Abstract
von Hippel-Lindau (VHL) disease is caused by germ-line mutations in the VHL tumor suppressor gene and is the most common cause of inherited renal cell carcinoma (RCC). Mutations in the VHL gene also occur in a large majority of sporadic cases of clear-cell RCC, which have high intrinsic resistance to chemotherapy and radiotherapy. Here we show that VHL-deficient RCC cells express lower levels of the pro-apoptotic Bcl-2 family protein BIMEL and are more resistant to etoposide and UV radiation induced death compared to the same cells stably expressing the wild type VHL protein (pVHL). Re-introducing pVHL into VHL-null cells increased the half-life of BIMEL protein without affecting its mRNA expression, and over-expressing pVHL inhibited BIMEL polyubiquitination. Suppressing pVHL expression with RNA interference resulted in a decrease in BIMEL protein and a corresponding decrease in the sensitivity of RCC cells to apoptotic stimuli. Directly inhibiting BIMEL expression in pVHL-expressing RCC cells caused a similar decrease in cell death. These results demonstrate that pVHL acts to promote BIMEL protein stability in RCC cells, and that destabilization of BIMEL in the absence of pVHL contributes to the increased resistance of VHL-null RCC cells to certain apoptotic stimuli.
Keywords: VHL, BIMEL, renal cell carcinoma, apoptosis, protein stability
Introduction
Renal cell carcinoma (RCC) is the most common form of kidney cancer arising from the renal tubule. The vast majority of cases are clear-cell RCC, and 75% of these are associated with mutations in the von Hippel-Lindau (VHL) gene (Cohen, 1999). Germ-line mutations in the VHL gene result in VHL disease, which is characterized by the development of clear-cell RCC, hemangioblastomas and pheochromocytomas (Kaelin, 2002; Nyhan et al., 2008). RCC associated with defects in VHL are highly resistant to conventional radiation and chemotherapies (Yagoda et al., 1995), which is recapitulated in vitro by the reduced susceptibility of VHL-deficient tumor cells to apoptotic stimuli (Qi and Ohh, 2003; Kim et al., 2004; Lee et al., 2005; Roe et al., 2006).
The VHL protein (pVHL) is best known as part of the E3 ubiquitin ligase that targets hypoxia-inducible factor (HIF) for degradation by the proteasome (Maxwell et al., 1999). HIF is a heterodimeric transcription factor comprised of an oxygen-sensitive α subunit and a constitutive β subunit that regulates the expression of genes that control angiogenesis, erythropoiesis, glycolysis and cell survival (Semenza, 2003). Under normoxic conditions, hydroxylation of proline in HIFα triggers its association with pVHL resulting in its polyubiquitination and degradation by the proteasome (Ivan et al., 2001; Jaakkola et al., 2001). Hypoxia suppresses the activity of the prolyl hydroxylases allowing HIFα to escape ubiquitination by pVHL and to become active as a transcription factor (Epstein et al., 2001; Bruick and McKnight, 2001). Besides HIFα proteins, a number of other proteins bind pVHL including p53, Card9, atypical protein kinase C (aPKC), multiple subunits of RNA polymerase II, myogenin, kinesin 2, and others (Frew and Krek, 2008). For some of these proteins, interaction with pVHL leads to their ubiquitination and degradation (Okuda et al., 2001; Fu et al., 2007). In other cases, pVHL serves as an adaptor linking proteins involved in transcription, cell adhesion, microtubule organization, and cell survival (Yang et al., 2007; Hergovich et al., 2003; Roe et al., 2006; Kurban et al., 2008).
Recently, Zantl et al. (2007) suggested that the poor apoptotic response of clear-cell RCC cells might be due to reduced expression of BIMEL, a pro-apoptotic BH3-only member of the Bcl-2 family (O’Connor et al., 1998). In their study (Zantl et al., 2007), BIMEL expression was readily detected in normal renal tubules but was markedly reduced in most clear-cell RCC samples analyzed. BIMEL is one of three major alternatively spliced forms of the BIM gene that collectively are important for cell death caused by many apoptotic stimuli (Bouillet et al., 1999). Regulation of BIMEL expression involves complex transcriptional and post-translational mechanisms, including phosphorylation-dependent regulation of its protein stability (Ley et al., 2005). Very little is known about the regulation of BIMEL in RCC cells and why its expression is frequently suppressed (Zantl et al., 2007). It is also not known whether the reduced levels of BIMEL in RCC are related to the status of pVHL in these cells.
Here we examined BIMEL expression in RCC cells with and without pVHL. Cells expressing pVHL had higher levels of BIMEL protein than the same cells lacking pVHL. Inhibiting pVHL expression with RNA interference led to a parallel decrease in BIMEL. Expression of pVHL reduced BIMEL polyubiquitination and extended its protein half-life, indicative of a role for pVHL in regulating BIMEL stability. pVHL-expressing RCC cells were also more sensitive to death induced by etoposide and UV radiation, which was reversed by inhibiting either pVHL or BIMEL with RNA interference. These results suggest that loss of pVHL may enhance the resistance of RCC to apoptosis-inducing therapies in part by destabilizing BIMEL.
Results
pVHL expression in RCC cells correlates with increased sensitivity to apoptotic stimuli
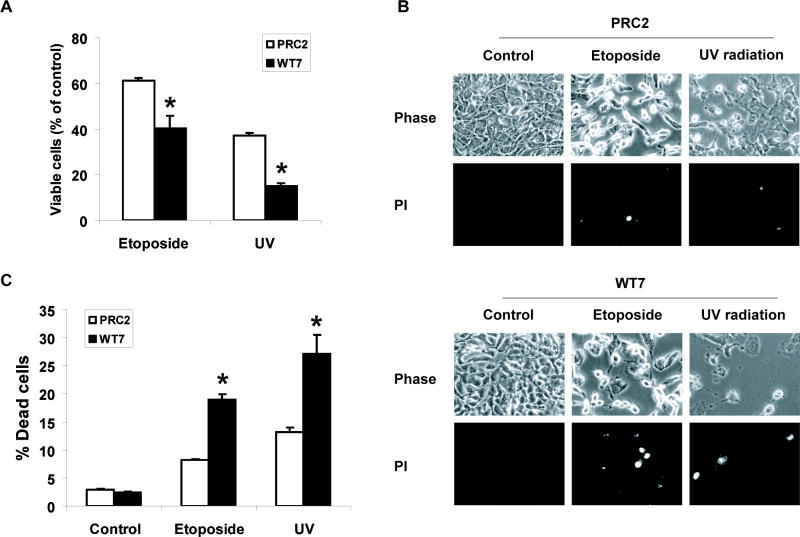
The RCC line 786-0 contains mutations in both alleles of the VHL gene resulting in the absence of pVHL expression (Gnarra et al., 1994). To determine if the status of pVHL in RCC cells influences their sensitivity to cytotoxic stimuli, we examined the effects of the chemotherapeutic drug etoposide on 786-0 derived cell lines stably expressing wild type pVHL (WT7) or empty vector (PRC2) (Iliopoulos et al., 1995). Initial experiments showed that 50–100 μM etoposide caused a 30–40% decrease in the number of viable PRC2 cells at 24 h. When PRC2 and WT7 cells were treated with etoposide (50 μM), significantly more PRC2 cells remained alive after 24 h (Figure 1A). When PRC2 and WT7 cells were exposed to a second apoptotic stimulus, UV radiation (1000 J/m2), there was a 60% decrease in the number of viable PRC2 cells after 24 h compared to an 85% decrease in WT7 cells. To determine if the greater loss of WT7 cells reflected an increase in cell death, PRC2 and WT7 cells were exposed to etoposide or UV and then stained with the membrane impermeable DNA-binding dye propidium iodide. In both cases, we observed a 2-fold increase in WT7 cells stained with propidium iodide compared to PRC2 cells (Figure 1B, C). Thus, pVHL-expressing WT7 cells are more sensitive to etoposide and UV radiation induced death than VHL-null PRC2 cells.
Figure 1. Effect of pVHL expression on the sensitivity of RCC cells to etoposide and UV radiation induced cell death.
(A) Equal numbers of PRC2 and WT7 cells were treated with etoposide (50 μM), exposed to UV radiation (1000 J/m2), or left untreated. After 24 h, the cells were fixed and stained with Hoechst dye in order to visualize nuclear morphology. Viability was assessed by first counting the number of drug or UV-treated cells that retained a normal cellular morphology (viewed under phase-contrast microscopy) and a rounded nucleus containing diffuse Hoechst-stained chromatin. This number was divided by the number of similarly appearing healthy cells counted in the corresponding untreated control, with the quotient converted to a percentage. Data are the mean ± SEM from 3 independent experiments. (B, C) PRC2 and WT7 cells were treated with etoposide or exposed to UV radiation as described above. Twenty-four hours later, the cells were stained with propidium iodide (PI) and digital images were captured from three fields of view in each dish. The percentage of propidium iodide stained cells was determined for each condition with the results representing the mean ± SEM from 3 independent experiments. Exposure to etoposide or UV resulted in a greater reduction in viable cell number and a larger number of dying cells in WT7 cells than in PRC2 cells (*, p<0.05 using ANOVA and Dunnett’s post-hoc test).
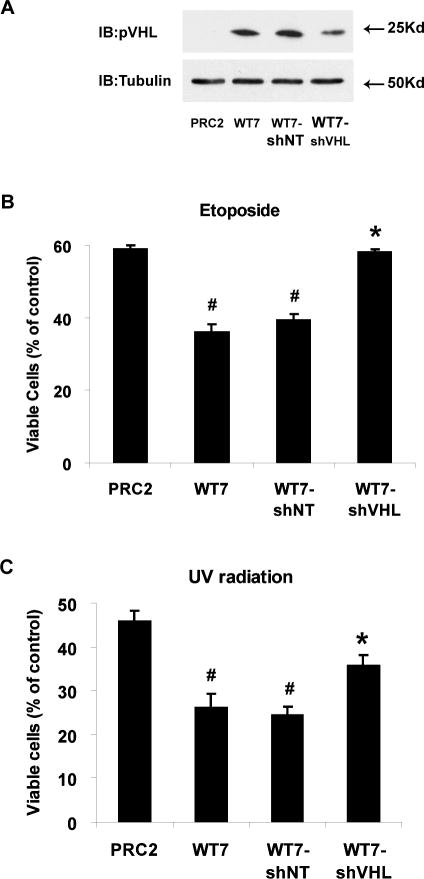
To determine if the increased death of WT7 cells is directly related to pVHL expression, we stably introduced a short hairpin RNA targeting VHL (shVHL) or a non-targeting short hairpin RNA (shNT) into WT7 cells. pVHL levels in WT7-shVHL cells were approximately 50% the levels in WT7 and WT7-shNT cells (Figure 2A). Exposing the cells to etoposide and UV radiation revealed that WT7 and WT7-shNT cells were equally sensitive to both treatments (Figure 2B, C). In contrast, WT7-shVHL cells were more resistant to etoposide and UV radiation. In the case of etoposide, the number of viable WT7-shVHL cells at 24 h was nearly the same as PRC2 cells. Thus the increased sensitivity of WT7 cells to apoptotic stimuli depends on maintaining a threshold of pVHL expression.
Figure 2. The enhanced sensitivity of WT7 cells to death-inducing stimuli can be reversed by inhibiting pVHL expression.
(A) pVHL levels in whole cell lysates from PRC2 cells, WT7 cells, and WT7 cells stably expressing shRNA targeting VHL (WT7-shVHL) or a non-targeting control (WT7-shNT) were analyzed by immunoblotting. Tubulin was immumoblotted as a control for protein loading. Similar results were obtained in 2 additional experiments (e.g., see figure 3C). (B) PRC2, WT7, WT7-shNT and WT7-shVHL cells were treated with etoposide (100 μM) or left untreated for 24 h. After fixing the cells and staining with Hoechst dye, the percentage of viable cells was determined as described in figure 1A and in Materials and methods. (C) RCC cells were exposed to UV radiation (1000 J/m2) or left untreated and 24 h later the percentage of viable cells was determined. Results in B and C represent the mean ± SEM from 3 independent experiments (#, p<0.05 compared with PRC2 cells; *, p<0.05 compared with WT7 and WT7-shNT cells).
BIMEL protein but not mRNA levels are increased in RCC cells expressing pVHL
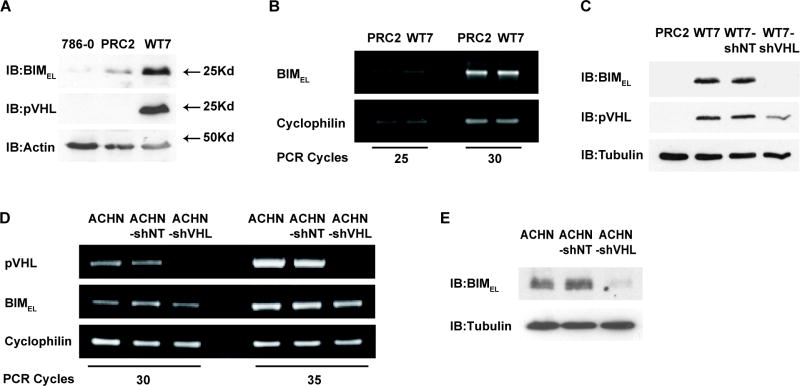
The intrinsic resistance of certain RCC cells to apoptosis was recently associated with reduced expression of BIMEL (Zantl et al., 2007). However, it is not known whether this is related to the status of the VHL gene. To determine if the different sensitivities of PRC2 and WT7 cells to etoposide and UV radiation might reflect a difference in BIMEL expression, we examined BIMEL protein levels in these cells and in VHL-deficient 786-0 cells. As expected, pVHL was detected in WT7 cells but not in PRC2 or 786-0 cells. BIMEL protein was also readily detectable in WT7 cells, but it was at much lower levels in PRC2 and 786-0 cells (Figure 3A). In contrast, BIMEL mRNA expression was not different suggesting that increased transcription or mRNA stability was not responsible for the higher levels of BIMEL protein (Figure 3B). To determine if the increased BIMEL protein in WT7 cells is a consequence of pVHL, we compared BIMEL levels in WT7-shNT and WT7-shVHL cells. BIMEL protein was at similar levels in WT7 and WT7-shNT cells but at much lower levels in WT7-shVHL cells and PRC2 cells (Figure 3C). Therefore, suppressing pVHL expression in WT7 cells leads to a marked decrease in BIMEL protein.
Figure 3. BIMEL protein levels in RCC cells correlates with expression of pVHL.
(A) Whole cell lysates from 786-0, PRC2, and WT7 cells were analyzed by immunoblotting with antibodies against BIM, pVHL and actin. Results shown are representative of blots from 4 independent experiments. (B) BIMEL mRNA expression in PRC2 and WT7 cells was analyzed by RT-PCR, starting with equal amounts of total RNA from each cell type. PCR products were compared after 25 and 30 cycles to avoid potential artifacts due to saturation of the PCR signal at high cycle number. Cyclophilin expression was analyzed to assess any variation in the amount of cDNA in each reaction. (C) Whole cell lysates from PRC2, WT7, WT7-shNT and WT7-shVHL cells were analyzed by immunoblotting for BIM, pVHL and tubulin. Results shown are representative of three independent experiments in which pVHL levels in WT7-shVHL cells are reduced an average of 50% (data not shown). (D) BIMEL and VHL expression in ACHN, ACHN-shNT, and ACHN-shVHL was analyzed by RT-PCR over 30 and 35 cycles of amplification. Cyclophilin expression was analyzed to assess potential variations in the amount of cDNA. (E) BIMEL protein levels in ACHN, ACHN-shNT, and ACHN-shVHL were analyzed by immunoblotting. Tubulin was immunoblotted as a control for protein loading. Results shown in B, D, and E are representative of results from 3 independent experiments.
To determine if inhibiting endogenous pVHL would cause a reduction in BIMEL protein, we stably introduced shNT and shVHL into ACHN cells, a renal adenocarcinoma cell line that retains the wild type VHL allele and produces wild type pVHL protein (Iliopoulos et al., 1995). Analysis of VHL mRNA levels by RT-PCR confirmed efficient knockdown in ACHN-shVHL cells but not in ACHN-shNT cells (Figure 3D). As in WT7 cells, the knockdown of VHL did not appear to affect BIMEL mRNA expression. However, ACHN-shVHL cells expressed much lower levels of BIMEL protein compared to either ACHN-shNT or parental ACHN cells (Figure 3E). Taken together, these data reveal that the absence or partial inhibition of pVHL expression results in a reduction in BIMEL protein but not mRNA expression.
BIMEL half-life is prolonged in RCC cells expressing pVHL
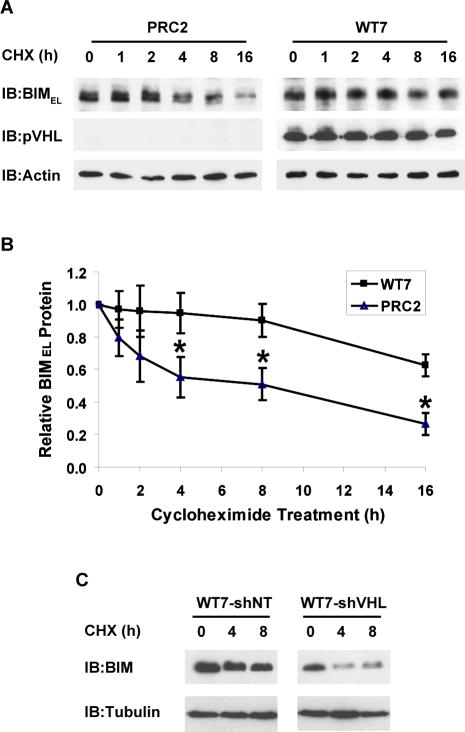
BIMEL protein levels are regulated in part by mechanisms that regulate its stability (Ley et al., 2005). To investigate whether pVHL affects BIMEL stability, we treated PRC2 and WT7 cells with a protein synthesis inhibitor for varying times and then analyzed BIMEL protein levels (Figure 4A). In PRC2 cells, BIMEL protein levels decreased 50% during the first 4-8 h after cycloheximide addition and another 50% by 16 h. In contrast, BIMEL protein levels were relatively constant in WT7 cells for the first 8 h after treatment and remained at nearly 70% of control levels at 16 h (Figure 4B). Results from 4 experiments revealed a half-life for BIMEL of ~5 h in PRC2 cells compared to >16 h in WT7 cells. pVHL levels in WT7 cells decreased only slightly over 16 h of cycloheximide treatment suggesting that it is relatively stable in these cells. BIMEL protein half-life was also markedly reduced in WT7-shVHL cells compared to WT7-shNT cells, confirming that its stability is closely linked to expression of pVHL (Figure 4C). These data strongly suggest that pVHL acts as a positive regulator of BIMEL protein stability.
Figure 4. BIMEL protein stability is enhanced in RCC cells expressing pVHL.
(A) PRC2 and WT7 cells were treated with cycloheximide (CHX, 10 μg/ml) for the indicated times. Whole cell lysates corresponding to equal numbers of cells were analyzed by immunoblotting with antibodies against BIM, pVHL and actin. (B) Cells were treated as described above and the abundance of BIMEL on the resulting immunoblots was determined relative to that in untreated controls after normalization to actin levels in the corresponding sample. Data represent mean ± SEM from 4 independent experiments (*, p<0.05 compared to corresponding PRC2 data point). (C) WT7 and WT7-shVHL cells were treated with cycloheximide for the indicated times and then analyzed for BIMEL and α-tubulin protein levels by immunoblotting. The more rapid decline in BIMEL expression after inhibiting protein synthesis in WH7-shVHL was confirmed in a second independent experiment.
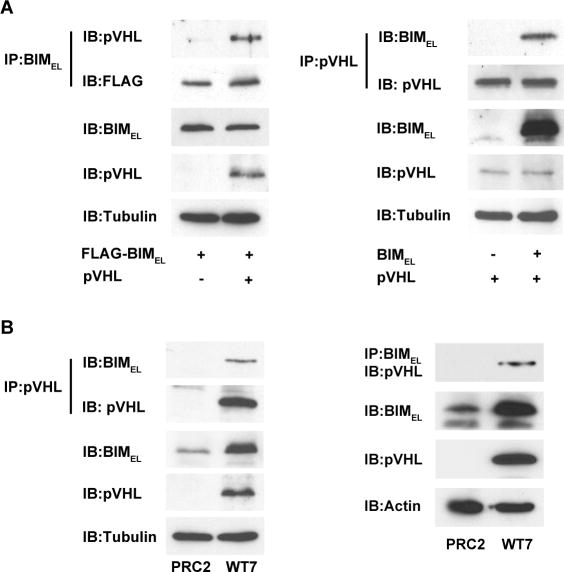
pVHL and BIMEL co-immunoprecipitate
To examine if pVHL can interact with BIMEL, we attempted co-immunoprecipitation experiments in COS-7 cells over-expressing pVHL and BIMEL. In these experiments, pVHL was detected in anti-BIM immunoprecipitates and BIMEL was detected in anti-pVHL immunoprecipitates, but only when both proteins were co-expressed (Figure 5A). We did similar co-immunoprecipitation experiments using WT7 and PRC2 cells. As observed previously (see figure 3), BIMEL protein levels were higher in WT7 cells than in PRC2 cells (Figure 5B). Furthermore, the endogenous BIMEL in WT7 cells co-immunoprecipitated with pVHL. From these experiments, we estimate that about 2-5% of the total BIMEL co-immunoprecipitated with pVHL. As expected, no BIMEL protein was detected in anti-pVHL immunoprecipitates from PRC2 cells. The specificity of the interaction was also demonstrated by the presence of pVHL in anti-Bim immunoprecipitates from WT7 but not PRC2 cells. These results, together with those described above, raise the possibility that an interaction between BIMEL and pVHL may contribute to the regulation of BIMEL stability.
Figure 5. Co-immunoprecipitation of pVHL and BIMEL.
(A) COS-7 cells were transiently transfected with 1 μg plasmid encoding FLAG-tagged BIMEL with or without 1 μg pVHL plasmid (left), or with 1 μg pVHL plasmid with or without 1 μg BIMEL plasmid (right). The next day, whole cell lysates were prepared and 500 μg of each were subjected to immunoprecipitation with anti-BIM polyclonal antibodies or anti-pVHL monoclonal antibody as indicated. The resulting immune complexes were split in half and analyzed by immunoblotting with anti-pVHL and anti-FLAG monoclonal antibodies (left) or anti-BIM and anti-pVHL polyclonal antibodies (right). A portion of each lysate (25 μg) taken prior to immunoprecipitation was blotted to determine total levels of BIMEL, pVHL and α-tubulin (to control for loading). The pVHL and BIMEL in the immunoprecipitates and in the whole cell lysates were analyzed on the same blots and for the same exposure times. (B) PRC2 and WT7 cell lysates (500 μg) were subjected to immunoprecipitation with anti-pVHL monoclonal antibody or anti-BIM polyclonal antibodies as indicated and the immunoprecipitated proteins were then immunoblotted with anti-BIM and anti-pVHL polyclonal antibodies (left) or with anti-pVHL monoclonal antibody (right). A portion (25ug) of each lysate was taken prior to immunoprecipitation to assess total levels of BIMEL, pVHL and either α-tubulin or actin. The pVHL and BIMEL in the immunoprecipitates and in the whole cell lysates were analyzed on the same blots and for the same exposure times. For both A and B, similar results were obtained in 3-4 independent experiments.
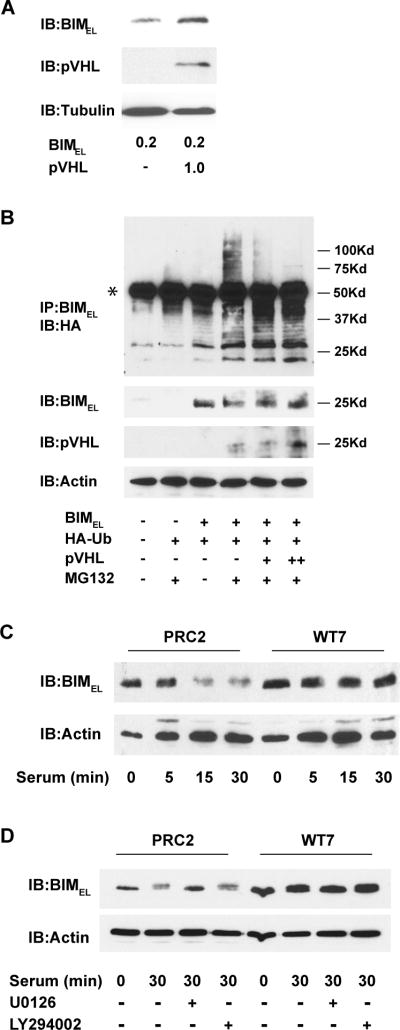
pVHL decreases BIMEL polyubiquitination and degradation
In response to serum or growth factor stimulation, BIMEL gets targeted for polyubiquitination and degradation by the proteasome (Ley et al., 2005). We wondered whether pVHL might prolong BIMEL half-life through inhibiting its polyubiquitination. Before addressing this question, we first determined whether pVHL could influence BIMEL steady-state levels in transfected COS-7 cells as was observed in RCC cells. When BIMEL was expressed at relatively low levels, co-expressing pVHL resulted in a corresponding increase in BIMEL protein (Figure 6A), suggesting that the presence of pVHL could stabilize BIMEL in this system. However, in subsequent experiments that included a proteasome inhibitor, we found we could only detect higher molecular weight polyubiquitinated forms of BIMEL if greater amounts of BIMEL plasmid were transfected (data not shown). Although co-expressing pVHL under these conditions no longer increased the net amount of BIMEL (for example, see figure 5A), we reasoned that if pVHL were to influence polyubiquitination of a subset of the BIMEL protein, it would still do so under these conditions.
Figure 6. Effects of pVHL on BIMEL polyubiquitination and its stability after serum-stimulation.
(A) COS-7 cells were transiently transfected with 0.2 μg of BIMEL plasmid with or without 1 μg of pVHL plasmid. The next day, whole cell lysates were prepared and immunoblotted for BIMEL, pVHL and α-tubulin. Note that co-expressing pVHL results in an increase in the levels of BIMEL under these conditions. (B) COS-7 cells were transfected with HA-ubiquitin (1 μg), BIMEL (1 μg) and either 1 or 3 μg (+ or ++, respectively) of pVHL expression plasmid. After 24 h, the cells were lysed and immunoprecipitated with anti-BIM polyclonal antibodies. Polyubiquitinated BIMEL was detected by immunoblotting the immune complexes with anti-HA monoclonal antibody (the asterisk identifies the IgG heavy chain from the anti-Bim antibody used for the immunoprecipitation). Where indicated, the proteasome inhibitor MG132 (50 μM) was added to the cells 6 h prior to lysis. A portion of each cell lysate taken prior to immunoprecipitation was analyzed by immunoblotting for BIMEL, pVHL and actin. This experiment has been repeated twice and in each case the presence of pVHL resulted in a decrease in the amount of high molecular weight polyubiquitinated BIMEL. (C) PRC2 and WT7 cells were cultured in serum-free media overnight and then stimulated with fresh serum-containing media for 0, 5, 15, or 30 minutes. Whole cell lysates were prepared and subjected to immunoblotting with antibodies against BIM and actin. (D) Serum-starved PRC2 and WT7 cells were stimulated with fresh serum-containing media or left in serum-free media for 30 min in the presence or absence of the MEK inhibitor U0126 (50 μM) or phosphatidylinositol 3-kinase inhibitor LY294002 (50 μM). At the end of the treatment period, whole cell lysates were prepared and subjected to immunoblotting with antibodies against BIM and actin. Note the MEK-dependent decrease in BIMEL levels after serum stimulation in PRC2 cells and the absence of any decrease in similarly treated WT7 cells. Results shown in C and D are representative of those obtained in 3 independent experiments.
To determine if pVHL can affect BIMEL polyubiquitination, COS-7 cells were co-transfected with BIMEL and HA-tagged ubiquitin, with or without pVHL. Before lysis, some of the cells were treated with the proteasome inhibitor MG132. Lysates were immunoprecipitated with anti-BIM antibody and the precipitated proteins immunoblotted with anti-HA antibody to detect ubiquitinated forms of BIMEL. Addition of the proteasome inhibitor to cells transfected with BIMEL and HA-ubiquitin resulted in the appearance of a high molecular mass smear not seen in the absence of MG132 or in cells lacking ectopic BIMEL (Figure 6B), consistent with the accumulation of polyubiquitinated BIMEL. Introducing exogenous pVHL resulted in a concentration dependent decrease in polyubiquitinated BIMEL, suggesting that pVHL may stabilize BIMEL by inhibiting its ubiquitination and subsequent degradation. While these results indicate that the polyubiquitination of BIMEL can be suppressed in transfected COS-7 cells by pVHL, it remains to be determined whether pVHL causes a similar decrease in the polyubiquitination of BIMEL in RCC cells.
We next examined if pVHL could inhibit pathways that lead to degradation of BIMEL in response to serum stimulation. In serum starved cells, BIMEL protein levels increase in part due to an increase in its stability. When the same cells are stimulated with serum, BIMEL is rapidly targeted for degradation by the proteasome (Ley et al., 2003). When PRC2 and WT7 cells were starved of serum overnight, BIMEL levels increased in both cell types (data not shown). Compared to our earlier experiments involving cells maintained in the presence of serum (see figure 3A and C), the levels of BIMEL in serum-starved WT7 cells were only slightly higher than those in PRC2 cells (Figure 6C and D, compare lanes 1 and 5). Presumably, this is because pVHL has little effect on BIMEL in the absence of signals that target BIMEL for polyubiquitination. When serum-starved PRC2 cells were stimulated with fresh serum, BIMEL protein levels decreased markedly within 15 min (Figure 6C, lanes 1 and 3). In contrast, BIMEL levels in WT7 cells remained unchanged even after 30 min of serum stimulation (Figure 6C, lanes 5 and 8), suggesting that the presence of pVHL may inhibit BIMEL degradation under these conditions. Growth factor stimulation activates extracellular signal-regulated kinases 1 and 2 (ERK1/2) and Akt protein kinase, both of which have been shown to phosphorylate BIMEL and stimulate its degradation by the proteasome (Ley et al., 2003; Qi et al., 2006). In PRC2 cells, however, the serum-induced decrease in BIMEL appears to be mediated by the MAP kinase kinase (MEK)/ERK pathway and not the phosphatidylinositol 3-kinase (PI3K)/Akt pathway since treatment with the MEK inhibitor U0126 and not the PI3K inhibitor LY294002 prevented the decrease in BIMEL (Figure 6D). Together these results suggest that the presence of pVHL can reduce BIMEL polyubiquitination and proteasomal degradation induced by ERK1/2 activation in response to growth factor stimulation.
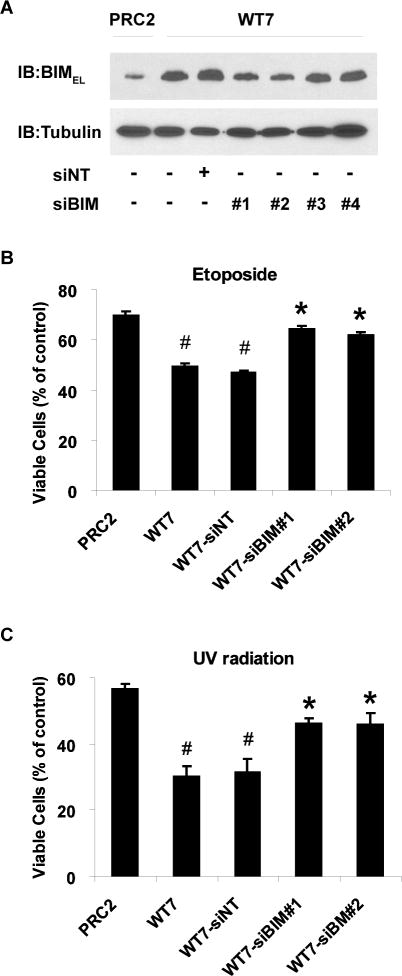
The increased sensitivity of RCC cells expressing pVHL to apoptotic stimuli is reversed by inhibiting BIMEL expression
The results described above raise the possibility that the different sensitivities of PRC2 and WT7 cells to apoptotic stimuli are related to their differential expression of BIMEL. To test this hypothesis, WT7 cells were transfected with 4 different siRNA duplexes targeting BIMEL or a non-targeting siRNA control. siBIM duplexes 1 and 2 reduced the protein level of BIMEL in WT7 cells by ~50%, closer to although still greater than the levels detected in PRC2 cells (Figure 7A). Control cells and cells transfected with siBIM or non-targeting siRNA were then treated with etoposide or subjected to UV radiation. As observed previously, WT7 cells were more sensitive to etoposide and UV radiation induced death than PRC2 cells. Inhibiting BIMEL expression with either siBIM-1 or siBIM-2 significantly reduced the sensitivity of WT7 cells to etoposide (Figure 7B) or UV radiation (Figure 7C). We conclude that the increased sensitivity of pVHL-expressing RCC cells to apoptotic stimuli is in part due to pVHL-mediated stabilization of BIMEL protein in these cells.
Figure 7. Reduced sensitivity of pVHL-expressing RCC cells to apoptotic stimuli after siRNA-mediated knockdown of BIMEL.
(A) WT7 cells were transfected with each of 4 distinct siRNA duplexes targeting BIMEL (siBIM) or with a control non-targeting siRNA duplex (siNT). Two days later the cells were lysed and the lysates immunoblotted with antibodies against BIM and tubulin. An equal amount of whole cell lysate from untransfected PRC2 cells was immunoblotted for comparison. Virtually identical results were obtained when the experiment was repeated. (B, C) WT7 cells were transfected with siBIM duplexes #1 and #2 or with siNT and 48 h later, transfected cells and untransfected controls were treated with etoposide (50 μM), exposed to UV radiation (1000 J/m2), or left untreated. PRC2 cells were treated in parallel for comparison. Viability was assessed 24 h after addition of etoposide or exposure to UV and is shown relative to untreated control cells as described in figure 1A. Data represent the mean ± SEM from 3 independent experiments (#, p<0.05 compared with PRC2 cells; *, p<0.05 compared with WT7 and WT7-siNT cells).
Discussion
The results described above establish a link between pVHL and BIMEL and the vulnerability of RCC cells to certain apoptotic stimuli. Initially, we observed that BIMEL protein levels were greater in RCC cells expressing wild type pVHL than in the same cells lacking pVHL. Inhibiting pVHL expression with RNA interference produced a corresponding decrease in BIMEL protein, while exogenous pVHL inhibited the accumulation of polyubiquitinated BIMEL in cells treated with a proteasome inhibitor. Consistent with these results, the presence of pVHL correlated with an increase in BIMEL protein half-life, which was reversed when pVHL expression was inhibited. Finally, siRNA-mediated suppression of BIMEL in RCC cells expressing pVHL reduced the death of these cells after exposure to etoposide or UV radiation.
The finding that loss of pVHL results in destabilization of the pro-apoptotic protein BIMEL provides a mechanistic explanation for a recent report that BIMEL expression is frequently suppressed in clear-cell RCC clinical samples and cell lines (Zantl et al., 2007). In their study, BIMEL expression was reduced in 35 of 43 clear-cell RCC samples and in 6 of 9 RCC cell lines. The projected incidence of reduced BIMEL expression in RCC is similar to estimates that 60–75% of sporadic clear-cell RCC harbor mutations in VHL (Cohen, 1999; van Houwelingen et al., 2005). Zantl et al. (2007) also described an inverse correlation between BIMEL levels and resistance of RCC cells to apoptosis. Our results demonstrating a role for pVHL in regulating BIMEL stability establish a potential cause and effect relationship between mutations in VHL and a reduction in BIMEL in RCC. It will be important to investigate whether reduced BIMEL expression is a more general characteristic of clear-cell RCC associated with defective VHL.
Regulation of BIMEL is complex and involves transcriptional and post-translational mechanisms. Data presented here show that pVHL can influence BIMEL protein levels without affecting its mRNA expression. While the precise mechanism is unknown, the ability of pVHL and BIMEL to co-immunoprecipitate raises the possibility that polyubiquitination of BIMEL may be influenced by its interaction with pVHL. Similar mechanisms were proposed to explain the stabilization of p53 and Jade-1 by pVHL (Roe et al., 2006; Zhou et al., 2002). Alternatively, pVHL could promote BIMEL stabilization by interfering with the function of a ubiquitin ligase that normally degrades BIMEL. Recently, an E3 ligase consisting of CIS (cytokine-inducible Src homology 2 domain-containing protein), elongins B and C, and Cul2 was shown to mediate the degradation of BIMEL in ovarian and breast cancer cells (Zhang et al., 2008). Elongins B and C and Cul2 are also part of the pVHL E3 ligase (Iwai et al., 1999; Stebbins et al., 1999). Thus, the abundance of pVHL relative to CIS may be important for determining the ubiquitin ligase activity of the latter and thus the stability of substrates such as BIMEL. Interestingly, the substrate-binding β-domain of pVHL and not its elongin C-binding α-domain co-immunoprecipitates with BIMEL (data not shown), raising the possibility that complexes containing pVHL and BIMEL could also contain elongin C.
In response to a variety of apoptotic stimuli, BIMEL and related BH3-only proteins act by sequestering anti-apoptotic Bcl-2 proteins at the mitochondria, thus enabling the pro-apoptotic proteins Bax and Bak to form complexes that promote release of cytochrome c into the cytoplasm and activation of the apoptosome (Willis et al., 2007). In the absence of death inducing stimuli, survival-promoting growth factors and cytokines stimulate phosphorylation of serine-65 in BIMEL by ERK1/2 leading to its polyubiquitination and degradation (Ewings et al., 2007). Our results raise the possibility that pVHL may act by inhibiting ERK-dependent phosphorylation of BIMEL or the interaction between ERK-phosphorylated BIMEL and its ubiquitin ligase. While these hypotheses remain to be tested, co-immunoprecipitation experiments with truncated forms of BIMEL reveal that pVHL can interact with residues 41-97 of BIMEL, which include the critical ERK phosphorylation site at serine-65 (data not shown).
The ability of pVHL to promote BIMEL stabilization is not the only means by which pVHL sensitizes cells to apoptosis. pVHL was recently shown to promote the stabilization and transcription factor activity of p53 (Roe et al., 2006), which correlated with the increased susceptibility of pVHL-expressing tumor cells to certain apoptotic stimuli. In another study, over-expression of pVHL in 786-0 cells with a recombinant adenovirus caused cell death which was associated with an increase in Bax expression (Kim et al., 2004). In addition to enhancing death pathways, pVHL may also suppress cell survival pathways including those mediated by NF-κB (Qi and Ohh, 2003; Yang et al., 2007), atypical PKC (Okuda et al., 2001), JunB (Lee et al., 2005) and HIF (Semenza, 2003). Despite these and other reports that pVHL can sensitize RCC and other tumor cells to apoptosis, expression of pVHL in RCC cells can also decrease sensitivity to cell death under certain conditions (Devarajan et al., 2001; Schoenfeld et al., 2000; Gorospe et al., 1999). Given the multiple functions of pVHL in the regulation of protein stability, microtubule stability, extracellular matrix signaling, and transcriptional regulation (Frew and Krek, 2008), it is perhaps not surprising that pVHL influences cell survival in diverse ways and through multiple mechanisms.
In summary, we have identified a mechanism by which loss of pVHL contributes to the desensitization of RCC cells to apoptotic stimuli that involves destabilization of the pro-apoptotic protein BIMEL. The high incidence of mutations in VHL in inherited and sporadic clear-cell RCC, together with the recent finding that BIMEL expression is suppressed in a similarly high percentage of clear-cell RCC, suggest that pVHL-dependent regulation of BIMEL stability may be an important factor in the development of many kidney tumors.
Materials and methods
Materials and plasmids
Cell culture reagents were from Invitrogen (Carlsbad, CA). Etoposide, propidium iodide, and cycloheximide were from Sigma-Aldrich (St. Louis, MO). MG132 and LY294002 were from Biomol Research Laboratories (Plymouth Meeting, PA) and U0126 was from Promega (Madison, WI).
Plasmids expressing shRNA targeting human VHL and a non-targeting (scrambled sequence) control shRNA were obtained from Thomas Benzing (University of Cologne, Koeln, Germany) and are described in Table 1 and Figure S3a of Schermer et al. (2006). The plasmid expressing HA-ubiquitin was a gift from Patricia Hinkle (University of Rochester, Rochester, NY). The full-length BIMEL coding region was amplified from rat cDNA using Pfu DNA polymerase (Stratagene, La Jolla, CA). The complete VHL open reading frame was amplified from human smooth muscle cell cDNA. BIMEL and VHL cDNAs were subcloned into pcDNA3 (Invitrogen) and validated by automated DNA sequencing.
Cell culture
RCC 786-0, PRC2 and WT7 cells were obtained from William Kaelin (Dana Farber Cancer Institute, Boston, MA) and are described elsewhere (Lonergan et al., 1998). ACHN cells (CRL-1611) were from American Type Culture Collection (Rockville, MD). All cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. The media for PRC2 and WT7 cells was supplemented with 10 μg/ml G418.
For irradiation, cells plated on 35 mm culture dishes in complete medium (>90% confluency) were exposed to UV (254 nm) at a dose of 1000 J/m2. After treatment, cells were returned to the incubator for 24 h before assessing viability. Transient transfections were done with LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s instructions. For experiments involving transfection of BIMEL plasmid, the caspase inhibitor boc-aspartyl(OMe)-fluoromethylketone (100 μM) (MP Biomedicals, Solon, OH) was added to the media in all wells to minimize cell death. For serum stimulation, fresh media containing 10% fetal bovine serum was added to cells previously cultured in the absence of serum for 12-14 h.
Viability assays
Equal numbers of cells were seeded into 35 mm dishes 24 h before initiating treatment. Twenty-four hours after addition of etoposide or irradiation with UV, cells were rinsed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. Cell nuclei were stained with Hoechst 33 342 (1 μg/ml in PBS, Invitrogen) and visualized under phase-contrast and fluorescence microscopy. For each treatment, three random fields of view encompassing a minimum of 200 cells were scored based on nuclear morphology as healthy (round nucleus containing diffusely stained chromatin) or dead/dying (pyknotic or misshapen nucleus with condensed chromatin). The number of healthy cells for a given treatment was normalized to the number of healthy cells in untreated controls. In some experiments, cells were stained with propidium iodide (1.5 μM in PBS) and then visualized under fluorescence microscopy. Cells that had lost plasma membrane integrity were able to take up propidium iodide and were presumed to be dead or dying. For a given treatment, cells in three fields of view were analyzed with the fraction of dead cells defined as the number of propidium iodide stained cells divided by the total number of cells. All viability experiments were repeated at least three times.
RNA interference
WT7 and ACHN cells were transfected with plasmids expressing shRNA targeting human VHL or a non-targeting scrambled control shRNA (plasmids described above). Stable transfectants were isolated by including 10 μg/ml blasticidin in the culture media. Drug-resistant colonies were pooled and expression of pVHL was assessed by immunoblotting.
siRNA oligonucleotides targeting human BIM and a non-targeting control siRNA were obtained from Thermo Fisher Scientific/Dharmacon (Waltham, MA). siRNAs (100nM) were transfected into WT7 cells with DharmaFECT according to the manufacturer’s instructions. Four different siRNAs targeting BIMEL were tested. siBIM 1 and 2 were most effective at decreasing BIMEL protein expression and thus were used in subsequent experiments.
Immunoblotting and immunoprecipitation
Cells were harvested in lysis buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA and a cocktail of protease inhibitors (Sigma-Aldrich). Cell lysates were clarified by centrifugation for 5 min and the protein concentration of the supernatants was determined using a modified Bradford assay (Bio-Rad, Hercules, CA). For immunoblotting, 25 μg of protein were separated by SDS-PAGE, transferred to nitrocellulose, and incubated as described previously (Xie et al., 2005) with the following primary antibodies and dilutions: anti-BIM, 1:1000 (cat. #AAP-330, Assay Designs, Ann Arbor, MI); anti-VHL monoclonal, 1:500 (cat. #556347, BD Biosciences, San Jose, CA); anti-VHL polyclonal, 1:1000 (cat. #2738, Cell Signaling, Danvers, MA); anti-actin, 1:1000 (cat. #A2066, Sigma-Aldrich); anti-HA, 1:1000 (cat. #1583816, Roche Diagnostics, Indianapolis, IN); anti-FLAG M2, 1:1000 (cat. #F3165, Sigma-Aldrich); anti-α-tubulin, 1:1000 (cat. #T5168, Sigma-Aldrich). Goat anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidase were used at a 1:10,000 dilution and immunoreactive bands were detected by chemiluminescence (SuperSignal, Thermo Fisher Scientific/Pierce, Rockford, IL). Densitometry was performed with NIH ImageJ software.
For immunoprecipitation experiments, 500 μg of clarified cell lysate were pre-absorbed with 50 μl of protein A Sepharose (GE Healthcare, Piscataway, NJ) for 1 h at 4°C. Lysates were then incubated overnight at 4°C with 1 μg of polyclonal anti-BIM antibody or monoclonal anti-pVHL antibody. Immune complexes were precipitated with 50 μl of Protein A Sepharose for 1 h and then washed three times with lysis buffer. Immunoprecipitated proteins were eluted in Laemmli sample buffer, separated by SDS-PAGE, and immunoblotted with the indicated antibodies as outlined above.
Reverse transcription—polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using an RNeasy Mini Kit (Qiagen, Valencia, CA). Reverse transcription reactions and PCR were performed as described previously (Lipscomb et al., 1999). PCR amplifications were done over a range of cycle numbers to ensure that the analyses were carried out under non-saturating conditions. PCR products were separated by electrophoresis on 1.0% agarose gels and analyzed after staining with ethidium bromide.
Statistical analysis
Quantitative data are reported as the mean ± standard error of the mean (SEM) from at least three independent experiments. The means for various treatment groups were compared using analysis of variance (ANOVA) and Dunnett’s post-hoc test.
Acknowledgments
We thank Dr. William Kaelin for providing cell lines and Drs. Thomas Benzing and Patricia Hinkle for providing plasmids used in this study. This work was supported by grants NS034400 and NS058868 from the National Institutes of Health.
References
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Cohen HT. Advances in the molecular basis of renal neoplasia. Curr Opin Nephrol Hypertens. 1999;8:325–331. doi: 10.1097/00041552-199905000-00008. [DOI] [PubMed] [Google Scholar]
- Devarajan P, De Leon M, Talasazan F, Schoenfeld AR, Davidowitz EJ, Burk RD. The von Hippel-Lindau gene product inhibits renal cell apoptosis via Bcl-2-dependent pathways. J Biol Chem. 2001;276:40599–40605. doi: 10.1074/jbc.M103424200. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Ewings KE, Wiggins CM, Cook SJ. Bim and the pro-survival Bcl-2 proteins: opposites attract, ERK repels. Cell Cycle. 2007;6:2236–2240. doi: 10.4161/cc.6.18.4728. [DOI] [PubMed] [Google Scholar]
- Frew IJ, Krek W. pVHL: a multipurpose adaptor protein. Sci Signal. 2008;1:pe30. doi: 10.1126/scisignal.124pe30. [DOI] [PubMed] [Google Scholar]
- Fu J, Menzies K, Freeman RS, Taubman MB. EGLN3 prolyl hydroxylase regulates skeletal muscle differentiation and myogenin protein stability. J Biol Chem. 2007;282:12410–12418. doi: 10.1074/jbc.M608748200. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Gorospe M, Egan JM, Zbar B, Lerman M, Geil L, Kuzmin I, et al. Protective function of von Hippel-Lindau protein against impaired protein processing in renal carcinoma cells. Mol Cell Biol. 1999;19:1289–1300. doi: 10.1128/mcb.19.2.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci U S A. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- Kim M, Yan Y, Lee K, Sgagias M, Cowan KH. Ectopic expression of von Hippel-Lindau tumor suppressor induces apoptosis in 786-O renal cell carcinoma cells and regresses tumor growth of 786-O cells in nude mouse. Biochem Biophys Res Commun. 2004;320:945–950. doi: 10.1016/j.bbrc.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Kurban G, Duplan E, Ramlal N, Hudon V, Sado Y, Ninomiya Y, et al. Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV alpha 2. Oncogene. 2008;27:1004–1012. doi: 10.1038/sj.onc.1210709. [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- Lipscomb EA, Sarmiere PD, Crowder RJ, Freeman RS. Expression of the SM-20 gene promotes death in nerve growth factor-dependent sympathetic neurons. J Neurochem. 1999;73:429–432. doi: 10.1046/j.1471-4159.1999.0730429.x. [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, et al. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Nyhan MJ, O’Sullivan GC, McKenna SL. Role of the VHL (von Hippel-Lindau) gene in renal cancer: a multifunctional tumour suppressor. Biochem Soc Trans. 2008;36:472–478. doi: 10.1042/BST0360472. [DOI] [PubMed] [Google Scholar]
- O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO Journal. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H, Saitoh K, Hirai S, Iwai K, Takaki Y, Baba M, et al. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J Biol Chem. 2001;276:43611–43617. doi: 10.1074/jbc.M107880200. [DOI] [PubMed] [Google Scholar]
- Qi H, Ohh M. The von Hippel-Lindau tumor suppressor protein sensitizes renal cell carcinoma cells to tumor necrosis factor-induced cytotoxicity by suppressing the nuclear factor-kappaB-dependent antiapoptotic pathway. Cancer Res. 2003;63:7076–7080. [PubMed] [Google Scholar]
- Qi XJ, Wildey GM, Howe PH. Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J Biol Chem. 2006;281:813–823. doi: 10.1074/jbc.M505546200. [DOI] [PubMed] [Google Scholar]
- Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Schermer B, Ghenoiu C, Bartram M, Muller RU, Kotsis F, Hohne M, et al. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J Cell Biol. 2006;175:547–554. doi: 10.1083/jcb.200605092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld AR, Parris T, Eisenberger A, Davidowitz EJ, De Leon M, Talasazan F, et al. The von Hippel-Lindau tumor suppressor gene protects cells from UV-mediated apoptosis. Oncogene. 2000;19:5851–5857. doi: 10.1038/sj.onc.1203985. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- van Houwelingen KP, van Dijk BA, Hulsbergen-van de Kaa CA, Schouten LJ, Gorissen HJ, Schalken JA, et al. Prevalence of von Hippel-Lindau gene mutations in sporadic renal cell carcinoma: results from The Netherlands cohort study. BMC Cancer. 2005;5:57. doi: 10.1186/1471-2407-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Xie L, Johnson RS, Freeman RS. Inhibition of NGF deprivation-induced death by low oxygen involves suppression of BIM(EL) and activation of HIF-1. J Cell Biol. 2005;168:911–920. doi: 10.1083/jcb.200407079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda A, Abi-Rached B, Petrylak D. Chemotherapy for advanced renal-cell carcinoma: 1983-1993. Semin Oncol. 1995;22:42–60. [PubMed] [Google Scholar]
- Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, et al. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26:7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- Zhang W, Cheng GZ, Gong J, Hermanto U, Zong CS, Chan J, et al. RACK1 and CIS Mediate the Degradation of BimEL in Cancer Cells. J Biol Chem. 2008;283:16416–16426. doi: 10.1074/jbc.M802360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MI, Wang H, Ross JJ, Kuzmin I, Xu C, Cohen HT. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem. 2002;277:39887–39898. doi: 10.1074/jbc.M205040200. [DOI] [PubMed] [Google Scholar]