disorganized (Fig. 3C and F). A superficial layer of periderm-like cells was present on the surface of the oral epithelia of Irf6+/R84C;Jag2+/ΔDSL mice, but they appeared irregular and lacked the flattened, elongated appearance characteristic of the periderm in wild-type embryos (arrowheads in Fig. 3F).
Figure 3.
Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL embryos display abnormal oral periderm. (A–F) Histological analysis of E13.5 wild-type (A), Irf6R84C/R84C (B) and Irf6+/R84C;Jag2+/ΔDSL mice (C) reveals abnormal lingual and palatal epithelia in mutant embryos (B, C). (D–F) Higher magnification views of the boxed areas in A, B and C. (D) In wild-type embryos, the epithelia consist of basal cuboidal cells and superficial, flattened periderm cells (D; arrowheads). (E) In contrast, Irf6R84C/R84C embryos display thickened oral epithelia with disorganized basal cells (arrows) and absence of periderm. (F) Irf6+/R84C;Jag2+/ΔDSL mice also exhibit disorganized basal cells but a superficial layer of rounded cells lacking the flattened appearance of normal periderm is observed (arrowheads). (G) At E13.5, keratin 17 is expressed in the periderm and tooth germs of wild-type embryos. (H) In Irf6R84C/R84C embryos, keratin 17 expression is reduced. (I) In Irf6+/R84C;Jag2+/ΔDSL mice, keratin 17 is expressed at similar levels to wild-type embryos, but has a patchy appearance especially in regions of epithelial adhesion (arrowed). (J–L) Deconvolved images of immunofluorescence for keratin 17 (green) and E-cadherin (red) at E13.5. (J) In wild-type mice, keratin 17-expressing periderm cells are distinguishable from the basal cells and, whereas dual labelling is evident on the basal surface of the periderm cells, E-cadherin expression is absent from their apical surface. (K) In contrast, Irf6R84C/R84C mice lack keratin 17 expression on the lingual and palatal epithelia and multiple regions of E-cadherin expression are present on the apical surface of the exposed basal cells (arrowed). (L) While the oral epithelia of Irf6+/R84C;Jag2+/ΔDSL mice are bi-layered, superficial cells lack normal peridermal morphology and dual labelling for keratin 17 and E-cadherin is apparent (asterisks). p, palatal shelf; t, tongue. Scale bars: A–C, 100 μm; D–F, 50 μm; G–I, 300 μm; J–L, 20 μm.
To analyse the periderm in greater detail, we investigated the expression of keratin 17 (34). At E13.5, keratin 17 expression was restricted to the periderm and the developing molar tooth germs of wild-type embryos (Fig. 3G). In Irf6R84C/R84C mice, keratin 17 expression was markedly reduced, residual expression appearing patchy (Fig. 3H). In contrast, although keratin 17 was expressed at similar levels in Irf6+/R84C;Jag2+/ΔDSL mice and their wild-type littermates, staining appeared patchy in the mutant embryos, particularly in the regions of abnormal epithelial attachment (arrows in Fig. 3I). During E14, keratin 17 continued to be expressed uniformly in the periderm with particularly strong expression observed in the developing molar tooth germs and in the epithelium of the dorsal surface of the tongue of wild-type embryos; however, keratin 17 expression was almost completely absent from the oral epithelia of their Irf6R84C/R84C littermates, although limited residual expression was observed on the dorsum of the tongue (data not shown). As observed at E13.5, keratin 17 expression appeared patchy and irregular in E14.5 Irf6+/R84C;Jag2+/ΔDSL mice with strong expression persisting in regions of abnormal epithelial adhesion between the developing maxilla and mandible and between the palatal shelves and the tongue (data not shown). To provide a more detailed understanding of the mechanisms underlying the intra-oral adhesions observed in Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL embryos, we performed dual labelling for keratin 17 and the cell adhesion molecule E-cadherin, images being acquired on a Delta Vision RT restoration microscope and analysed using deconvolution software. While a distinct layer of keratin 17-positive periderm cells whose apical surfaces were negative for E-cadherin were present in E13.5 wild-type embryos (Fig. 3J), their Irf6R84C/R84C littermates lacked this layer and E-cadherin staining was clearly visible on the apical surfaces of the exposed abnormal cells (arrows in Fig. 3K). In the case of Irf6+/R84C;Jag2+/ΔDSL embryos, while a layer of keratin 17-positive cells was clearly visible, these lacked the characteristic flattened morphology of the wild-type periderm cells and many of the cells abnormally expressed E-cadherin on their apical surfaces, particularly in regions of inter-epithelial adhesion (Fig. 3L).
Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice exhibit abnormal oral epithelial differentiation and altered notch signalling
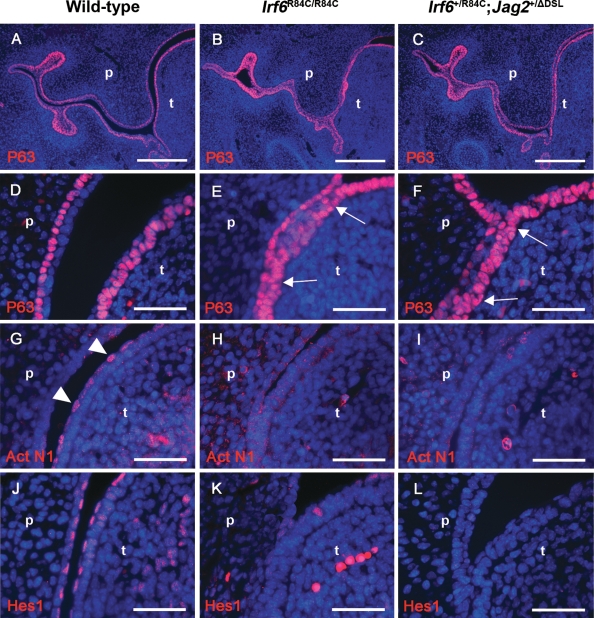
As a consequence of the abnormal oral periderm formation observed in both the Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice, we analysed oral epithelial differentiation in the mutant mice in greater detail. As p63 is known to be a key regulator of squamous epithelial development and has been shown to be vital for normal palatal fusion (35), we analysed p63 expression in the oral epithelia of E13.5 Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice (Fig. 4A–F). While superficially the overall pattern appeared to be similar in wild-type, Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice (Fig. 4A–C), closer examination revealed that although p63 was confined to the basal cells of the palatal and lingual epithelia with no expression in the overlying periderm cells in wild-type mice (Fig. 4D), multiple regions were observed in Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL embryos where p63 expression was expanded into the superficial cell layers, particularly in the regions of abnormal epithelial adhesion (arrows in Fig. 4E and F). As previous results have indicated that the levels of activated Notch1 are down-regulated in the oral epithelia of Jag2ΔDSL/ΔDSL mice (33), similar analyses were performed on Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL embryos. At E13.5, activated Notch1 was localized primarily within the nuclei of the periderm cells of the lingual epithelium (arrowheads in Fig. 4G); in contrast, activated Notch1 staining was absent from the oral epithelia of Irf6R84C/R84C embryos, presumably as a consequence of failure of periderm formation in mice of this genotype (Fig. 4H), and down-regulated in the abnormal periderm of Irf6+/R84C;Jag2+/ΔDSL embryos (Fig. 4I). Similar expression patterns for Hes1, a down-stream target of Notch signalling, were observed in E13.5 Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice compared to their wild-type littermates (Fig. 4J–L).
Figure 4.
Irf6+/R84C;Jag2+/ΔDSL and Irf6R84C/R84C mice exhibit abnormal oral epithelial differentiation and altered Notch signalling. Immunofluorescence analysis of E13.5 wild-type (A, D, G, J), Irf6R84C/R84C (B, E, H, K) and Irf6+/R84C;Jag2+/ΔDSL (C, F, I, L) embryos. (A–C) Immunofluorescence analysis reveals apparently normal levels of p63 expression throughout the oral epithelia of Irf6R84C/R84C (B) and Irf6+/R84C;Jag2+/ΔDSL (C) embryos compared with wild-type littermates (A). (D–F) Higher magnification views indicate that p63 expression is restricted to the basal cells of the lingual and palatal epithelial in wild-type mice (D); in contrast, in Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL embryos, p63 staining is expanded into the superficial layers of the oral epithelia (arrows in E and F, respectively). (G–I) Immunofluorescence analysis of the activated form of Notch1 (Act N1) reveals expression predominantly in the periderm of the lingual epithelia of E13.5 wild-type mice (arrowheads in G). Similar analyses of Irf6R84C/R84C (H) and Irf6+/R84C;Jag2+/ΔDSL (I) embryos reveals a complete loss of activated Notch1 expression; a similar staining pattern is observed for the Notch target, Hes1 (J–L). t, tongue; p, palate. Scale bars: A–C, 300 μm; D–L, 50 μm.
Irf6+/R84C;Jag2+/ΔDSL but not the Irf6R84C/R84C mice demonstrate medial edge seam degeneration
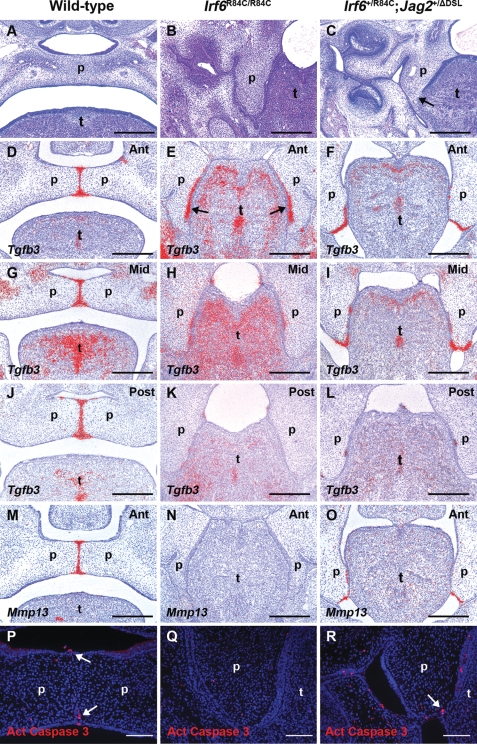
Despite the strong similarities between the phenotypes of Irf6R84C/R84C, Jag2ΔDSL/ΔDSL and Irf6+/R84C;Jag2+/ΔDSL mice, one striking difference between them was observed (Fig. 5 and Supplementary Material, Fig. S1). While partial breakdown of the epithelial seam between the medial surface of the vertical palatal shelves and the lateral surface of the tongue was observed in regions of the presumptive palatal MEE in Jag2ΔDSL/ΔDSL and Irf6+/R84C;Jag2+/ΔDSL mice by E14.5 (Fig. 5C and Supplementary Material, Fig. S1B and C) (33), it remained intact along the entire antero-posterior axis of the secondary palate in all Irf6R84C/R84C embryos examined (Fig. 5B; n = 14). In light of this difference, we analysed the expression of Tgfb3 and its downstream target Mmp13 which have been implicated in degeneration of the MEE during normal palatogenesis (21). Section in situ hybridization revealed strong uniform expression of both Tgfb3 and Mmp13 throughout the MEE seam along the entire antero-posterior axis in wild-type mice at E14.5 (Fig. 5D, G, J and M and Supplementary Material, Fig. S1G and J). Analysis of Tgfb3 expression in Irf6R84C/R84C mice revealed relatively normal levels in the anterior secondary palate; however, expression appeared to be shifted to the future nasal surface of the palate (arrows in Fig. 5E). In the mid- and posterior regions of the secondary palate of Irf6R84C/R84C mice, Tgfb3 expression was drastically reduced with only small patches remaining in the adherent epithelium between the nasal surface of the palatal shelves and the lateral surface of the tongue (Fig. 5H and K). No expression of Mmp13 was detected along the entire antero-posterior axis of the secondary palate at E14.5 in Irf6R84C/R84C mice (Fig. 5N and data not shown). In Irf6+/R84C;Jag2+/ΔDSL mice, Tgfb3 and Mmp13 were expressed in the epithelia of the anterior and middle regions of the secondary palatal shelves where fusion to the lateral aspect of the tongue had occurred (Fig. 5F, I and O and Supplementary Material, Fig. S1H, I, K and L); however, expression of both molecules was virtually absent from the posterior region (Fig. 5L and data not shown). To investigate whether cell death correlated with the regions of epithelial breakdown in Irf6+/R84C;Jag2+/ΔDSL compound heterozygous mice, we used immunostaining for activated caspase-3. Activated caspase-3-positive cells were observed in the MES and the oral and nasal epithelial triangles of wild-type embryos at E14.5 (arrows in Fig. 5P and Supplementary Material, Fig. S1D); similarly, multiple activated caspase-3-positive cells were observed in the epithelial seam between the palatal shelves and the tongue of Irf6+/R84C;Jag2+/ΔDSL mice at the same gestational age (arrows in Fig. 5R and Supplementary Material, Fig. S1E and F). In contrast, no cell death was detected in the region of adhesion between the palatal shelves and the tongue in Irf6R84C/R84C mice, nor in the areas of epithelial adhesion between the maxilla and mandible in the region of the molar tooth germs of Irf6R84C/R84C and Irf6+/R84C;Jag2+/ΔDSL mice, regions in which the epithelia remain intact (Fig. 5Q and data not shown).
Figure 5.
Irf6+/R84C;Jag2+/ΔDSL but not Irf6R84C/R84C mice demonstrate MES degeneration. Analysis of wild-type (A, D, G, J, M, P), Irf6R84C/R84C (B, E, H, K, N, Q) and Irf6+/R84C;Jag2+/ΔDSL (C, F, I, L, O, R) embryos. (A–C) At E15.5, the wild-type secondary palate has fused (A), whereas in Irf6R84C/R84C (B) and Irf6+/R84C;Jag2+/ΔDSL embryos (C), it remains cleft as a consequence of inter-epithelial adhesions. While partial breakdown of the epithelia between the tongue and palate occurs in Irf6+/R84C;Jag2+/ΔDSL mice (arrow in C), the equivalent region remains intact in Irf6R84C/R84C embryos (B). At E14.5, Tgfb3 is expressed throughout the wild-type MEE (D, G, J); in contrast, while Tgfb3 is detected in the anterior region of the secondary palate of Irf6R84C/R84C mice where it fuses to the lateral aspect of the tongue (arrows in E), it is down-regulated in the mid- (H) and posterior (K) regions. (F, I, L) Tgfb3 is also expressed in the presumptive MEE of the secondary palate in Irf6+/R84C;Jag2+/ΔDSL mice; however, isolated areas of residual Tgfb3 expression are observed in the posterior region (L). Whereas Mmp13 expression is present in the MEE of E14.5 wild-type (M) and Irf6+/R84C;Jag2+/ΔDSL mice (O), no Mmp13 expression is observed in the secondary palate of Irf6R84C/R84C mice (N). At E14.5, immunofluorescence reveals activated caspase 3-positive cells in the MES and oral/nasal epithelial triangles of wild-type embryos and in the epithelia between the tongue and the palate in Irf6+/R84C;Jag2+/ΔDSL (P and R, arrows); similar analyses in Irf6R84C/R84C mice fail to reveal activated caspase 3-positive cells in the region of the MEE (Q). t, tongue; p, palate. Scale bars: A–O, 300 μm; P–R, 100 μm.
DISCUSSION
Previous research has shown that Irf6R84C/R84C mice exhibit cleft palate as a consequence of inappropriate intra-oral epithelial adhesions (31,32); however, detailed analyses of these mutant mice and the role of IRF6 in specifying normal palatogenesis have not been undertaken. Interestingly, Irf6R84C/R84C mice partially phenocopy mice carrying mutations in the notch ligand, Jagged2, which also exhibit cleft palate as the result of abnormal intra-oral adhesions (26,33). In the current study, we have demonstrated that IRF6 and Jagged2 interact genetically during development of the secondary palate. Comparison of the phenotypes displayed by the mutant mice revealed that the oral adhesions in Irf6R84C/R84C mice were apparent from the earliest stages of, and persisted throughout, development of the secondary palate. These adhesions were severe in that they involved the entire oral epithelia resulting in almost total occlusion of the oral cavity. In contrast, no obvious differences in palatal shelf development were observed between either Jag2ΔDSL/ΔDSL or Irf6+/R84C;Jag2+/ΔDSL mice and their wild-type littermates until E12 when the palatal shelves had extended vertically down the sides of the tongue, at which point the aberrant oral adhesions involved only the palate/tongue and the maxilla/mandible in the region of the molar tooth germs. As such, the intra-oral adhesions arose earlier and were more severe in Irf6R84C/R84C than in either Jag2ΔDSL/ΔDSL or Irf6+/R84C;Jag2+/ΔDSL mice. In general, the Irf6+/R84C;Jag2+/sm mice exhibited even less extensive oral epithelial adhesions, supporting the hypothesis that the sm mutation, which is a Gly267Ser mis-sense change in the first EGF repeat of Jagged2, is a hypomorphic allele of Jag2 (26,33,36).
As we have been unable to obtain any evidence of a direct interaction between IRF6 and Jagged2, we propose that they function in convergent developmental pathways. During E11–E15, the oral epithelia are composed of a basal layer of cuboidal ectodermal cells and a surface layer of flattened periderm cells. The periderm, which forms from the basal cell layer in a patterned manner, persists throughout embryogenesis and is removed shortly before birth once terminal epithelial differentiation has occurred (37–39). While little is known about the function of the periderm, it has been suggested to act as a protective mediator to the amniotic fluid, a physical barrier or a contributor to formation of the keratinized layer (37,40,41). Previously, it has been demonstrated that the tongue of Jag2ΔDSL/ΔDSL mutant mice displays abnormalities of oral epithelial differentiation particularly of periderm development (33). Casey et al. (33) suggested that Jagged2-mediated Notch signalling was not required for initial periderm formation as the Jag2ΔDSL/ΔDSL homozygous mice exhibited a stratified oral epithelium; rather they hypothesized that Jagged2–Notch1 signalling is required for normal periderm differentiation and/or function as the periderm cells in the Jag2ΔDSL/ΔDSL homozygous mice displayed abnormal morphology and down-regulation of the activated, nuclear form of Notch1. These results are supported by our findings in Irf6+/R84C;Jag2+/ΔDSL embryos, which also exhibit formation of a bi-layered oral epithelium consisting of a disorganized basal layer and overlying periderm cells that lack the usual flattened morphology (Fig. 3). In contrast, similar analyses in Irf6R84C/R84C homozygous mice failed to reveal any periderm formation within the oral epithelia suggesting that normal IRF6 function may be an absolute requirement for oral periderm formation from the basal cells; indeed, our initial analyses suggest that IRF6 may play a central role in the formation of the periderm more widely. This differential effect of Jagged2–Notch1 signalling on one hand versus IRF6 signalling on the other is supported by the range of severity of oral adhesions across the various mouse mutants noted above. Taken together, our results indicate that the phenotype observed in Irf6+/R84C;Jag2+/ΔDSL embryos is unlikely to result from disruption of a protein–protein interaction between IRF6 and Jagged2, or from a failure of one molecule to activate transcription of the other. We therefore speculate that the mechanism underlying the genetic interaction between Jag2 and Irf6 is the consequence of their combined effects on periderm formation, maintenance and function. In this context, the p63 and activated Notch1 expression patterns observed in Irf6+/R84C;Jag2+/ΔDSL embryos suggest that IRF6 affects Jagged2–Notch1 signalling during periderm maintenance. Although the loss of activated Notch1 staining observed in Irf6R84C/R84C mice is likely to be a secondary effect resulting from the absence of periderm (Fig. 3), this is not the case in Irf6+/R84C;Jag2+/ΔDSL embryos where keratin17-positive, p63-negative cells with nuclei characteristic of abnormal periderm cells are observed (Figs 3L and 4F); in wild-type embryos, these cells are activated Notch1 and Hes1-positive (Fig. 4G and J). Together, our results suggest that IRF6 functions in two phases: it is initially required for periderm formation and subsequently functions to maintain periderm integrity through the Jagged2–Notch1 pathway.
Importantly, our data lead us to hypothesize that the function of oral periderm may be to prevent adhesion of intimately apposed epithelia during development. This may be of relevance to the molecular pathogenesis of the intra-oral adhesions and skin folds observed in VWS and PPS. A role for periderm in the prevention of inappropriate epithelial adhesions is particularly important in the oral cavity where the palatal shelves initially develop lateral to the tongue prior to elevating to a horizontal position above it; during this process, the oral epithelia are in close apposition with little intra-oral space (42) and it is therefore imperative that the apposed epithelia do not adhere inappropriately. Despite the mechanisms that ensure pathological adhesions do not occur, the palatal MEE must rapidly acquire fusion capability if the palate is not to remain cleft. Given our hypothesis that one of the functions of the periderm is to prevent inappropriate adhesion of intimately apposed epithelia during development, this cell layer must be removed to allow fusion of the palatal shelves. In this context, data from the rat have suggested that the periderm covering the MEE sloughs away from the basal cells after the palatal shelves assume a horizontal position (7); however, more recent data have suggested that periderm cells are removed from the MEE by migration to the oral and nasal epithelial triangles (16). Regardless of how periderm cells are removed from the midline seam, completion of palatal fusion is known to be critically dependent on TGFβ signalling acting via Smad-dependent and P38 MAPK-dependent pathways (17,18,30,43). Interestingly, homozygous Jag2ΔDSL/ΔDSL and Jag2sm/sm mice exhibit partial MEE seam degeneration between the cleft palatal shelves and the lateral aspect of the tongue suggesting that the signalling mechanisms which allow seam degeneration remain functional in these mice (26,33). Further support for this hypothesis was provided by the observation that palatal shelves dissected from either mutant Jag2 strain are capable of fusing in vitro when placed in apposition indicating that the reason that the palatal shelves of these mice remain cleft is the result of their failure to come into contact rather than a failure of the fusion mechanisms themselves. Indeed, Casey et al. (33) demonstrated clearly that the expression patterns of Tgfb3 and its known downstream target Mmp13, and the patterns of apoptosis, were normal in the secondary palate of Jag2ΔDSL/ΔDSL homozygous mice suggesting that the deletion of Jag2 and its effect on Notch signalling has no effect on TGFβ3 signalling. In the current study, we have demonstrated that partial breakdown of the epithelial seam between the palate and the tongue is also observed in Irf6+/R84C;Jag2+/ΔDSL mice; although this is generally less complete than that occurring in Jag2ΔDSL/ΔDSL mice. Correspondingly, Tgfb3 and Mmp13 are expressed in the presumptive MEE in the anterior and middle regions of the secondary palate, expression in the posterior region being patchier. In contrast, degeneration of the palate/tongue epithelial seam was never observed at any point along the antero-posterior axis of Irf6R84C/R84C mice (n = 14), indicating that IRF6 function is essential for this process. Previous results have demonstrated that Irf6 shares a similar expression pattern in the MEE to Tgfb3 during normal secondary palate development and that Irf6 may function downstream of TGFβ3 signalling in the palate (29,30). Intriguingly, Tgfb3 is only expressed in the anterior region of the palate in Irf6R84C/R84C mice suggesting that IRF6 may exert reciprocal control over TGFβ3 signalling, at least in the posterior two-thirds of the secondary palate. In contrast, Mmp13 expression was absent from the presumptive MEE in all regions of the secondary palate of Irf6R84C/R84C mice, even in the anterior region where down-regulation of Tgfb3 was not observed strongly suggesting that Mmp13 lies downstream of Irf6 function in the palate. The recent delineation of the IRF6 consensus binding site (44) will facilitate the analysis of whether Mmp13 is a direct or indirect target of IRF6 signalling.
In summary, we have demonstrated that the integration of IRF6 and Jagged2 signalling is essential for the control of palatal adhesion and fusion competence via a combined role in the control of oral periderm formation/differentiation. Importantly, our results in Irf6R84C/R84C mice provide a potential explanation for the molecular pathogenesis of VWS and PPS and suggest a wider role for the periderm in preventing self-adhesion of the developing epidermis as observed in conditions such as Bartsocas–Papas syndrome (45).
MATERIALS AND METHODS
Generation of mutant mice
Generation and maintenance of mice carrying the targeted Irf6+/R84C, Jag2+/ΔDSL and Jag2+/sm mutant alleles on C57BL6/J backgrounds have been described previously (26,31,33). Heterozygous Irf6+/R84C mice were inter-crossed with either Jag2+/ΔDSL or Jag2+/sm heterozygotes to generate mice that were heterozygous for both mutant alleles. All experiments were performed in accordance with the Animals (Scientific Procedures) Act, 1986.
Histological, section in situ hybridization and immunofluorescence analyses
For histological analysis, embryos were dissected from time-mated pregnant female mice, the morning on which the vaginal plug was detected being designated embryonic day (E) 0.5. The embryos were staged on the basis of external morphological characteristics, fixed in Bouin's solution, dehydrated through a graded ethanol series, cleared in chloroform, embedded in wax, sectioned at 6 µm and stained with haematoxylin and eosin. Radioactive section in situ hybridization was performed as described by Wilkinson (46) with the modifications described by Tucker et al. (47). All sections were counterstained with haematoxylin and the silver grains pseudocoloured red using Adobe Photoshop 6.0. The Irf6, Jag2, Tgfβ3 and Mmp13 probes have been described previously (29,33). For immunofluorescence analyses, 4% paraformaldehyde or Carnoy's (for cleaved Notch1 staining) fixed sections were treated with 10 mm citrate buffer at 96°C for 10 min for antigen retrieval. Sections were incubated overnight at 4°C with antibodies against E-cadherin (1:200; BD Biosciences), keratin 17 (1:1000) (34), cleaved Notch1 (Val1744; 1:10; Cell Signalling), Hes1 (1:1000) (48), p63 (4A4; 1:50) (49) and activated caspase 3 (1:750; R & D Systems). Sections were counterstained with DAPI and visualized using a Leica DMRB microscope. For keratin 17 and E-cadherin dual immunolocalization, images were acquired on a Delta Vision RT restoration microscope (Applied Precision) using a 60x/1.40 PlanApo objective and the Sedat filter set (Chroma 89000). Images were collected using a Coolsnap HQ camera (Photometrics) with a Z optical spacing of 0.2 μm. Raw images were deconvolved using the Softworx software and maximum intensity projections of these deconvolved images are shown in the results.
Co-immunoprecipitation
For in vitro co-immunoprecipitation experiments, the cDNAs encoding IRF6 and Jagged2 were cloned into the vectors pSG5-FLAG and pSG5-HA and sequence verified. Constructs were transfected into HEK293 cells and immunoprecipitation experiments performed as described previously (50).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The financial support of Wellcome Trust (082868), the Medical Research Council (G0400264) and the National Institutes of Health (P50-DE016215) is gratefully acknowledged. M.J.D. is supported by the NIHR Manchester Biomedical Research Centre. Funding to pay the Open Access charge was provided by Wellcome Trust.
ACKNOWLEDGEMENTS
We thank Pierre Coulombe, Nadeen Brown and Frank McKeon for providing the anti-keratin 17, Hes1 and P63 antibodies, respectively, and Gian-Paolo Dotto for appraising the manuscript. The assistance of Peter March with the imaging is gratefully acknowledged. The Bio-imaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome Trust and the University of Manchester Strategic Fund.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Vanderas A.P. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- 2.Murray J.C., Daack-Hirsch S., Buetow K.H., Munger R., Espina L., Paglinawan N., Villanueva E., Rary J., Magee K., Magge W. Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate Craniofac. J. 1997;34:7–10. doi: 10.1597/1545-1569_1997_034_0007_caesoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 3.Murray J.C. Gene/environment causes of cleft lip and/or palate. Clin. Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 4.Dixon M.J., Murray J.C. Nature Encyclopaedia of the Human Genome. Nature Publishing Group; 2003. Orofacial clefting. [Google Scholar]
- 5.Gritli-Linde A. Molecular control of secondary palate development. Dev. Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro B.L., Sweney L. Electron microscopic and histochemical examination of oral epithelial-mesenchymal interaction (programmed cell death) J. Dent. Res. 1969;48:652–660. doi: 10.1177/00220345690480050801. [DOI] [PubMed] [Google Scholar]
- 7.Fitchett J.E., Hay E.D. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev. Biol. 1989;131:455–474. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- 8.Ito Y., Yeo J.Y., Chytil A., Han J., Bringas P., Jr, Nakajima A., Shuler C.F., Moses H.L., Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 9.Carette M.J., Ferguson M.W. The fate of medial edge epithelial cells during palatal fusion in vitro: An analysis by DiI labelling and confocal microscopy. Development. 1992;114:379–388. doi: 10.1242/dev.114.2.379. [DOI] [PubMed] [Google Scholar]
- 10.Griffith C.M., Hay E.D. Epithelial-mesenchymal transformation during palatal fusion: carboxyfluorescein traces cells at light and electron microscopic levels. Development. 1992;116:1087–1099. doi: 10.1242/dev.116.4.1087. [DOI] [PubMed] [Google Scholar]
- 11.Shuler C.F., Halpern D.E., Guo Y., Sank A.C. Medial edge epithelium fate traced by cell lineage analysis during epithelial-mesenchymal transformation in vivo. Dev. Biol. 1992;154:318–330. doi: 10.1016/0012-1606(92)90071-n. [DOI] [PubMed] [Google Scholar]
- 12.Cecconi F., Alvarez-Bolado G., Meyer B.I., Roth K.A., Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Alvarez C., Tudela C., Perez-Miguelsanz J., O’Kane S., Puerta J., Ferguson M.W.J. Medial edge epithelial cell fate during palatal fusion. Dev. Biol. 2000;220:343–357. doi: 10.1006/dbio.2000.9644. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Alvarez C., Bonelli R., Tudela C., Gato A., Mena J., O’Kane S., Ferguson M.W.J. Bulging medial edge epithelial cells and palatal fusion. Int. J. Dev. Biol. 2000;44:331–335. [PubMed] [Google Scholar]
- 15.Cuervo R., Valencia C., Chandraratna R.A., Covarrubias L. Programmed cell death is required for palate shelf fusion and is regulated by retinoic acid. Dev. Biol. 2002;245:145–156. doi: 10.1006/dbio.2002.0620. [DOI] [PubMed] [Google Scholar]
- 16.Cuervo R., Covarrubias L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development. 2004;131:15–24. doi: 10.1242/dev.00907. [DOI] [PubMed] [Google Scholar]
- 17.Proetzel G., Pawlowski S.A., Wiles M.V., Yin M., Boivin G.P., Howles P.N., Ding J., Ferguson M.W.J., Doetschman T. Transforming growth factor-β3 is required for secondary palate fusion. Nat. Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaartinen V., Voncken J.W., Shuler C., Warburton D., Bu D., Heisterkamp N., Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-β3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 19.Kaartinen V., Cui X.M., Heisterkamp N., Groffen J., Shuler C.F. Transforming growth factor-β3 regulates transdifferentiation of medial edge epithelium during palatal fusion and associated degradation of the basement membrane. Dev. Dyn. 1997;209:255–260. doi: 10.1002/(SICI)1097-0177(199707)209:3<255::AID-AJA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 20.Taya Y., O’Kane S., Ferguson M.W.J. Pathogenesis of cleft palate in TGF-β3 knockout mice. Development. 1999;126:3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- 21.Blavier L., Lazaryev A.L., Groffen J., Heisterkamp N., DeClerk Y.A., Kaartinen V. Tgf-β3-induced palatogenesis requires matrix metalloproteinases. Mol. Biol. Cell. 2001;12:1457–1466. doi: 10.1091/mbc.12.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gato A., Martinez M.L., Tudela C., Alonso I., Moro J.A., Formoso M.A., Ferguson M.W.J., Martinez-Alvarez C. TGF-β3-induced chondroitin suphate proteoglycan mediates palatal shelf adhesion. Dev. Biol. 2002;250:393–405. [PubMed] [Google Scholar]
- 23.Humphrey T. Palatopharyngeal fusion in a human fetus and its relation to cleft formation. Alabama J. Med. Sci. 1970;7:398–429. [PubMed] [Google Scholar]
- 24.Mato M., Uchiyama Y. Ultrastructures of glosso-palatal fusion after treatment of meclozine-hydrochloride. Virch. Archs. A Path. Anat. Histol. 1975;369:7–17. doi: 10.1007/BF00432457. [DOI] [PubMed] [Google Scholar]
- 25.Shah R.M. Palatomandibular and maxillo-mandibular fusion, partial aglossia and cleft palate in a human embryo. Report of a case. Teratology. 1977;15:261–272. doi: 10.1002/tera.1420150308. [DOI] [PubMed] [Google Scholar]
- 26.Jiang R., Lan Y., Chapman H.D., Shawber C., Norton C.R., Serreze D.V., Weinmaster G., Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., Howard E., Ferreira de Lima R.L.L., Daack-Hirsch S., Sander A., et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Woude A. Fistula labii inferioris congenita and its association with cleft lip and palate. Am. J. Hum. Genet. 1954;6:244–256. [PMC free article] [PubMed] [Google Scholar]
- 29.Knight A.S., Schutte B.C., Jiang R., Dixon M.J. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev. Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- 30.Xu X., Han J., Ito Y., Bringas P., Jr, Urata M.M., Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev. Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Richardson R.J., Dixon J., Malhotra S., Hardman M.J., Knowles L., Boot-Handford R.P., Shore P., Whitmarsh A., Dixon M.J. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- 32.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M., et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat. Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casey L.M., Lan Y., Cho E.S., Maltby K.M., Gridley T., Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGowan K.M., Coulombe P.A. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomason H.A., Dixon M.J., Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev. Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 36.Sidow A., Bulotsky M.S., Kerrebrock A.W., Bronson R.T., Daly M.J., Reeve M.P., Hawkins T.L., Birren B.W., Jaenisch R., Lander E.S. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- 37.Holbrook K.A., Odland G.F. The fine structure of developing human epidermis: light, scanning, and transmission electron microscopy of the periderm. J. Invest. Dermatol. 1975;65:16–38. doi: 10.1111/1523-1747.ep12598029. [DOI] [PubMed] [Google Scholar]
- 38.Sanes J.R., Rubenstein J.L.R., Nicholas F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M’Boneko V., Merker H.J. Development and morphology of the periderm of mouse embryos (days 9–12 of gestation) Acta Anat. 1988;133:325–336. doi: 10.1159/000146662. [DOI] [PubMed] [Google Scholar]
- 40.Hayward A.F. The permeability of the epithelium of the skin of fetal rats demonstrated with a lanthanum-containing solution. J. Anat. 1983;136:379–388. [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama M., Smith L.T., Yoneda K., Holbrook K.A., Hohl D., Shimizu H. Periderm cells form cornified cell envelope in their regression process during human epidermal development. J. Invest. Dermatol. 1999;112:903–909. doi: 10.1046/j.1523-1747.1999.00592.x. [DOI] [PubMed] [Google Scholar]
- 42.Greene R.M., Kocchar D.M. Palatal closure in the mouse as demonstrated in frozen sections. Am. J. Anat. 1973;137:477–482. doi: 10.1002/aja.1001370409. [DOI] [PubMed] [Google Scholar]
- 43.Xu X., Han J., Ito Y., Bringas P., Jr, Deng C., Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev. Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Little H.J., Rorick N.K., Su L-I., Baldock C., Malhotra S., Jowitt T., Gakhar L., Subramanian R., Schutte B.C., Dixon M.J., et al. Mis-sense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6. Hum. Mol. Genet. 2009;18:535–545. doi: 10.1093/hmg/ddn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartsocas C.S., Papas C.V. Popliteal pterygium syndrome: evidence for a severe autosomal recessive form. J. Med. Genet. 1972;9:222–226. doi: 10.1136/jmg.9.2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson D.G. In situ Hybridisation: A Practical Approach. Oxford: IRL press; 1995. [Google Scholar]
- 47.Tucker A.S., Al Khamis A., Sharpe P.T. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 48.Lee H.Y., Wroblewski E., Philips G.T., Stair C.N., Conley K., Reedy M., Mastick G.S., Brown N.L. Multiple requirements for Hes 1 during early eye formation. Dev. Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 50.Mooney L.M., Whitmarsh A.J. Docking interactions in the c-Jun N-terminal pathway. J. Biol. Chem. 2004;279:11843–11852. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.