Abstract
Mitochondrial biogenesis is controlled by signaling networks that relay information to and from the organelles. However, key mitochondrial factors that mediate such pathways and how they contribute to human disease are not understood fully. Here we demonstrate that the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 are key downstream effectors of mitochondrial biogenesis that perform unique, yet cooperative functions. The primary function of h-mtTFB2 is mtDNA transcription and maintenance, which is independent of its rRNA methyltransferase activity, while that of h-mtTFB1 is mitochondrial 12S rRNA methylation needed for normal mitochondrial translation, metabolism and cell growth. Over-expression of h-mtTFB1 causes 12S rRNA hypermethylation, aberrant mitochondrial biogenesis and increased sorbitol-induced cell death. These phenotypes are recapitulated in cells harboring the pathogenic A1555G mtDNA mutation, implicating a deleterious rRNA methylation-dependent retrograde signal in maternally inherited deafness pathology and shedding significant insight into how h-mtTFB1 acts as a nuclear modifier of this disease.
INTRODUCTION
A fundamental question in eukaryotic cell biology is how the biogenesis of structurally and functionally distinct organelles is achieved and regulated. This requires establishment and maintenance of complex, organelle-specific macromolecular environments that impart separate functions to each compartment. In the case of mitochondria, this issue is complicated further by the presence of and strict requirement for mtDNA, which in humans encodes 13 essential protein components of the ATP-producing oxidative phosphorylation (OXPHOS) complexes. Therefore, mitochondrial biogenesis requires additional factors for expression and maintenance of the mitochondrial genome (1). Furthermore, because the majority of the ∼1500 mitochondrial proteins are encoded by nuclear genes, mitochondria are an amalgam of proteins encoded by two genomes, the regulation of which must be intricately regulated.
Signaling pathways that control mitochondrial biogenesis and function in mammalian cells have begun to be elucidated. These include both anterograde and retrograde pathways that relay information to and from mitochondria, respectively (2). One of the best-characterized anterograde pathways involves the PGC family of proteins, PGC-1α, PGC-1β and PRC (3,4). These are transcriptional co-activators for a number of nuclear DNA-binding transcription factors, including the nuclear respiratory factors (NRFs) that engage the promoters of many nuclear genes that encode mitochondrial proteins, including a subset of those required for mtDNA transcription and replication (1,3,5). Thus, in collaboration with the NRFs, the PGC family of co-activators induce expression of nuclear-encoded factors required for mitochondrial function and mtDNA expression/replication in response to changing cellular needs or environmental stimuli. For example, PGC-1α was originally identified as a factor required for increased mitochondrial biogenesis during adaptive thermogenesis (6) and promotes exercise-induced mitochondrial biogenesis (7). While the PGC pathway paradigm is now well established and often cited as the ‘master’ regulatory pathway for mitochondrial biogenesis, it is becoming clear that other pathways can act independently or in concert to regulate mitochondrial biogenesis and function (8). For example, the cell cycle regulatory factors cyclin D1, E2F1 and c-Myc have been identified as modulators of mitochondrial biogenesis and activity (9–13). In the case of cyclin D1, this involves phosphorylation and negative regulation of NRF-1 (13). Thus, a reduction in cyclin D1 yields up-regulation of certain NRF-1 target genes and enhances mitochondrial function and biogenesis (12).
While signaling pathways that regulate mitochondrial biogenesis and function are being elucidated at an unprecedented rate, key downstream mitochondrial factors that execute the actual mitochondrial changes in response to these pathways have received considerably less attention. However, the mitochondrial transcription machinery appears to be one class of mitochondrial proteins that is relevant in this regard. The core machinery required for human mitochondrial transcription has been elucidated and comprises a dedicated single-subunit mitochondrial RNA polymerase, POLRMT, the high-mobility-group box protein h-mtTFA (or Tfam), and two transcriptional co-activators, h-mtTFB1 and h-mtTFB2, that stimulate transcription by POLRMT in conjunction with h-mtTFA (1,14,15). The first of these to be identified was h-mtTFA, which is a DNA-binding protein required for efficient transcription initiation at mtDNA promoters (16,17). It was also one of the first mitochondrial genes in the nucleus shown to contain NRF sites and hence be subject to trans-activation by these factors (18). Given this property and its established roles in transcription, transcription-primed mtDNA replication and mtDNA packaging, h-mtTFA has been assumed to be a key regulatory target for mitochondrial biogenesis pathways such as those under control of the PGC family (3,19,20). While h-mtTFA is a critical molecule for mtDNA expression and maintenance, is a bona fide NRF target, and certainly necessary for mitochondrial biogenesis, it is by no means sufficient and whether it is regulatory at all in this regard remains unclear (21,22). In contrast, h-mtTFB1 and h-mtTFB2, which are also NRF targets (23), can induce increases in mitochondrial mass, activity and gene expression when over-expressed in HeLa cells (24), properties consistent with a direct role in controlling mitochondrial biogenesis per se.
As already noted, mitochondria are unique in that they contain proteins derived from two genomes, the nuclear genome and mtDNA. This is perhaps most relevant with regard to the synthesis of the OXPHOS complexes, which comprise 13 mtDNA-encoded subunits around which ∼70 nucleus-encoded subunits assemble to form four of the five enzymatic complexes. This situation clearly dictates some form of communication between the nuclear and mitochondrial genomes to achieve proper regulation of such a complex system. Also noteworthy is the fact that the mtDNA-encoded OXPHOS subunits are translated by a dedicated set of mitochondrial ribosomes in the organelle matrix. Other than the OXPHOS complexes themselves, mitochondrial ribosomes are the only other entity in mitochondria derived from components encoded by both nuclear and mtDNA genomes. The two mitochondrial rRNAs (12S and 16S subunits) are mtDNA-encoded, whereas all the ribosomal proteins are encoded by nuclear genes and imported into the matrix where they associate with the rRNAs to form functional ribosomes. Given that ribosome assembly in bacteria and in the eukaryotic cytoplasm is an expensive and stringently regulated process (25,26), it stands to reason that mitochondrial ribosome assembly may likewise be a primary regulatory control point for mitochondrial biogenesis. Interestingly, the h-mtTFB1 and h-mtTFB2 transcription factors are homologous to the KsgA class of site-specific, rRNA N6-adenine-dimethyltransferases and both exhibit this activity on a heterologous bacterial rRNA substrate (27–29). In our previous study, in which we documented a role for these factors as regulators of mitochondrial biogenesis and gene expression, we could not distinguish the relative contribution of the transcription factor and putative 12S rRNA methyltransferase functions of these proteins. Therefore, that rRNA methylation and/or mitochondrial ribosome assembly was a key contributor to the mitochondrial biogenesis effects we were observing remained an intriguing possibility.
Disruptions in mitochondrial homeostasis and biogenesis are increasingly implicated in human disease and aging (30). Inherited mitochondrial diseases caused by mutations in either mtDNA (maternally inherited) or nuclear genes that encode mitochondrial factors are now estimated to occur in ∼1/5000 live births (31). The pathogenicity of dysfunctional mitochondria can involve a number of principle underlying mechanisms, including disruptions of energy metabolism, perturbed reactive oxygen species generation causing signaling defects or oxidative stress, and altered apoptotic responses and cell death pathways. Relevant to the current study is the deafness-associated A1555G point mutation in mtDNA that causes non-syndromic or antibiotic-induced deafness (32,33). This mutation occurs in the mitochondrial 12S rRNA gene and is postulated to cause a mild mitochondrial translation defect that can be exacerbated by aminoglycosides (34). This presumably contributes to loss of cochlear hair cell function that may also involve irreversible hair cell death. However, the precise mitochondrial defects that cause the tissue-specific deafness pathology are currently unknown. Furthermore, the phenotypic expression of deafness in patients and of the mitochondrial translation-associated defects in patient-derived primary cells and cybrid cell lines is markedly dependent on nuclear modifying loci (35). One such nuclear modifier is the TFB1M gene, encoding h-mtTFB1, polymorphisms near which are associated with a protective effect in A1555G pedigrees (36). While the mechanism through which h-mtTFB1 acts as a nuclear modifier of this disease is currently unknown, the fact that the A1555G mutation occurs in the 3′-end of the 12S rRNA very near the conserved stem-loop that is its predicted methylation substrate (27,28), strongly suggests that the methylation of the 12S rRNA by h-mtTFB1 could be involved (37).
RESULTS
rRNA methyltransferase activity is necessary for induction of mitochondrial biogenesis parameters by h-mtTFB1, but not h-mtTFB2
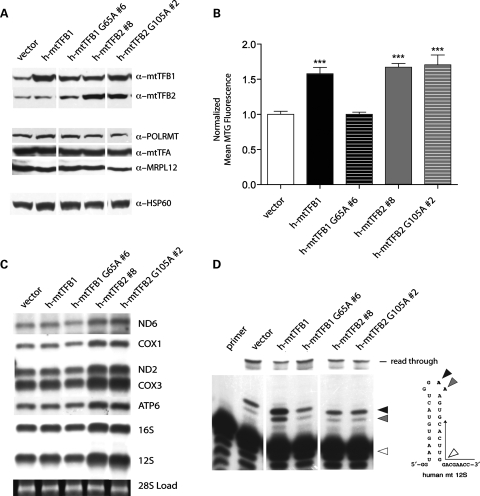
We demonstrated previously that individual over-expression of the human mitochondrial transcription factors/rRNA methyltransferases, h-mtTFB1 or h-mtTFB2, in HeLa cells results in unique influences on mitochondrial biogenesis, gene expression and activity (24). Specifically, over-expression of h-mtTFB1 affects an increase in mitochondrial mass, indicating it regulates key aspects of mitochondrial biogenesis on its own, whereas increased expression of h-mtTFB2 not only increases mitochondrial mass but also results in increased mitochondrial membrane potential, mtDNA copy number, mtDNA-encoded transcripts and mitochondrial translation rates. However, because a retrograde signal to the nucleus leads to coordinate up-regulation of h-mtTFB1 expression in response to h-mtTFB2 over-expression (24), the individual contributions of each of these factors to the observed changes in mitochondrial function in the latter case could not be assigned. Given that these two mitochondrial transcription factors also have rRNA methyltransferase activity (27,28), we determined the degree to which the above-mentioned mitochondrial phenotypes are linked to this enzymatic activity by over-expressing methyltransferase-deficient, point-mutant versions of each: h-mtTFB1 (G65A) and h-mtTFB2 (G105A). We obtained HeLa cell lines that stably over-express each of these mutant proteins (Fig. 1A). Analysis of these lines for changes in mitochondrial mass, membrane potential, mtDNA copy number and steady-state levels of mtDNA-encoded transcripts and proteins revealed that the h-mtTFB2-dependent increase in these parameters was not curtailed by loss of its rRNA methyltransferase activity (Fig. 1B and C and data not shown). That is, similar increases were observed in the wild-type and methyltransferase-deficient h-mtTFB2 over-expression lines. In contrast, over-expression of a methyltransferase-deficient h-mtTFB1 completely abrogated the increase in mitochondrial mass associated with over-expression of the wild-type protein (Fig. 1B). Thus, the function of h-mtTFB1 in modulating mitochondrial biogenesis is completely dependent on its rRNA methyltransferase activity. We also note that the mutant h-mtTFB1 cell lines contained slightly higher levels of h-mtTFB2, but the steady-state levels of other known transcription components POLRMT, h-mtTFA and MRPL12 (38) remained minimally affected in all of the cell lines tested. Finally, the up-regulation of h-mtTFB1 due to h-mtTFB2 over-expression we reported previously (24) was still observed in the h-mtTFB2 methyltransferase-deficient cell lines (Fig. 1A, compare lane 1 to lanes 4 and 5), leaving open the likely possibility that the increase in mitochondrial mass observed in cells that over-express h-mtTFB2 is h-mtTFB1-dependent.
Figure 1.
Ribosomal RNA methyltransferase activity is required for the functions of h-mtTFB1, but not h-mtTFB2 in HeLa cells. (A) Shown is a western blot of mitochondrial extracts from HeLa cells that over-express wild-type (h-mtTFB1 and h-mtTFB2 #8) and methyltransferase-deficient forms (h-mtTFB1 G65A #6 and h-mtTFB2 G105A #2) of h-mtTFB1 and h-mtTFB2. Blots were probed for h-mtTFB1 and h-mtTFB2, other components of the mitochondrial transcription machinery (h-mtTFA, POLRMT and MRPL12) and a mitochondrial loading control (HSP60) as indicated. A cell line transformed with an empty-vector (vector) served as the negative control line to which all others were compared. (B) Mitotracker Green fluorescence (a measure of mitochondrial mass) values in the cell lines described in (A). Shown is the mean fluorescence, normalized to that observed in the vector control cell line, which was given a value of 1.0. (C) Northern blots of total RNA from the cell lines in (A) using the mitochondrial gene probes indicated to the right. Ethidium bromide staining of the 28S cytoplasmic rRNA (at the bottom) indicates similar loading in the lanes. (D) Results of methylation-specific primer extension analysis of mitochondrial 12S rRNA from the cell lines in (A). A primer anti-sense to the 3′-end of the 12S rRNA (diagrammed to the right) was end-labeled and incubated with total mitochondrial RNA. Extension from this primer by AMV reverse transcriptase is blocked at dimethylated adenines, resulting in shorter products (solid triangles). If no methylation is present, extension reads through the stem-loop region and produces longer products (indicated at the top). The position of the unextended primer is indicated by an open triangle. The ratio of truncated (methylated)/extended (read through) products indicates of the relative methylation status of 12S rRNA. In this and all remaining figures t-tests were performed where appropriate. The error bars indicate ± 1 SD from the mean, and statistical significance is indicated as follows: *P < 0.05, **P < 0.005 and ***P < 0.0005.
To determine if the mitochondrial mass changes observed in the wild-type and mutant h-mtTFB1 over-expression lines correlated with the methylation status of the 12S rRNA, we performed methylation-sensitive primer extension assay as described previously (28). As expected, we observed a significant increase in 12S methylation in the wild-type h-mtTFB1 over-expression lines, evidenced as the increased ratio of methylated/unmethylated species detected in the wild-type over-expression cell lines (Fig. 1D, compare lane 3 to lane 2) that was not observed in corresponding methyltransferase-deficient over-expression cell lines (Fig. 1D, compare lane 4 to lane 2). In contrast, no changes in the 12S rRNA methylation ratio were observed in any of the wild-type or methyltransferase-deficient h-mtTFB2 over-expression lines (Fig. 1D, lanes 5 and 6), despite the fact that there is an increase in 12S rRNA levels in these cells (which was accounted for in these experiments by loading the same amount of total mitochondrial RNA in the assays). These data indicate that there is a functional linkage between 12S rRNA methylation by h-mtTFB1 and the regulation of mitochondrial mass.
Coordinate up-regulation of h-mtTFB1 is required for robust mitochondrial biogenesis and functional changes induced by h-mtTFB2
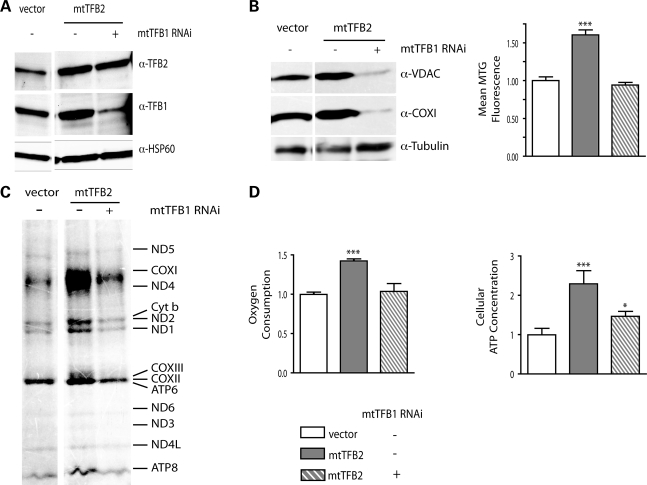
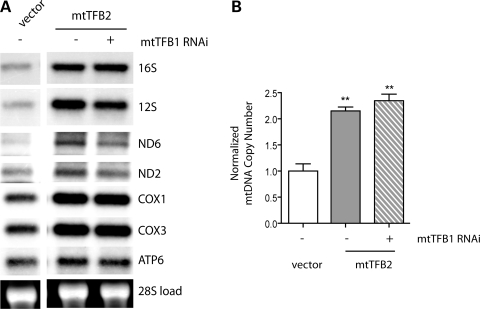
Having shown that elevated levels of h-mtTFB1 and its methyltransferase activity are correlated with increased mitochondrial mass (Fig. 1B), we next tested the hypothesis that the increase in mitochondrial mass observed in h-mtTFB2 over-expression lines (Fig. 1B) was due to the coordinated up-regulation of h-mtTFB1 (as opposed to h-mtTFB2 per se). To accomplish this we knocked down expression of h-mtTFB1 by shRNA in h-mtTFB2 over-expression lines. Transfection of a scrambled shRNA was also analyzed as an appropriate negative control. We were successful in knocking down h-mtTFB1 by ∼90% in the h-mtTFB2 over-expression line, in which the expression of h-mtTFB2 was unaffected (Fig. 2A). Analysis of these lines clearly demonstrates that up-regulation of h-mtTFB1 is indeed required for the increase in mitochondrial biogenesis in the h-mtTFB2 over-expression cell line, as evidenced by decreased levels of COX1 and VDAC in response to h-mtTFB1 knock-down compared with the tubulin loading control, as well as reduced Mitotracker Green staining (Fig. 2B). However, the influence of h-mtTFB1 went beyond its effects on mitochondrial mass, as the increases in mitochondrial translation, oxygen consumption and cellular ATP associated with h-mtTFB2 over-expression were also eliminated when h-mtTFB1 was knocked down (Fig. 2C and D). However, mtDNA copy number and the steady-state levels of mtDNA-encoded transcripts in h-mtTFB2 over-expression lines were largely unaffected by knock-down of h-mtTFB1 (Fig. 3), indicating that these parameters are primarily under direct control of h-mtTFB2. These results demonstrate an intricate interplay between h-mtTFB1 and h-mtTFB2 to orchestrate mitochondrial biogenesis, gene expression and homeostasis.
Figure 2.
Increased mitochondrial mass, translation and activity in h-mtTFB2 over-expressing cells requires the coordinate up-regulation of h-mtTFB1. (A) Immunoblot analysis of mitochondrial extracts from HeLa cell lines over-expressing h-mtTFB2, with (+) or without (−) stable h-mtTFB1 knock-down by shRNA. Empty-vector transformed cells (vector) are also shown as negative controls. The blots were probed for h-mtTFB1, h-mtTFB2 and HSP60 (as a mitochondrial loading control). (B) Immunoblot analysis of whole-cell extracts from the cell lines in (A). Blots were probed for COXI (a representative mtDNA-encoded protein), VDAC (a nucleus-encoded mitochondrial protein that can be taken as one indicator of mitochondrial mass), and tubulin (loading control). The graph on the right shows mean Mitotracker Green fluorescence values from the same cell lines as a separate measure of mitochondrial mass. (C) Mitochondrial translation profiles of the cell lines described in (A). A representative autoradiogram of the indicated radiolabeled mtDNA-encoded translation products is shown. (D) Mitochondrial oxygen consumption and total cellular ATP in the cell lines described in (A). Both graphs represent measurements that were normalized to the values obtained in the empty-vector control cell lines, which were given a value of 1.0.
Figure 3.
The increase in mtDNA and mitochondrial transcripts caused by h-mtTFB2 over-expression does not require simultaneous up-regulation of h-mtTFB1. (A) Northern analysis of HeLa cell lines over-expressing h-mtTFB2, with (+) or without (−) stable h-mtTFB1 knock-down by shRNA. Empty-vector transformed cells (vector) are also shown as negative controls. The blots were probed for the mtDNA transcripts indicated on the right and nuclear 28S is shown as a loading control. (B) Real-time PCR analysis of mtDNA copy number from the same cell lines as in (A).
Requirement for h-mtTFB1 in mitochondrial translation, 12S rRNA accumulation and normal cell proliferation
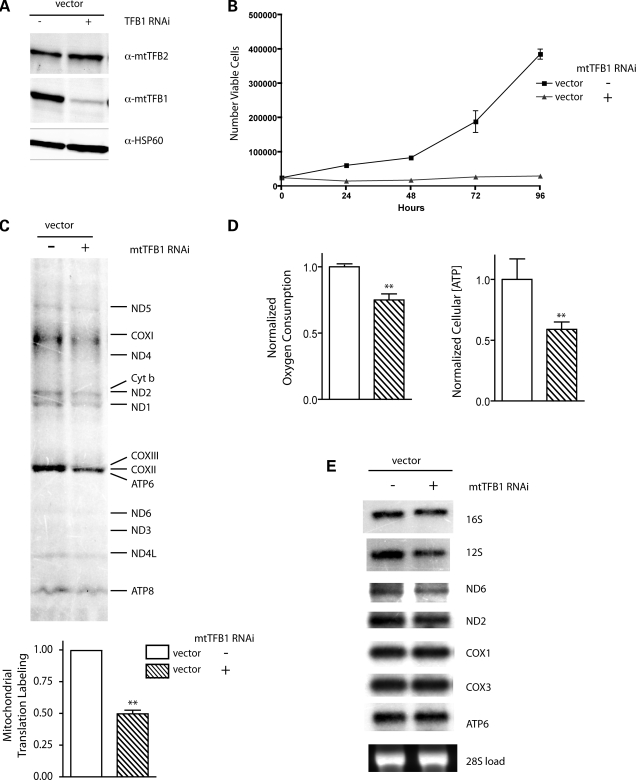
To gain additional insight into the function of h-mtTFB1, we also knocked down its expression by shRNA in normal HeLa cells (i.e. those that are not over-expressing h-mtTFB2) (Fig. 4A). Stable depletion of h-mtTFB1 resulted in a severe reduction of cell growth rates (Fig. 4B) that, interestingly, did not result from perturbation of any specific stage of the cell cycle (Supplementary Material, Fig. S1). There appeared to be no major cell death occurring, as evidenced by lack of floating cells or cellular debris (data not shown), and only a slight increase the amount of apoptotic cells (i.e. annexin V+, propidium iodide+ cells), the latter of which could not account for the major increase in doubling time observed (Supplementary Material, Fig. S1). Analysis of mitochondrial parameters in the h-mtTFB1 knock-down cell lines revealed a significant reduction in mitochondrial translation rates (Fig. 4C), oxygen consumption and cellular ATP (Fig. 4D), but no global changes in mitochondrial transcripts (Fig. 4E), mass or mtDNA (data not shown). However, there was a reproducible decrease in the steady-state level of the 12S rRNA (the substrate for the h-mtTFB1 methyltransferase), suggesting that methylation by h-mtTFB1 is required to stabilize this RNA or that h-mtTFB1 is involved specifically in its transcription or processing (Fig. 4E). Similar effects on 12S steady-state levels were also seen when h-mtTFB1 was knocked down in h-mtTFB2 over-expression lines (Fig. 3A). Altogether, these results demonstrate that h-mtTFB1 is required for optimal cell growth due to positive roles in mitochondrial translation and ribosome biogenesis, OXPHOS activity and cellular ATP production.
Figure 4.
h-mtTFB1 is required for optimal mitochondrial translation and activity, 12S rRNA accumulation, and normal cell growth. (A) Western blot of mitochondrial extracts from empty-vector transformed HeLa cell lines, with (+) or without (−) h-mtTFB1 knock-down by shRNA. Blots were probed for h-mtTFB1, h-mtTFB2 and HSP60 (a mitochondrial loading control). (B) Growth curves of the cell lines described in (A), plotted as the number of viable cells as a function of time in hours. (C) Mitochondrial translation profiles of the cell lines described in (A). (D) Mitochondrial oxygen consumption and total cellular ATP in the cell lines described in (A). (E) Northern blot of total RNA from the cell lines in (A).
12S rRNA hypermethylation induces aberrant mitochondrial biogenesis in cybrid cell lines harboring the mitochondrial deafness-associated A1555G mtDNA mutation
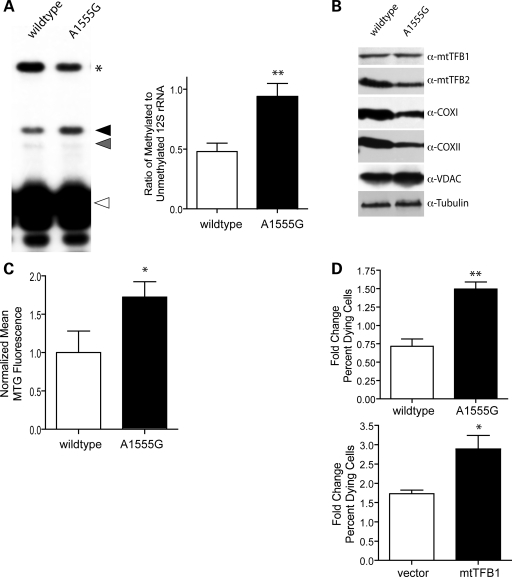
The A1555G mtDNA point mutation causes maternally inherited, non-syndromic deafness on its own or in response to aminoglycoside antibiotic exposure (39). Penetrance of this mutation can be significantly influenced by nuclear genetic background and h-mtTFB1 has been identified as one potential nuclear modifier of this disease (36). The A1555G mutation occurs very near the site for methylation by h-mtTFB1 in an evolutionarily conserved stem-loop at the extreme 3′-end of the 12S rRNA (40). Therefore, we entertained the possibility that this mutation may alter the secondary or tertiary structure of this region of the mitochondrial ribosome so that methylation by h-mtTFB1 is affected. This would shed light onto how h-mtTFB1 might act a nuclear modifier of this mtDNA mutation. To address this, we performed primer extension analysis of the A1555G patient-derived cybrids to assess their 12S rRNA methylation status. There was a clear difference in 12S rRNA methylation status in the mutant cybrids compared with wild-type controls, with the A1555G mutation resulting a hypermethylation of the 12S rRNA. That is, wild-type cybrids had a ∼2:1 ratio of unmethylated/methylated 12S rRNA, whereas the A1555G cybrids have roughly equal amounts of methylated and unmethylated 12S rRNA (Fig. 5A). Western blot analysis revealed similar amounts of h-mtTFB1 and slightly decreased amounts of h-mtTFB2 in the cybrid lines (Fig. 5B), indicating that the increase in methylation was not due to increased expression of h-mtTFB1 or h-mtTFB2. The mutant cybrids also showed no significant change in h-mtTFA or POLRMT levels (data not shown).
Figure 5.
Cells carrying the deafness-associated A1555G mtDNA mutation, like those that over-express h-mtTFB1, exhibit mitochondrial 12S hypermethylation, aberrant mitochondrial biogenesis, and increased susceptibility to cell death. (A) Shown are the results of methylation-sensitive 12S rRNA primer extension reaction from total RNA of wild-type and A1555G mutant cybrids, exactly as described in the legend to Figure 1, except in this case read-through products (*) were terminating due the presence of ddGTP (to yield a product of known length for quantification purposes, see Materials and Methods). The right panel shows quantification of results with means and standard deviations calculated from six separate measurements. (B) Immunoblots of whole cell extracts from wild-type and A1555G mtDNA mutant cybrids were serially probed for mitochondrial h-mtTFB1, h-mtTFB2, COX1, COXII and VDAC and tubulin (as a loading control) as indicated. (C) Mitotracker Green fluorescence values from three separate samples from wild-type and A1555G cybrids. (D) Results of sorbitol-induced cell death assays of wild-type and A1555G cybrids (upper graph) and h-mtTFB1-overexpression and empty-vector (vector) control cell lines. Graphed are the mean fold changes in percent dying cells due to sorbitol treatment of the indicated cell lines.
The fact that 12S rRNA methylation was increased in the A1555G cybrids, without an increase in h-mtTFB1 or h-mtTFB2, provided an opportunity to address the role of 12S rRNA methylation per se as a signal for mitochondrial biogenesis. Remarkably, as was the case with the h-mtTFB1 over-expression cell lines (which have increased rRNA methylation and biogenesis, Fig. 1B and D), the A1555G cybrids had increased mitochondrial biogenesis as signified by increased levels of VDAC (compared to tubulin; Fig. 5B) and Mitotracker Green staining (Fig. 5C). As was the case for over-expression of h-mtTFB1, this increase in mitochondrial biogenesis was aberrant, as evidenced by the decrease of mtDNA-encoded COX1 and COX2 subunits (Fig. 5B), despite there being more organelle mass. Altogether, these results point to 12S rRNA methylation by h-mtTFB1 as a novel retrograde signal for mitochondrial biogenesis and suggest that hypermethylation of the 12S rRNA resulting from the A1555G mutation may contribute to the pathogenicity of this mutation via disruption of this newly identified biogenesis signal.
Aberrant mitochondrial biogenesis due to 12S rRNA hypermethylation increases susceptibility to cell death
The results so far establish a normal homeostatic role for 12S rRNA methylation by h-mtTFB1, however, when this signal is initiated out of context (e.g. via the overexpression of h-mtTFB1 alone or by the A1555G mutation), we propose that a retrograde response is enacted that leads to uncoordinated mitochondrial biogenesis. One predicted outcome of such a scenario is that the susceptibility to cell death could be affected. To test this hypothesis, we determined the susceptibility of h-mtTFB1 over-expressing cells and cybrids harboring the A1555G deafness-associated mutation to cell death induced by osmotic stress (41). This method of induction was chosen because it is linked to Jun N-terminal kinase (JNK) activation in tumor cells and JNK inhibition protects hair cells from cell death during aminoglycoside treatment and acoustic-trauma (42–44). Both the h-mtTFB1 over-expression cell line and the A1555G mutant cybrids exhibited significantly enhanced sorbitol-induced cell death compared with vector-only and wild-type cybrids (Fig. 5D). Since both of these cell lines exhibit hypermethylation of the 12S rRNA (but for different reasons), these data strongly indicate that inappropriate activation of the proposed 12S methylation-induced retrograde signal induces uncoordinated mitochondrial biogenesis that, in turn, increases susceptibility to cell death in response to certain stresses.
DISCUSSION
In this study we focused our attention on deciphering the precise individual functions of two important and related mitochondrial regulatory proteins, h-mtTFB1 and h-mtTFB2, in mitochondrial biogenesis and homeostasis and exploring their potential linkage to the A1555G mtDNA mutation that causes maternally inherited deafness (36). From our results, we draw four primary conclusions: (i) that h-mtTFB1 and h-mtTFB2 are key downstream regulators of mitochondrial biogenesis, gene expression and metabolism that have unique independent, but cooperative functions, (ii) that the primary functions of h-mtTFB1 are dependent on its rRNA methyltransferase activity, while those of h-mtTFB2 are not, (iii) that h-mtTFB1 is not only essential for 12S rRNA methylation, efficient mitochondrial translation and normal cell growth, but, when over-expressed, elicits a rRNA hypermethylation-dependent retrograde signal that induces an aberrant mitochondrial biogenesis response and (iv) that the A1555G mtDNA mutation, like h-mtTFB1 over-expression, causes 12S rRNA hypermethylation, induces a similar faulty retrograde mitochondrial biogenesis signal and predisposes cells to stress-induced cell death. We propose that these new molecular phenotypes are likely relevant to the deafness pathology of the A1555G mutation in hair cells. The rationale for these conclusions is discussed below.
We discovered previously that over-expression of h-mtTFB1 alone is capable of increasing mitochondrial mass in HeLa cells (24). This occurs without a coordinate increase in mitochondrial DNA, transcripts or translation and, therefore, results in organelles with a depressed number of OXPHOS complexes and lower membrane potential per unit mass (i.e. an aberrant mitochondrial biogenesis response). In contrast, over-expression of h-mtTFB2 not only increases mitochondrial mass, but also mtDNA, transcription, translation and membrane potential (24), which is more characteristic of a typical mitochondrial biogenesis profile. However, in that initial study, we were unable to dissect which phenotypes were due specifically to the transcription factor versus the putative rRNA methyltransferase functions of each. Furthermore, because h-mtTFB1 is coordinately up-regulated when h-mtTFB2 is over-expressed (24), assigning specific phenotypes to over-expression of h-mtTFB2 alone was not possible.
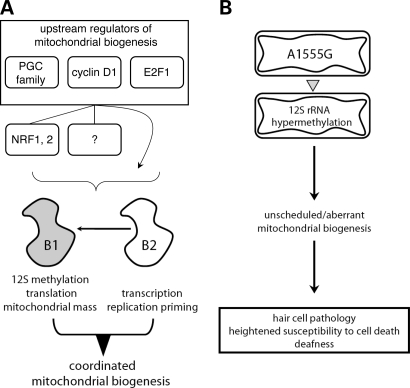
Our results clearly demonstrate that the mitochondrial mass (Fig. 1B) and 12S rRNA methylation (Fig. 1D) increases imparted by h-mtTFB1 over-expression are completely dependent on its rRNA methyltransferase activity (24,28). In contrast, all of the typical increases in mitochondrial biogenesis and activity were observed when a methyltransferase-deficient form of h-mtTFB2 is over-expressed (Fig. 1B and C). However, in this case, the increase in mitochondrial mass, translation, oxygen consumption and ATP production observed in the h-mtTFB2 over-expression lines is lost when h-mtTFB1 expression is knocked down by shRNA (Fig. 2), while the increase in mtDNA copy number and transcript levels is not (Fig. 3). Thus, h-mtTFB1 and h-mtTFB2 have specific roles in mitochondrial biogenesis, gene expression and function, but usually cooperate in order to coordinate all the intended changes in these mitochondrial parameters (Fig. 6A). Conversely, it follows that expression of either alone would result in aberrant modulation of mitochondrial biogenesis and/or function. We also conclude that the primary function of h-mtTFB1 is the 12S rRNA methylation event we documented previously (24,28), which secondarily affects ribosome biogenesis and translation; whereas the primary function of h-mtTFB2 is to facilitate transcription and transcription-primed mtDNA replication (Fig. 6A). These conclusions are consistent with those reached by Kaguni and colleagues in their studies of dm-mtTFB1 and dm-mtTFB2 in Drosophila Schneider cells (45,46), but our results link h-mtTFB1 to 12S rRNA methylation activity directly and to the subsequent regulation of mitochondrial mass. In addition, unlike what we observe here with the knock-down of h-mtTFB1 (Fig. 3B), these investigators did not observe a major effect on cell growth upon knock-down of dm-mtTFB1, likely indicating salient functional differences between the human and fly factors. Finally, a recent knock-out study of mouse mtTFB1 (47) demonstrates that it is essential for mouse development and normal heart function because of a critical role in ribosome biogenesis, which is entirely consistent with our functional assignments for h-mtTFB1 and h-mtTFB2 documented herein and in our previous study (24).
Figure 6.
Models depicting the independent, yet coordinated roles of h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and the proposed involvement of aberrant mitochondrial biogenesis due to 12S rRNA hypermethylation in the deafness-associated pathology of the A1555G mtDNA mutation. Based on the results of this study and published information, we propose (A) that h-mtTFB1 and h-mtTFB2 are key regulators of mitochondrial biogenesis, each with distinct, but normally coordinated functions (h-mtTFB1: 12S methylation, translation and mass regulation; h-mtTFB2: transcription and transcription-primed DNA replication). Thus, h-mtTFB1 and h-mtTFB2 are likely key downstream mediators of characterized mitochondrial biogenesis signaling pathways (e.g. those mediated by PGC-1 family of regulators, cyclin D1 and E2F1). As h-mtTFB1 and h-mtTFB2 are known targets of nuclear-respiratory factors (NRF-1 and NRF-2), signaling can go through these factors. However, we acknowledge the possibility of other mediators (?) or direct effects from these factors (arrow bypassing intermediaries), and likely also other biogenesis pathways, such as those mediated by the PGC family and NRFs (left). The arrow between h-mtTFB2 and h-mtTFB1 represents the retrograde signal we have identified that allows up-regulation of h-mtTFB1 in response to increased h-mtTFB2. Thus, in principle, both factors could be activated by pathways that initially only stimulate h-mtTFB2. In (B) we postulate that the A1555G mtDNA mutation results in hypermethylation the mitochondrial 12S rRNA (most likely due to change in the conformation of the substrate) that elicits a retrograde signal that increases mitochondrial mass without coordinately up-regulating mitochondrial gene expression and mtDNA replication (i.e. unscheduled/aberrant mitochondrial biogenesis). This, in turn, results in heightened susceptibility to stress-induced hair cell pathology and cell death that contributes to the increased incidence in irreversible deafness in patients with the A1555G mutation.
In addition to the cooperative role with h-mtTFB2 in the induction of mitochondrial biogenesis and activity described above, our results from knocking down h-mtTFB1 in isolation also demonstrate a critical homeostatic function for h-mtTFB1 in mitochondrial translation, OXPHOS activity and normal cell growth (Fig. 4). Consistent with the results of knocking down h-mtTFB1 in the context of h-mtTFB2 over-expression (Fig. 3), there was little effect on the steady-state levels of mitochondrial transcripts when h-mtTFB1 alone was depleted (Fig. 4E). The one exception is the 12S rRNA transcript itself, which was reproducibly decreased in the h-mtTFB1 knock-down cells (Figs 3A and 4E). Since this is the substrate for h-mtTFB1 and this methylation event is hypothesized to occur during the assembly of ribosomes in bacteria (48), it stands to reason that the reduced 12S methylation in the face of h-mtTFB1 depletion results in instability of the 12S rRNA inherently or because it is not properly assembled into mitochondrial ribosomes. However, loss of a methyltransferase-independent role for h-mtTFB1 in the transcription and/or processing of the 12S rRNA cannot be excluded outright from our results or from those of the recent mouse mtTFB1 knockout study as is claimed by Larsson and colleagues (47). Regardless of the mechanism, the decreased levels of the 12S rRNA is expected to contribute substantially to the observed translation defect in the h-mtTFB1 depleted cells (Figs 2C and 4C).
The strict dependence of the mitochondrial mass increase observed in the h-mtTFB1 and h-mtTFB2 over-expression lines on 12S methylation (i.e. h-mtTFB1 methyltransferase activity) strongly suggests that this event is a novel signal for some aspect of increased mitochondrial biogenesis. One prediction of this hypothesis is that conditions that lead to 12S hypermethylation in the absence of h-mtTFB1 over-expression would also result in increased mitochondrial biogenesis. Our analysis of cybrid cell lines harboring the deafness-associated A1555G mtDNA mutation fulfills this prediction (Fig. 5). The increase in 12S methylation cannot be attributed to increased expression of h-mtTFB1 or h-mtTFB2 since western analysis reveals no change in h-mtTFB1 and a small reduction in h-mtTFB2 in these cells (Fig. 5B). Therefore, the most likely explanation is that the A1555G mutation (which occurs very near the methylated positions 1583 and 1584 in the 12S rRNA) results in a conformational change in the ribosome that increases the accessibility of the adenines in the conserved stem-loop for h-mtTFB1 methylation. For example, the new base pair that occurs as a result of the A1555G mutation (in the stem of the nearby penultimate stem-loop) could allow the terminal substrate stem-loop to rotate more freely and hence be more readily accessible to h-mtTFB1. Independent of the exact mechanism of 12S hypermethylation, the inescapable conclusion is that increased adenine methylation at the conserved stem-loop of the mitochondrial 12S is part of a retrograde signal that elicits an increase in mitochondrial mass. However, as we will endeavor to explain below, this signal alone likely results in an aberrant up-regulation of mitochondrial biogenesis that has negative consequences.
Though we have identified 12S hypermethylation as a novel retrograde mitochondrial biogenesis signal, we can only speculate on how and why this event is monitored in this way. The amount or status of mitochondrial ribosome biogenesis is a logical process that a cell could use to simultaneously gauge the quality of nuclear and mitochondrial gene expression since, other than the OXPHOS complexes themselves, mitochondrial ribosomes are the only entity derived from products from both genomes (see Introduction). Mitochondrial ribosomes are also the largest and most energetically expensive complex of this type to be assembled in the organelle. Given that methylation of the 12S rRNA is thought to occur during the actual assembly of the small subunit based on studies in bacteria (48), this may serve as a checkpoint for the rate of ribosome biogenesis. Furthermore, that mitochondrial ribosomes are membrane-associated may provide a mechanism to survey available mitochondrial membrane surface area, which is a parameter that could be ‘sensed’ in terms of the need for more overall mitochondrial mass. In such a scenario, more methylation could trigger a change in the ribosome/membrane ratio that triggers the need for increased mitochondrial biogenesis. Alternatively, it is also possible that it is the methylation status of the ribosomes per se that elicits a signal via the rate of synthesis of mtDNA-encoded subunits of the OXPHOS system. There are two adenines that are each potentially mono- or di-methylated by h-mtTFB1. It is possible that ribosomes that have different patterns of methylation have different activity or localization. The existence of such a ‘code’ may underlie why this methylation event is the most evolutionarily conserved rRNA post-translational modification known.
Finally, we propose that our results are relevant to the hair cell pathology caused by the deafness-associated A1555G mtDNA mutation. This mutation occurs near the 3′-end of the mitochondrial 12S rRNA and the primary defect postulated based on in vitro studies of patient-derived primary lymphoblasts and cybrids is a reduced mitochondrial translation. However, the severity of the translation defect in the absence of aminoglycosides antibiotics (which induces deafness in some patients with A1555G) is minimal (34,39) if existent at all (49) in these cultured cell lines. While our results clearly demonstrate a decrease in the steady-state levels of mtDNA-encoded COXI and COX II in the A1555G cell line (Fig. 5C), consistent with a translation and/or assembly defect, they also demonstrate for the first time that these cells have an increase in aberrant mitochondrial biogenesis. As already discussed, this phenotype is likely attributable to hypermethylation of the mitochondrial 12S rRNA causing a retrograde signal that promotes an aberrant mitochondrial biogenesis response (Fig. 6B). Importantly, this scenario results in increased susceptibility to sorbitol-induced cell death (Fig. 5D). Human hair cells are incapable of regenerating and, therefore, cell death is thought to be a major contributor to the irreversible hearing loss associated with the A1555G mutation and in stress-induced deafness in general. Thus, the increased susceptibility of the A1555G cybrids to cell death due to the signaling and mitochondrial biogenesis perturbations elucidated here may provide new molecular insight into why hair cells are more susceptible to cell death when harboring the A1555G mutation alone or in the presence of added stress (e.g. from exposure to aminoglycosides). We postulate that it is this mitochondrial biogenesis retrograde signaling defect elicited by 12S hypermethylation that is equally, or perhaps even more, relevant to the deafness phenotype than the previously proposed translation defect per se. Furthermore, the link we have established between the A1555G and 12S hypermethylation provides obvious mechanistic insight into how h-mtTFB1 (as the primary 12S rRNA methyltransferase) could act as a nuclear modifier of maternally inherited deafness as reported (36). That is, mutations that reduce expression of h-mtTFB1 (or its activity) could provide a protective effect by returning the level of 12S methylation closer to normal.
In conclusion, this study has provided significant new molecular insight into the regulation of mitochondrial biogenesis and metabolism and how its deregulation can contribute to mitochondrial pathogenesis. A deeper understanding of how h-mtTFB1 and h-mtTFB2 regulate mitochondrial biogenesis, gene expression and function, as well as, the precise signaling pathways involved may provide new ways to target these factors/pathways in the treatment of human mitochondrial diseases and age-related pathology. Furthermore, identification of 12S hypermethylation, aberrant mitochondrial biogenesis and enhanced susceptibility to cell death as molecular phenotypes of the A1555G deafness-associated mtDNA mutation may likewise provide development of treatment or preventative measures for this form of hearing loss.
MATERIALS AND METHODS
Stable inhibition of h-mtTFB1 expression by retroviral-delivered shRNA
Vectors for producing shRNA directed against h-mtTFB1 were constructed as follows. Sequences predicted to target h-mtTFB1 mRNA for degradation were obtained using Block-IT RNAi Designer for shRNA (Invitrogen). Accession number NM_016020.2 was input and pENTR/U6 and ‘Sense-loop-antisense’ options were selected. The top three highest ranked hits were selected. The CGAA loop sequence was added and the subsequent sequences were used to generate inserts for pSiren-RetroQ (Clonetech). ‘Sense-loop-antisense’ sequences obtained above were copied and linker sequences were added: To the top strand, GATCC was added to the 5′-end and TTTTTCTCGAGG to the 3′-end. To the bottom strand, AATTCCTCGAGAAAAA was added to the 5′-end and a G to the 3′-end. The final sequences obtained were: TFB1M sh#1 top 5′-GAT CCG GAC TTG AGG CTG ACA GAT AAC GAA TTA TCT GTC AGC CTC AAG TCC TTT TTC TCG AGG-3′, bottom 5′-AAT TCC TCG AGA AAA AGG ACT TGA GGC TGA CAG ATA ATT CGT TAT CTG TCA GCC TCA AGT CCG-3′; TFB1M sh#2 top 5′-GAT CCG CAA TCT GAC AAA TGC TTA TGC GAA CAT AAG CAT TTG TCA GAT TGC TTT TTC TCG AGG-3′, bottom 5′-AAT TCC TCG AGA AAA AGC AAT CTG ACA AAT GCT TAT GTT CGC ATA AGC ATT TGT CAG ATT GCG-3′; TFB1M sh#3 top 5′-GAT CCG CAA TGT TCG ACA CAT CTT TAC GAA TAA AGA TGT GTC GAA CAT TGC TTT TTC TCG AGG-3′, bottom 5′-AAT TCC TCG AGA AAA AGC AAT GTT CGA CAC ATC TTT ATT CGT AAA GAT GTG TCG AAC ATT GCG-3′. A negative control was constructed using sequences from Clonetech: neg top 5′-GAT CCG TGC GTT GCT AGT ACC AAC TTC AAG AGA TTT TTT ACG CGT G-3′, neg bottom 5′-AAT TCA CGC GTA AAA AAT CTC TTG AAG TTG GTA CTA GCA ACG CAC G-3′. Top and bottom strands were annealed and ligated with pSiren-RetroQ that had been digested with BamHI and EcoRI for TFB1M sequences or MluI for negative control sequences. The resulting vectors were sequenced to confirm the correct insert.
To generate the retrovirus carrying the shRNA sequences, HEK293T cells in 10 cm dishes were transfected with pVSVg, pCL-Eco, and the appropriate pSiren vector using Fugene 6. After 24 h, the transfection media was removed and replaced with 5 ml of OptiMEM. Every 24 h for 4 days, the media was collected, stored at 4°C and replaced with fresh OptiMEM. After the last day, all media containing virus was pooled, sterilized with a 0.45 µm filter and concentrated to 3 ml using a Centricon-20 spin column (Millipore). Virus was aliquoted and stored at −80°C until needed. To infect cells with virus, growth media was replaced with fresh media containing 4 µg/ml polybrene. Negative control virus (500 µl) was diluted to 2 ml in OptiMEM and added directly to the plate. For h-mtTFB1 knock-down, 250 µl of each TFB1M shRNA virus was pooled and diluted to 2 ml with OptiMEM. This mixture was then added directly to each desired plate. Infected cells were allowed to grow for 24 h, after which media was removed and replaced with normal growth media plus 0.6 µg/ml puromycin. Cells were grown for an additional 48 h and then split 1:5 with continued puromycin selection. The resulting cells were tested for knock-down and used in experiments described below.
Cell culture, growth curves and cell cycle analysis
HeLa cells were cultivated as described previously (24). Cybrid cell lines were obtained as a kind gift from Min Guan and grown as described (35). For growth curve experiments, 25 000 cells were plated into individual wells of 12-well plates. Every 24 h, cells were harvested by trypsinization, stained with trypan blue and viable cells were counted with a hemacytometer. For cell cycle analysis, cells were fixed with ethanol, stained with propidium iodide and a profile was obtained with a BD FACSCalibur. For measuring markers of cell death, cells were stained with the Annexin V-FITC Apoptosis detection kit II (BD Pharmigen) according to the manufacturer's recommendations. Stained cells were then analyzed with a BD FACSCalibur.
Mitochondrial translation assays
Labeling of mitochondrial translation products was performed essentially as described (24). Empty-vector containing cells with h-mtTFB1 knock-down were plated at 15 000 cells/cm2 due to their much slower growth rate. Only 25 µg of mitochondrial protein could be obtained from these cells, so all other samples were adjusted to match this amount when loaded on the linear gradient gel.
Oxygen consumption, ATP, mitochondrial mass and membrane potential measurements
Oxygen consumption measurements were made using a YSI 5300A Oxygen Monitor with 5 × 106 cells resuspended in 5 ml of DMEM + 10% bovine growth serum and added to the sample chamber. Oxygen consumption was recorded for 10 min at 37°C with stirring. Sodium azide was then added to 5 mm and oxygen consumption was measured for 5 min to determine the amount of mitochondrial oxygen consumption. ATP measurements were obtained from 1 × 105 cells using the ATP Bioluminescence CLS II kit (Roche) according to the manufacturers recommendation and luminescence was measured using a plate reader. Measurements were compared to an ATP standard curve to determine ATP concentration in each sample. Measurement of mitochondrial mass and membrane potential was performed using Mitotracker Green staining as described previously (24).
Immunoblot, northern and 12S methylation-sensitive primer extension analysis
Immunoblots and northern blots were performed as described previously (24). Primers for generating double-stranded probes for COX1, COX3 and ATP6 were as follows: COX1-5′, 5′-CGG CGC ATG AGC TGG AGT CC-3′; COX1-3′, 5′-GAG AGA TAG GAG AAG TAG G-3′; COX3-5′, 5′-CCT AAT GAC CTC CGG CCT AG-3′; COX3-3′, 5′-GAG CCG TAG ATG CCG TCG G-3′; ATP6-5′, 5′-CCA CAA TCC TAG GCC TAC C-3′ and ATP6-5′, 5′-GCA TGA GTA GGT GGC CTG C-3′. Mitochondrial RNA was harvested by first isolating mitochondria (24) then processing the resulting mitochondrial pellet with the RNEasy Kit (Qiagen). For primer extension analysis of 12S rRNA, a primer designed to bind the 3′-terminus (5′-CTGGTTCGTCCAAGTG-3′) of the 12S rRNA was labeled with 32P by T4 polynucleotide kinase as described previously (28). Total RNA (10 µg) or mitochondrial RNA (2 µg) were incubated with 200 fmol of labeled primer in 1× AMV RT buffer (Roche) with either 1 mm dNTPs or 1 × dNTP mix (1 mm dATP, dTTP, dCTP and 250 uM ddGTP) in a 25 µl reaction volume for 20 min at 55°C. The tubes were allowed to cool to room temperature for 5 min then 10 µl of AMV RT mix was added (1 × AMV RT buffer, 20 units AMV RT, 1 mm dNTPs of 1 × dNTP mix) and samples were incubated at 42°C for 30 min. After elongation had occurred an equal volume of formamide loading buffer was added to each sample and loaded on a 15% acrylamide:7 M urea sequencing gel. Samples were electrophoresed at 25 mA for ∼2.5 h. The gel was then placed on filter paper, dried and exposed to film.
Cell death assays
Cells were seeded at 5000 cells/cm2 in 12-well dishes and grown for 72 h in DMEM with serum and additives. Osmotic shock and subsequent cell death were induced essentially as described (50). Briefly, media was changed to RPMI with 10% fetal calf serum and 0.4 M sorbitol for 4 h to induce cell death. To assay for cell death, cells were rinsed with phosphate-buffered saline, dislodged with TryPLE and pelleted. Cell pellets were then resuspended and labeled with FITC-Annexin V and propidium iodide according to the manufacturer's instructions (BD Bioscience, Catalog #556547). Labeled cells were analyzed on a FACSCalibur, fluorescence data were acquired using Cell Quest and analyzed with FlowJo software. The amount of cell death was defined as the proportion of the total cell population that stained positive for both Annexin V and propidium iodide. The fold change in the percentage of cell death in each cell line due to sorbitol exposure is plotted in Figure 5D.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by NIH grant HL-059655 from the National Heart, Lung, and Blood Institute and the ‘2007 Dawn and Brook Lenfest Grant in Auditory Science’ from the National Organization for Hearing Research Foundation, both awarded to G.S.S. The authors wish to thank Min Guan for the kind gift of patient-derived A1555G and wild-type cybrid cell lines.
REFERENCES
- 1.Bonawitz N.D., Clayton D.A., Shadel G.S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Butow R.A., Avadhani N.G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 3.Scarpulla R.C. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 4.Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 5.Asin-Cayuela J., Gustafsson C.M. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 7.Irrcher I., Adhihetty P.J., Sheehan T., Joseph A.M., Hood D.A. PPARgamma coactivator-1alpha expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. Am. J. Physiol. Cell. Physiol. 2003;284:C1669–C1677. doi: 10.1152/ajpcell.00409.2002. [DOI] [PubMed] [Google Scholar]
- 8.D'souza A.D., Parikh N., Kaech S.M., Shadel G.S. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luciakova K., Barath P., Li R., Zaid A., Nelson B.D. Activity of the human cytochrome c1 promoter is modulated by E2F. Biochem. J. 2000;351:251–256. doi: 10.1042/0264-6021:3510251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto Y., Hayashi R., Kang D., Yoshida K. Acute loss of transcription factor E2F1 induces mitochondrial biogenesis in HeLa cells. J. Cell Physiol. 2006;209:923–934. doi: 10.1002/jcp.20802. [DOI] [PubMed] [Google Scholar]
- 11.Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O'Donnell K.A., Kim J.W., Yustein J.T., Lee L.A., Dang C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamaki T., Casimiro M.C., Ju X., Quong A.A., Katiyar S., Liu M., Jiao X., Li A., Zhang X., Lu Y., et al. Cyclin D1 determines mitochondrial function in vivo. Mol. Cell. Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Li Z., Lu Y., Du R., Katiyar S., Yang J., Fu M., Leader J.E., Quong A., Novikoff P.M., et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc. Natl. Acad. Sci. USA. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCulloch V., Shadel G.S. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 16.Fisher R.P., Parisi M.A., Clayton D.A. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev. 1989;3:2202–2217. doi: 10.1101/gad.3.12b.2202. [DOI] [PubMed] [Google Scholar]
- 17.Fisher R.P., Clayton D.A. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virbasius J.V., Scarpulla R.C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baar K. Involvement of PPAR gamma co-activator-1, nuclear respiratory factors 1 and 2, and PPAR alpha in the adaptive response to endurance exercise. Proc. Nutr. Soc. 2004;63:269–273. doi: 10.1079/PNS2004334. [DOI] [PubMed] [Google Scholar]
- 20.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Maniura-Weber K., Goffart S., Garstka H.L., Montoya J., Wiesner R.J. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 23.Gleyzer N., Vercauteren K., Scarpulla R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotney J., Wang Z., Shadel G.S. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granneman S., Baserga S.J. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Wilson D.N., Nierhaus K.H. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 27.Cotney J., Shadel G.S. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 28.Seidel-Rogol B.L., McCulloch V., Shadel G.S. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 29.McCulloch V., Seidel-Rogol B.L., Shadel G.S. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shadel G.S. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am. J. Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorburn D.R. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 2004;27:349–362. doi: 10.1023/B:BOLI.0000031098.41409.55. [DOI] [PubMed] [Google Scholar]
- 32.Shohat M., Fischel-Ghodsian N., Legum C., Halpern G.J. Aminoglycoside-induced deafness associated with the mitochondrial DNA mutation A1555G. Am. J. Otolaryngol. 1999;20:64–67. doi: 10.1016/s0196-0709(99)90054-6. [DOI] [PubMed] [Google Scholar]
- 33.Prezant T.R., Chaltraw W.E., Fischel-Ghodsian N. Identification of an overexpressed yeast gene which prevents aminoglycoside toxicity. Microbiology. 1996;142:3407–3414. doi: 10.1099/13500872-142-12-3407. [DOI] [PubMed] [Google Scholar]
- 34.Guan M.X., Fischel-Ghodsian N., Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 35.Guan M.X., Fischel-Ghodsian N., Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- 36.Bykhovskaya Y., Mengesha E., Wang D., Yang H., Estivill X., Shohat M., Fischel-Ghodsian N. Human mitochondrial transcription factor B1 as a modifier gene for hearing loss associated with the mitochondrial A1555G mutation. Mol. Genet. Metab. 2004;82:27–32. doi: 10.1016/j.ymgme.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Shadel G.S. A dual-function mitochondrial transcription factor tunes out deafness. Mol. Genet. Metab. 2004;82:1–3. doi: 10.1016/j.ymgme.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z., Cotney J., Shadel G.S. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan M.X., Fischel-Ghodsian N., Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum. Mol. Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- 40.Prezant T.R., Agapian J.V., Bohlman M.C., Bu X., Oztas S., Qiu W.Q., Arnos K.S., Cortopassi G.A., Jaber L., Rotter J.I. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 41.Criollo A., Galluzzi L., Maiuri M.C., Tasdemir E., Lavandero S., Kroemer G. Mitochondrial control of cell death induced by hyperosmotic stress. Apoptosis. 2007;12:3–18. doi: 10.1007/s10495-006-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Van De Water T.R., Bonny C., de Ribaupierre F., Puel J.L., Zine A. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J. Neurosci. 2003;23:8596–8607. doi: 10.1523/JNEUROSCI.23-24-08596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsui J.I., Gale J.E., Warchol M.E. Critical signaling events during the aminoglycoside-induced death of sensory hair cells in vitro. J. Neurobiol. 2004;61:250–266. doi: 10.1002/neu.20054. [DOI] [PubMed] [Google Scholar]
- 44.Wang J., Ruel J., Ladrech S., Bonny C., van de Water T.R., Puel J.L. Inhibition of the c-Jun N-terminal kinase-mediated mitochondrial cell death pathway restores auditory function in sound-exposed animals. Mol. Pharmacol. 2007;71:654–666. doi: 10.1124/mol.106.028936. [DOI] [PubMed] [Google Scholar]
- 45.Matsushima Y., Adan C., Garesse R., Kaguni L.S. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 46.Matsushima Y., Garesse R., Kaguni L.S. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 47.Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Thammana P., Held W.A. Methylation of 16S RNA during ribosome assembly in vitro. Nature. 1974;251:682–686. doi: 10.1038/251682a0. [DOI] [PubMed] [Google Scholar]
- 49.Giordano C., Pallotti F., Walker W.F., Checcarelli N., Musumeci O., Santorelli F., d'Amati G., Schon E.A., DiMauro S., Hirano M., et al. Pathogenesis of the deafness-associated A1555G mitochondrial DNA mutation. Biochem. Biophys. Res. Commun. 2002;293:521–529. doi: 10.1016/S0006-291X(02)00256-5. [DOI] [PubMed] [Google Scholar]
- 50.Bilney A.J., Murray A.W. Pro- and anti-apoptotic effects of K+ in HeLa cells. FEBS Lett. 1998;424:221–224. doi: 10.1016/s0014-5793(98)00172-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.