Abstract
QT interval duration reflecting myocardial repolarization on the electrocardiogram is a heritable risk factor for sudden cardiac death and drug-induced arrhythmias. We conducted a meta-analysis of 3 genome-wide association studies in 13,685 individuals of European ancestry from the Framingham Heart Study, the Rotterdam Study and the Cardiovascular Health Study. We observed associations at P < 5×10−8 with variants in NOS1AP, KCNQ1, KCNE1, KCNH2 and SCN5A, known to be involved in myocardial repolarization and Mendelian Long QT Syndromes. Associations at five novel loci included 16q21 near NDRG4 and GINS3, 6q22 near PLN, 1p36 near RNF207, 16p13 near LITAF and 17q12 near LIG3 and RIFFL. Collectively, the 14 independent variants at these 10 loci explain 5.4–6.5% of variation in QT interval. Identifying the causal variants and defining their impact on myocardial repolarization may add incrementally to the prevention of sudden cardiac death and drug-induced arrhythmias.
Sudden cardiac death (SCD) and drug-induced ventricular arrhythmia, a major barrier to drug development, are poorly predicted.1 Prolongation of electrocardiographic QT interval duration, a measure of myocardial repolarization time, is a risk factor for drug-induced arrhythmias and SCD. Continuous QT interval duration is heritable2 (h2 ≈0.35) and has multiple environmental and genetic contributors. Its genetic determinants in populations are poorly characterized.3 Congenital Long and Short QT Syndromes of ventricular arrhythmias and SCD due to extremes of QT interval duration are often caused by mutations with large effect sizes, commonly in ion channels involved in myocardial repolarization. These mutations are typically private to specific families and individually explain little of the population variation in QT interval duration or SCD risk.4 The few common variants in candidate genes associated with QT interval duration thus far reported5–9 leave much of its heritability unexplained. Genome-wide association studies covering the majority of common variation in the human genome can be used in large samples to identify genetic variants that typically confer modest effect sizes for quantitative complex traits such as QT interval duration. We completed a meta-analysis of three genome-wide association studies of QT interval duration in 13,685 self-identified white individuals of European ancestry drawn from three prospective cohort studies: the Framingham Heart Study (FHS, n = 7,650), the Rotterdam Study (RS, n = 4,606) and the first and second rounds of genotyping in the Cardiovascular Health Study (CHS, n = 1,429).10 We used genotypes from the Affymetrix 500K chip and 50K gene-centered MIP, the Illumina 550K and Illumina 370CNV arrays, respectively, to impute genotypes for 2,543,686 autosomal SNPs with reference to HapMap CEU linkage disequilibrium patterns.
Results
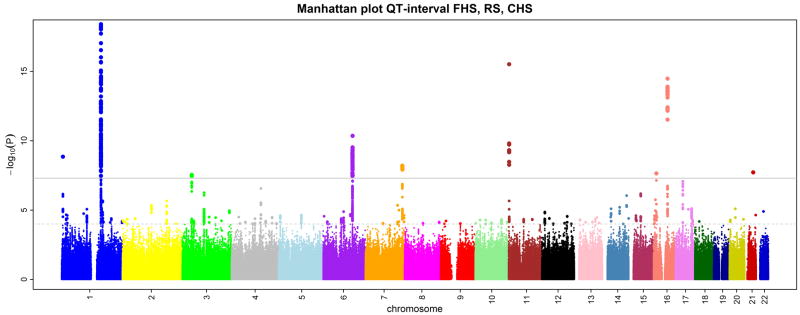
The mean ages of the individuals in the FHS, RS and CHS cohorts were 40, 69, and 73 years, respectively. Additional clinical characteristics are shown in Table 1. QT interval measures were adjusted for age and RR-interval (inverse heart rate) using cohort- and sex-specific linear regression, and the standardized residuals served as the phenotype for the association analysis. We started with a set of SNPs passing study-specific quality control filters: in FHS, 378,163 SNPs from the Affymetrix 500K chip + 50K gene-centered MIP; in RS, 512,349 SNPs from the Illumina 550K array; and in CHS, 332,946 SNPs from the Illumina 370CNV array. We imputed genotypes with reference to phased chromosomes from HapMap CEU (see Methods).11 After quality control filtering, we used genotypes from up to 2,543,686 SNPs to test for association in cohort-specific analyses. Genomic control was used to adjust for test-statistic inflation,12 which was minimal, with λgc ranging from 1.02 to 1.04. Cohort-specific quantile-quantile plots of p-value distributions are shown in Supplementary Figure 1. Using inverse variance weights, we combined genomic-controlled association results under an additive model from the three cohorts in fixed effects meta-analysis (overall λgc = 1.012). Nine loci were independently associated at a genome-wide P < 5×10−8 (Table 2, Figure 1, Figure 2). An additional locus was borderline significant (P = 8×10−8), but was externally validated. Five of the ten associated loci are related to the myocardial repolarization genes previously known to be associated with QT interval duration in the general population or in Mendelian conditions: NOS1AP, KCNQ1, KCNH2, KCNE1 and SCN5A. We observed an excess number of associations compared with the expectation under the null. For a nominal P < 10−5 we observed 568 associations compared with 25 expected under the null hypothesis (P ≪ 10−4, Supplementary Table 1). This finding suggests that among the many false positives at less stringent statistical thresholds additional truly associated variants may exist.
Table 1.
Clinical characteristics by cohort and by sex.
| Framingham Heart Study | Rotterdam Study | Cardiovascular Health | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| men n = 3,440 | women n = 4,210 | men n = 1,854 | women n = 2,752 | men n = 802 | women n = 627 | |
| Age (years) | 40 (10) | 40 (11) | 68 (8) | 69 (9) | 73 (6) | 73 (5) |
| Body mass index | 27 (4) | 25 (5) | 26 (3) | 27 (4) | 26 (3) | 26 (5) |
| Hypertension | 23% | 14% | 34% | 41% | 54% | 60% |
| Diabetes | 2.3% | 1.6% | 10% | 10% | 16% | 11% |
| Raw QT (msec) | 390 (35) | 391 (38) | 398 (29) | 399 (29) | 417 (35) | 413 (31) |
| Heart rate (beats/minute) | 67 (13) | 72 (14) | 68 (12) | 71 (12) | 66 (11) | 69 (10) |
| RR interval (msec) | 943 (167) | 880 (164) | 904 (155) | 865 (140) | 979 (167) | 928 (140) |
| QTc (msec)* | 404 (22) | 419 (22) | 421 (23) | 431 (22) | 423 (19) | 431 (22) |
| Standard deviation of QT residuals (msec)† | 20.6 | 20.8 | 18.3 | 17.9 | 17.3 | 17.3 |
Bazett’s correction for heart rate: QTc = QT/sqrt(RR interval).
Residuals are from sex-specific linear regression models adjusting for age and RR-interval.
Table 2.
SNPs with evidence for independent association at 10 loci with P < 5×10−8. A SNP at the LIG3 locus met our significance threshold in an interim analysis and was confirmed in the QTSCD consortium study. Chromosomal positions and coded alleles are given relative to forward strand of NCBI build 36. Effect sizes are shown as beta estimates from linear regression models for increasing copy of the coded allele and are on the standard deviation scale (1 SD ≈ 17.5 msec). A beta estimate of 0.08 SD is equivalent to a change in QT interval of 1.4 msec and an effect of 0.48 SD is equivalent to an 8.4 msec change. The effective sample size reflects the power relative to the total sample size of 13,685 with imputed data resulting from variation in imputation quality (see Methods). Selected genes from each locus are shown for reference. Results using the same coded allele from the QTSCD study (reported separately) and meta-analysis of the QTGEN and QTSCD study using inverse variance weighting are shown (n ≤ 29,539). Chr = chromosome. SE = standard error.
| QTGEN | QTSCD | Meta-analysis QTGEN + QTSCD |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||||
| SNP | Chr | Function/gene | Other genes within 500kb at novel loci |
Coded allele |
Allele frequency |
Effective sample size |
Beta estimate |
SE | P-value | Beta estimate |
SE | P-value | Beta estimate |
SE | P-value |
| rs12143842 | 1q | upstream NOS1AP | T | 0.26 | 13,241 | 0.21 | 0.02 | 8×10−46 | 0.16 | 0.01 | 5×10−36 | 0.18 | 0.01 | 2×10−78 | |
| rs12029454 | 1 | intron NOS1AP | A | 0.15 | 12,172 | 0.21 | 0.02 | 6×10−28 | 0.15 | 0.02 | 3×10−20 | 0.17 | 0.01 | 3×10−45 | |
| rs16857031 | 1 | intron NOS1AP | G | 0.14 | 13,154 | 0.19 | 0.02 | 3×10−23 | 0.12 | 0.02 | 1×10−14 | 0.15 | 0.01 | 1×10−34 | |
| rs2074238 | 11p | intron KCNQ1 | T | 0.06 | 2,888 | −0.47 | 0.06 | 3×10−16 | −0.33 | 0.14 | 0.02 | −0.45 | 0.05 | 3×10−17 | |
| rs37062 | 16q | intron CNOT1 | GINS3, NDRG4, SLC38A7, GOT2 | G | 0.24 | 13,440 | −0.12 | 0.02 | 3×10−15 | −0.09 | 0.01 | 5×10−12 | −0.10 | 0.01 | 3×10−25 |
| rs11756438 | 6q | intron c6orf204 | SLC35F1, PLN, ASF1A | A | 0.47 | 12,707 | 0.09 | 0.01 | 4×10−11 | 0.08 | 0.01 | 2×10−12 | 0.08 | 0.01 | 5×10−22 |
| rs12576239 | 11p | intron KCNQ1 | T | 0.13 | 13,211 | 0.12 | 0.02 | 2×10−10 | 0.08 | 0.02 | 3×10−7 | 0.10 | 0.01 | 1×10−15 | |
| rs846111 | 1p | 3′ UTR RNF207 | NPHP4, CHDS, ACOT7, PLEKHG5, KLH21 | C | 0.28 | 6,480 | 0.12 | 0.02 | 1×10−9 | 0.08 | 0.01 | 4×10−9 | 0.10 | 0.01 | 1×10−16 |
| rs4725982 | 7q | downstream KCNH2 | T | 0.22 | 13,706 | 0.09 | 0.02 | 6×10−9 | 0.08 | 0.01 | 1×10−8 | 0.09 | 0.01 | 5×10−16 | |
| rs8049607 | 16p | upstream LITAF | CLEC16A,SNN, ZC3H7A, TNFRSF17 | T | 0.49 | 10,543 | 0.08 | 0.01 | 2×10−8 | 0.07 | 0.01 | 4×10−8 | 0.07 | 0.01 | 5×10−15 |
| rs1805128 | 21q | missense KCNE1 | A | 0.010 | 7644 | 0.48 | 0.09 | 2×10−8 | −0.06 | 0.04 | 0.16 | 0.05 | 0.04 | 0.22 | |
| rs12053903 | 3p | intron SCN5A | C | 0.34 | 13,491 | −0.08 | 0.01 | 3×10−8 | −0.06 | 0.01 | 6×10−8 | −0.07 | 0.01 | 1×10−14 | |
| rs2074518 | 17q | intron LIG3 | RFFL | T | 0.46 | 13,488 | −0.07 | 0.01 | 8×10−8 | −0.05 | 0.01 | 7×10−6 | −0.06 | 0.01 | 6×10−12 |
| rs2968864 | 7q | downstream KCNH2 | C | 0.25 | 12,932 | −0.08 | 0.02 | 1×10−7 | −0.08 | 0.01 | 1×10−9 | −0.08 | 0.01 | 8×10−16 | |
Figure 1.
QT interval association results for 2,543,686 imputed SNPs in 13,685 individuals from 3 cohorts. Results are shown on the −log10(P) scale and are truncated at −log10(P) = 18 for display purposes. The solid bar corresponds to the genome-wide significance threshold of 5×10−8.
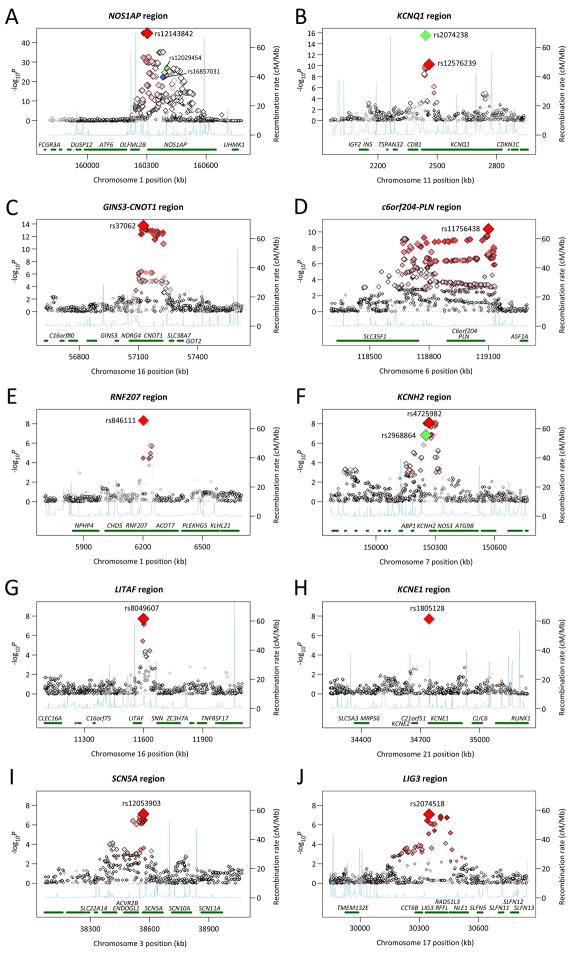
Figure 2.
Regional association plots for one megabase surrounding each associated locus. Statistical significance of associated SNPs at each locus are shown on the -log(P) scale as a function of chromosomal position (NCBI Build 36). The primary associated SNP at each locus is shown in red. The correlation of the primary SNP to other SNPs at the locus is shown on a scale from minimal (white) to maximal (bright red). The quality of imputation as assessed by the observed/expected variance on allele dosage is represented by the darkness of the diamond outline ranging from maximal (black) to minimal (light gray). Estimated recombination rates from HapMap and RefSeq annotations are shown. The loci shown include: (a) 1q23.3 including NOS1AP with three independent associations, rs12143842 in red, rs12029454 in green and rs16857031 in blue, (b) 11p15.5 including KCNQ1 with two independent associations, rs2074238 in green and rs12576329 in red, (c) 16q21 including GINS3, NDRG4 and CNOT1, (d) 6q22.31 including c6orf204 and PLN, (e) 1p36.31 including RNF207, (f) 7q36.1 including KCNH2 with two independent associations, rs4725982 in red and rs2968864 in green, (g) 16p13.3 including LITAF, (h) 21q22.12 including KCNE1, (i) 3p22.2 including SCN5A and (j) 17q12 including LIG3 and RFFL.
We had the opportunity to compare our top results with the QTSCD consortium, which included 15,854 individuals of European ancestry.13 All associations but one were confirmed at 2-sided P < 0.05 (Table 2).
Genes known to be involved in myocardial repolarization
At the NOS1AP locus, we observed the strongest association in the genome for rs12143842, 6kb 5′ of NOS1AP, with 0.21 SD QT increase per minor allele copy (minor allele frequency, MAF = 0.26, P = 8×10−46, Table 2, Figure 2a). All results are shown on the standard deviation scale (1 SD ≈ 17.5 msec). Two additional independent signals were observed at rs12029454 (MAF 0.15) and rs16857031 (MAF 0.14) in intron 2 and intron 1, respectively, all with r2 to each other <0.05 in HapMap and with p<0.05 when entered into a single regression model in FHS and RS (CHS with a smaller sample is underpowered, Supplementary Table 2). We have previously reported association in FHS and RS samples of rs10494366 at NOS1AP (P = 5×10−30 in the current report) and this association has been widely replicated.5,14–17 This SNP is not significant in models adjusting for the 3 SNPs identified in the current study (P > 0.05) and it shows some degree of correlation to each of the 3 SNPs (to rs12043842 r2 = 0.46–0.47 in FHS and RS and r2 = 0.11 in HapMap CEU; to rs12029454 r2 = 0.17 in RS and HapMap CEU; to rs16857031 r2 = 0.17 in HapMap CEU).18 We conclude that there are three independent signals at the locus and that rs10494366 captures the association signal from at least one of these 3 SNPs.
We identified two common variants in intron 1 of KCNQ1 that were associated with QT interval duration (Table 2, Figure 2b). Rare mutations in KCNQ1, a potassium channel involved in myocardial repolarization, have been associated with Long QT Syndrome type 1 and Short QT Syndrome type 2.4 In this meta-analysis, SNP rs2074238 (MAF = 0.06) was associated with 0.47 SD shorter QT interval for each minor allele (P = 3×10−16) and rs12576239 (MAF = 0.13) was associated with 0.12 SD longer QT interval for each minor allele (P = 2×10−10). The two SNPs were independently associated with QT in models that included both SNPs (P = 6×10−5, P = 1×10−4, respectively in FHS and P = 3×10−10, P = 0.03 in RS, Supplementary Table 2). Coupled with the low correlation of the two SNPs (HapMap CEU r2=0.009, FHS r2=0.014, RS r2=0.011) these findings support two independent association signals at the locus. Pfeufer et al. previously reported association with QT interval of rs757092 (MAF=0.38) which lies 3kb away from rs12576239 in intron 1 and to which it is partially correlated (r2 = 0.31 HapMap CEU) and 14kb away from rs2074238 to which it is not correlated (r2 = 0.005 HapMap CEU).7 We did not find supportive evidence of association of rs757092, which was well imputed, with QT interval in QTGEN (P = 0.11).
The common SNP rs4725982, 3′ of KCNH2, was associated with QT interval (+0.09 SD/minor allele, MAF=0.22, P = 6×10−9, Table 2, Figure 2f). A second SNP at KCNH2, rs2968864 was associated with shorter QT interval duration for increasing minor allele count, but did not reach our pre-specified genome-wide significance threshold for unselected genetic variants in the QTGEN samples (−0.08 SD/minor allele, MAF=0.25, P = 1×10−7, Table 2, Figure 2f). Rare mutations in KCNH2, a potassium channel involved in myocardial repolarization and drug-induced arrhythmias, are known to underlie congenital Long QT Syndrome type 2 and Short QT Syndrome type 1.4 The two SNPs were significant or nearly so when entered into a single regression model (P = 4×10−3, P = 3×10−4, respectively in FHS and P = 4×10−3, P = 0.16, respectively in RS, Supplementary Table 2). Coupled with the low correlation between the SNPs in HapMap CEU (r2=0.09), the two SNPs thus appear to represent independent signals of association. The missense variant rs1805123 (K897T) has been associated with QT interval in most studies, including our own,6–9,19 and is perfectly correlated with rs2968864 (r2=1.0 in FHS, data not shown), which is thus not a novel finding.8 An intronic SNP has been previously reported by Pfeufer et al. to be associated with QT interval (rs3815459), is poorly correlated with the currently reported rs4725982 or rs2968864/rs1805123 variants (r2 = 0.08, r2 = 0.08, respectively in KORA, personal communication, Arne Pfeufer) and could not be imputed because it is not represented in HapMap. Another variant previously reported by us (rs3807375)27 has limited correlation with rs2968864 (r2=0.21 HapMap CEU) and rs4725982 (r2 = 0.39 HapMap CEU) and is not significant in models containing rs2968864 and rs4725982, suggesting that it does not represent an independent signal of association.
SNP rs1805128 was associated with QT interval duration (+0.48 SD/minor allele, MAF = 0.01, P = 2×10−8, Table 2, Figure 2h). This SNP encodes a change from aspartate to asparagine at amino acid 85 (D85N) in KCNE1, a potassium channel involved in myocardial repolarization in which rare mutations result in Long QT Syndrome type 5.4 D85N is poorly covered by the fixed genotyping arrays used here, but was included on the supplemental Affymetrix 50K array used in FHS, for which results are presented. Association of rs1805128 has been reported by Gouas et al. with extremes of QT interval duration in 398 individuals from the DESIR cohort (P = 0.02),9 and by us in 4,487 CHS participants (P = 0.003),20 and more recently in 5043 individuals from the Health2000 study (P = 4×10−11).21 Importantly, the D85N variant has also been related to drug-induced arrhythmia22,23 and Long QT Syndrome.24
In the published literature, no common variants in SCN5A have been convincingly associated with QT interval duration in European-derived individuals. We observed an association of rs12053903 in intron 27 of SCN5A with QT interval (−0.08 SD/minor allele, MAF = 0.34, P = 3×10−8, Table 2, Figure 2i). Rare mutations in SCN5A, the cardiac sodium channel, result in Long QT Syndrome type 3.25 A common missense variant in SCN5A, S1102Y, is associated with QT prolongation, ventricular arrhythmias and sudden cardiac death in African Americans,25,26 but is nearly monomorphic (MAF <0.01) in individuals of European ancestry.
The finding of 9 associated common variants in 5 genes known to influence myocardial repolarization and cardiac arrhythmias at the top of our list of results, 8 achieving a stringent genome-wide significance threshold in our meta-analysis of three independent cohorts, gave us confidence in the validity of the five novel loci that exceeded this threshold.
QT interval associations with novel loci
The first novel locus on chromosome 16q21 near NDRG4, SETD6, CNOT1, SLC38A7 and GINS3 included SNP rs37062 (MAF = 0.24) which falls in intron 40 of CNOT1, a regulator of RNA transcription, and was associated with 0.12 SD reduced QT per minor allele (P = 3×10−15, Table 2, Figure 2c). None of the nearby genes is known to modulate myocardial repolarization in humans, although recent experiments in zebrafish suggest potential candidates at the locus. Milan et al tested zebrafish mutants generated by Amsterdam et al. in an insertional mutagenesis screen27 for altered response to challenge with dofetilide, a QT-interval prolonging medication used in humans that prolongs cardiomyocyte action potential duration in humans and zebrafish. They found that a mutant with an insertion in intron1 of GINS complex subunit 3 (GINS3) was resistant to the QT-prolonging effects of exposure to dofetilide (personal communication, David Milan). The GINS complex is involved in the establishment of DNA replication forks. In humans, GINS3 falls near the 16q21 interval associated with QT interval in our meta-analysis (127kb from rs37062). In addition, a recent report on NDRG4 (N-myc downstream-regulated gene family member 4, 56kb from rs37062) in zebrafish observed expression restricted to the central nervous system and the heart starting at 24 hours post-fertilization.28 Morpholino knockdown of NDRG4 was associated with hypoplastic hearts with pericardial edema, dilated atria, looping defects and slower heart rates compared to controls. Zebrafish respond to exposure to QT-prolonging drugs with heart rate slowing,29 although this may be a non-specific finding in the NDRG4 morphants. While we cannot exclude a source of the association in the many other genes at this locus, GINS3 and NDRG4 are promising candidates for further work.
The second novel locus is on chromosome 6q22.31. SNP rs11756438 (MAF = 0.47) lies in an intron of a predicted gene of unknown function c6orf204 and near SLC35F1 and PLN and was associated with 0.09 SD higher QT interval per minor allele (P = 4×10−11, Table 2, Figure 2d). Interestingly, PLN (122kb away from this SNP) encodes phospholamban, an inhibitor of cardiac sarcoplasmic reticulum Ca++-ATPase (SERCA2a). Phospholamban knockout mice show enhanced myocardial contractility in response to beta adrenergic agonists.30 Cardiomyocyte dysregulation of Ca++ handling due to increased phospholamban activity has been linked to dilated cardiomyopathy and heart failure in mouse models31 and in a human family with cardiomyopathy and ventricular tachycardia.32 While more work will be required to localize the source of the signal of association reported here, it is interesting to note that NOS1AP activates neuronal nitric oxide synthase 1,5 a regulator of calcium cycling in the sarcoplasmic reticulum, and that rare variants in CACNA1C, a subunit of the L-type voltage-dependent calcium channel, cause congenital Long QT Syndrome.33 These observations suggest a unifying hypothesis that genetic variation influencing calcium cycling in cardiac myocytes influences repolarization and, when altered, contributes to arrhythmogenesis.
The third novel locus was on chromosome 1p36.31 near several genes including CHD5, RPL22, RNF207, ICMT, HES3, GPR153, and ACOT7. The top SNP at the locus, rs846111 (MAF = 0.28), lies in the 3′ untranslated region of RNF207 and was associated with 0.12 SD higher QT interval for each copy of the minor allele (P = 1×10−9, Table 2, Figure 2e). RNF207, which encodes ring finger protein 207, is of unknown function, in a family of molecules involved generally in protein-protein interaction and ubiquitination.
The fourth novel locus was on chromosome 16p13.3 upstream of LITAF, encoding lipopolysaccharide induced tumor necrosis factor, which has no known relationship to myocardial repolarization but missense mutations in this gene have been related to Charcot-Marie-Tooth, a hereditary motor and sensory neuropathy.34 The top SNP at the locus, rs8049607 (MAF = 0.49), was associated with 0.08 SD higher QT interval for each minor allele copy (P = 2×10−8, Table 2, Figure 2g).
The fifth novel locus was on chromosome 17q12 near LIG3 and RFFL. The top SNP at the locus, rs2074518 (MAF = 0.46) in intron 11 of LIG3, was associated with 0.07 lower QT interval for each minor allele copy (P = 8×10−8, Table 2, Figure 2j). While the result did not achieve genome-wide significance in our data alone, replication was observed in the QTSCD consortium (P = 7×10−6, joint P = 6×10−12). LIG3 encodes DNA ligase III, is involved in DNA base-excision repair and is not an obvious candidate to modulate myocardial repolarization. The nearby gene RFFL encodes the rififylin protein and is involved in the endocytic recycling compartment.
Estimates of the coverage by the imputed SNPs of SNPs found in HapMap CEU at each of the novel loci are shown in Supplementary Table 3.
Technical validation of poorly imputed SNPs and secondary analysis
Because the imputation quality of individual SNPs varied due to variation in coverage of SNPs by fixed genotyping arrays, we directly genotyped 3 sentinel SNPs in the entire RS sample and in a subset of the FHS sample (total n 7,000), as well as three additional SNPs only in the FHS subset. For example, rs2074238 in intron 1 of KCNQ1 had an effective sample size (see Methods) in 6,975 individuals examined of only 2,359 when accounting for the relatively low imputation quality in Framingham (observed/expected variance = 0.10) and Rotterdam (0.45). The SNP was filtered out in the CHS analysis by the imputation QC thresholds. We compared the significance of imputed association results in the directly genotyped subsample to that of the direct genotyping results. Direct genotyping confirmed the association of rs2074238 with QT interval with a substantially stronger significance, consistent with the rise in effective sample size from 2,359 to 6,975 individuals (P = 1×10−13 imputed vs P = 6×10−23 directly genotyped, Table 3). The appropriate filters to be applied to imputed genotypes of varying quality are a matter of debate, but it was certainly encouraging that no association based on imputed results failed to be supported by directly genotyped SNP results (Table 3). Including poorly imputed variants in analyses may be valuable even though they have substantially reduced power to detect truly associated variants.
Table 3.
QT interval association results of directly genotyped SNPs compared to imputed SNPs. Shown are meta-analysis of genotype-phenotype association results using imputed and directly genotyped SNPs in 1) a subset of the Framingham sample and the entire Rotterdam Study (n ≤ 6,975) or 2) the Framingham subset only (n ≤ 2,566). Three SNPs were genotyped only in the Framingham Heart Study (and did not specifically have low imputation quality). Effects are shown on the standard deviation (SD) scale. For SNPs that were less well imputed (small effective sample size) the increase in significance tracks with the fall in standard error and rise in effective sample size. N_effective is the sample size (N) * (observed/expected variance) [see Methods].
| Imputed SNP genotype association | Directly genotyped SNP association | Sample | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| SNP | Beta (SD) | SE | P | N_effective | Beta (SD) | SE | P | N | |
| rs2074238 | −0.47 | 0.06 | 1×10−13 | 2,359 | −0.31 | 0.03 | 6×10−23 | 6,975 | FHS+RS |
| rs846111 | 0.09 | 0.03 | 4×10−4 | 3,709 | 0.10 | 0.02 | 2×10−7 | 6,825 | FHS+RS |
| rs8049607 | 0.09 | 0.02 | 1×10−6 | 5,964 | 0.08 | 0.02 | 4×10−6 | 6,921 | FHS+RS |
|
| |||||||||
| rs12576239 | 0.13 | 0.04 | 2×10−3 | 2,560 | 0.13 | 0.04 | 1×10−3 | 2,566 | FHS only |
| rs4725982 | 0.10 | 0.03 | 3×10−3 | 2,556 | 0.11 | 0.03 | 1×10−3 | 2,556 | FHS only |
| rs12053903 | −0.04 | 0.03 | 0.17 | 2,385 | −0.04 | 0.03 | 0.24 | 2,465 | FHS only |
Because of the strong effect of sex on QT interval variation, which explains approximately 5% of its variability, we tested for but observed no significant interaction of sex with the SNP-QT associations.
Discussion
Confirmation of association in an independent sample
Examination of results for 14 SNPs at 10 loci identified by the QTGEN consortium in the QTSCD consortium data, demonstrated strong confirmation of results for 12 of the 14 associations (Table 2).13 One SNP that only weakly replicated is the less common KCNQ1 SNP (rs2074238), which is poorly imputed from Affymetrix arrays used in QTSCD and is likely due to low power given the strong association in our data (P = 6×10−23 upon direct genotyping in 6,975 individuals). A SNP that did not replicate is the low frequency (MAF = 0.01) missense SNP rs1805128 (D85N) in KCNE1, which is poorly imputed from Affymetrix genotypes and is well replicated in external studies with direct genotyping.9,20,35 Both SNPs thus had limited power to be replicated due to genotype imprecision from poor imputation quality. Additionally, genetic variants reaching genome-wide significance in the QTSCD consortium were strongly confirmed in the QTGEN consortium, including rs10919071 at a locus containing ATP1B1 (QTGEN P = 4×10−5) and rs17779747 at a locus containing KCNJ2, a Long QT Syndrome gene (QTGEN P = 4×10−5).
Population impact of 14 variants associated in QTGEN
In summary, the QTGEN meta-analysis of three genome-wide association studies detected nine common variants at five known candidate genes and an additional five common variants at novel loci not previously recognized to modulate myocardial repolarization. In total, these variants explain a substantial proportion of variation in QT interval (Framingham 5.4%, Rotterdam 6.5%, CHS 2.3%, Supplementary Table 2). These variants in aggregate explain more of QT interval variation than any other covariate (excluding heart rate) including female sex, a known risk factor for QT prolongation and drug-induced arrhythmia.
To assess the potential clinical impact of the genetic variants examined here, we constructed a QT genotype score using the allele copy number and the effect estimates for the 14 SNPs from our meta-analysis and tested the score in the FHS and RS samples (the larger samples). The top quintile of QT genotype score was associated with 9.7 msec and 12.4 msec higher Bazett-corrected QTc (the heart rate correction used in clinical settings) compared to the bottom quintile in FHS and RS samples (P = 5×10−28, P = 1×10−31, respectively). A prolonged QTc ≥450 msec in men and ≥470 msec in women has previously been shown to be associated with 2.5-fold increased hazard of sudden cardiac death in the Rotterdam Study.1 The top quintile of QT genotype score was associated with odds ratios for prolonged QTc of 2.6 and 3.1 in FHS and RS (P = 3×10−5, P = 6×10−7, respectively, Supplementary Table 4). The finding that the top 20% of genotype score in the population has a QTc increase compared to the bottom 20% in excess of the QT-prolonging effect of some drugs causing arrhythmias (as little as 8 msec), and that one of the SNPs (D85N KCNE1) is associated with the congenital Long QT Syndrome24 and drug-induced arrhythmias,22,23 further supports the clinical relevance of the genetic variants identified in the current report. Tests of the hypothesis that these variants, individually or in aggregate, contribute to risk of sudden cardiac death or drug-induced arrhythmias will require additional work in well-powered samples.
Although genetic effects have been thought to be weaker at older ages, striking associations were identified among these populations of middle aged and older adults. This may be expected for common variants of modest effects that elude negative selection and modulate traits in which environmental factors play only a modest role.
Additional fine mapping with direct genotyping to refine the signal of association and to identify the specific genes involved will be required. As illustrated by the number of common variants in Long and Short QT Syndrome genes, the spectrum of allele frequencies and effect sizes for the variants at many genes ranges from rare variants of strong effect underlying Mendelian forms of disease to less common variants with intermediate effects to highly polymorphic variants with comparatively modest effects. Certainly, this study will have missed variants that have even more modest effects, that are poorly captured by fixed genotyping arrays including those in the minor allele frequency range from 0.5–5%, and that due to random sampling variation failed to rise to the top of our results but might in an equally sized independent sample. Resequencing of each gene will be needed to fully characterize the allelic architecture of QT variation in the general population as well as its relevance to the approximately 25% of LQTS families without recognized mutations in known genes and the great majority of those who die of sudden cardiac death in the general population without recognized genetic risks.
METHODS
Study samples
The QTGEN consortium includes European-derived samples from three cohorts. The Framingham Heart Study (FHS) is a community-based, longitudinal cohort study comprising three generations of individuals in multigenerational pedigrees and additional unrelated individuals. The current study included individuals from Generation 1 (11th examination), Generation 2 (1st examination) and Generation 3 (1st examination).36–38 The Rotterdam Study (RS) is a prospective population-based cohort study of chronic diseases begun in 1990.39,40 The current RS study sample included data from one of 4 examination cycles at which the first eligible electrocardiogram was available for each individual. The Cardiovascular Health Study (CHS) is a prospective, cohort study of risk factors for heart disease and stroke begun in 1989 and included 4,925 self-described white participants.41 The CHS study sample used in this analysis included self-identified whites from the first two rounds of genotyping in a nested case-cohort study of myocardial infarction. Data on QT interval and covariates came from the baseline examination, at which prevalent cardiovascular disease was an exclusion criterion in the parent case-cohort study. All studies were approved by local institutional review boards and written informed consent was given.
Individuals were excluded for bundle branch block or QRS duration >120msec, atrial fibrillation or flutter, pacemaker activity, or use of a QT-altering medication (not applied to FHS Generation 3). After exclusions there were 7,650 FHS, 4,606 RS and 1,429 CHS individuals with phenotype and genotype data who contributed to genotype-phenotype association analyses.
QT measurement methods
In FHS, paper electrocardiograms were scanned and digital caliper measurements were made using proprietary software. In the Rotterdam Study, digital measurements of the QT interval were made using the Modular ECG Analysis System (MEANS).42 In the Cardiovascular Health Study, the electrocardiograms were recorded on MAC PC-DT ECG recorder (Marquette Electronics Inc, Milwaukee, WI, USA) machines and measurements of QT interval made using the Marquette 12SL algorithm (see Supplementary Methods for full details).
Phenotype modeling
The overall strategy involved linear regression to adjust QT interval for effects of age, sex, and RR interval (inverse heart rate) and residuals were used in genotype-phenotype association testing. In FHS, cohort-, sex- and cardiac cycle-specific regression models were created to adjust for age and RR interval. Residuals from these regression models were standardized to mean 0, standard deviation (SD) 1 and then averaged across up to 4 cardiac cycles. The averaged residuals were then restandardized to mean 0, SD 1 in cohort- and sex-specific samples. These residuals were then used in genotype-phenotype association testing. In RS, sex-specific regression models were constructed to adjust for RR interval and age and generate residuals. In CHS, the adjustment for age, sex, RR interval and study site was performed in the genotype-phenotype association step (see below).
Genotyping
In FHS, genotyping was performed using the Affymetrix 500K GeneChip array, called using the BRLMM algorithm,43 and a custom-designed gene-centric 50K MIP. In RS, genotyping was performed using the Infinium II HumanHap550K Genotyping BeadChip version 3. In CHS, genotyping was performed using the Illumina 370CNV BeadChip system. Associations of poorly imputed SNPs were validated by re-genotyping FHS samples using Sequenom and RS samples using Taqman. See Supplementary Methods for cohort-specific genotyping details including filters.
Imputation
We imputed estimated allele dosage, defined as the expected number of copies of the minor allele (a fractional value between 0 and 2), of all autosomal SNPs using MACH44 (HapMap CEU release 22, build 36) in FHS and RS and using BIMBAM11 (release 21a, build 35) in CHS (see Supplementary Methods for details).
Genotype-phenotype association method
In FHS, standardized QT residuals were tested for association with imputed allele dosage under an additive genetic model using the linear mixed effects model of the kinship package in R to account for relatedness.45,46 In the Rotterdam Study, QT residuals were tested for association using MACH2QTL, which uses dosage value (0 2, continuous) as a predictor in a linear regression framework.44 In CHS, QT interval was linearly regressed on age, sex, clinic, RR-interval and SNP dosage using R.45 The regression was weighted to reflect case-cohort sampling probabilities.
Meta-analysis
The minor allele from HapMap CEU genotypes was used to define the coded allele in all analyses, regardless of frequency in individual cohorts. For an A/T SNP, the T allele is the “coded allele” under the following coding: AA=0, AT=1, TT=2. To implement genomic control, the lambda value was used to correct the standard error as follows, SE_corrected=SE*sqrt(lambda). Each effect estimate (beta) was standardized to the standard deviation of the cohort-specific adjusted residuals to put all results on the SD scale. The ratio of the observed to the expected variance of the imputed allele frequency was used as the quality metric for the imputation of a given SNP. To account for the difference in power, and thus the interpretation of resulting p-values for each SNP, we created a variable N_effective which discounted the total sample size by imputation quality as follows: N_effective= N*(observed/expected variance). We used inverse variance-weighted fixed effects meta-analysis of the beta estimates from linear regression as the primary meta-analysis method. Weighting by the square root of the N_effective resulted in very similar -log(p-value) for results with P<0.01 (r=0.9980). The scripts developed for this project are freely available at http://www.broad.mit.edu/~debakker/meta.html. We a priori declared results significant at α =5×10−8, based on estimates adjusting for all common variant tests in the human genome of European ancestry individuals for a target genome-wide P<0.05.47
Exchange with QTSCD consortium
We submitted to the QTSCD consortium a list of our top SNP associations (one SNP per signal of association) and received QTSCD results for those SNPs. A reciprocal exchange was performed. We performed meta-analysis of the QTGEN meta-analysis results (n=13,685) with the QTSCD results (n=15,854) using inverse variance weights.
QT genotype score
A QT genotype score was calculated using the effect estimates from the QTGEN meta-analysis for the coded allele of each of 14 SNPs. The score, on the standard deviation scale, was calculated as follows for each individual: QT genotype score= beta1*allele_copy_number1 + beta2*allele_copy_number2 + … beta14*allele_copy_number14
Supplementary Material
Acknowledgments
The Framingham Heart Study work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine (Contract No. N01-HC-25195), its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278), and the Doris Duke Charitable Foundation (C.N.-C.) and Burroughs Wellcome Fund (C.N.-C.), based on analyses by Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. The measurement of ECG intervals in Framingham Heart Study generation 1 and 2 samples was performed by eResearchTechnology and was supported by an unrestricted grant from Pfizer. The measurement of ECG intervals in the Framingham Heart Study generation 3 sample was completed by Alim Hirji and Sirisha Kovvali using AMPS software provided through an unrestricted academic license by AMPS, LLC (New York, NY, USA). The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO Groot, 175.010.2005.011). The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The authors acknowledge the essential role of the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium in development and support of this manuscript. CHARGE members include the Netherland’s Rotterdam Study, the NHLBI’s Atherosclerosis Risk in Communities (ARIC) Study, Cardiovascular Health Study (CHS) and Framingham Heart Study (FHS), and the NIA’s Iceland Age, Gene/Environment Susceptibility (AGES) Study. C.N.-C. is supported by NIH K23-HL-080025, a Doris Duke Charitable Foundation Clinical Scientist Development Award, and a Burroughs Wellcome Fund Career Award for Medical Scientists. M.E is funded by the Dutch Heart Foundation 2007B221. J.I.R. is supported by the Cedars-Sinai Board of Governors’ Chair in Medical Genetics. The authors wish to thank the following people: Gabriel Crawford and Candace Guiducci (Broad Institute of Harvard and Massachusetts Institute of Technology) who completed the Sequenom-based technical validation genotyping of the Framingham Heart Study samples; Pascal Arp and Mila Jhamai (Erasmus Medical Center) for Illumina array genotyping and Taqman-based technical validation genotyping of the Rotterdam Study samples; and Michael Moorhouse and Marijn Verkerk (Erasmus Medical Center) for database management in the Rotterdam Study. The QTGEN consortium would like to thank the QTSCD consortium for the opportunity to exchange top results pre-publication.
Footnotes
Detailed QTGEN investigator list by cohort/center (alphabetical)
Framingham Heart Study: Martin G. Larson, Christopher Newton-Cheh, Peter A. Noseworthy, Christopher J. O’Donnell, Xiaoyan Yin.
Rotterdam Study: Mark Eijgelsheim, Karol Estrada, Albert Hofman, Jan A. Kors, Fernando Rivadeneira, Bruno H.Ch. Stricker, Andre G. Uitterlinden, Jacqueline C.M. Witteman.
Cardiovascular Health Study: Joshua Bis, Susan R. Heckbert, Thomas Lumley, Kristin Marciante, Christopher Newton-Cheh, Bruce M. Psaty, Kenneth M. Rice, Jerome I. Rotter, Nicholas L. Smith, Nona Sotoodehnia.
Broad Institute of Harvard and Massachusetts Institute of Technology: Paul I.W. de Bakker, Christopher Newton-Cheh.
QTGEN author contributions (alphabetical)
Design of QTGEN study: P.I.W.dB., M.E., M.G.L., T.L., C.N.-C., C.J.O., B.M.P., K.M.R., B.H.Ch.S. Genotyping: Affymetrix, C.N.-C., F.R., J.I.R., A.U. Statistical Analysis and Informatics: J.B., P.I.W.dB., M.E., K.E., T.L., K.M., C.N.-C., K.M.R., F.R., A.U., X.Y. Drafting of manuscript: C.N.-C. Critical revision of manuscript: J.B., P.I.W.dB., M.E., K.E., S.R.H., A.H., J.A.K., P.A.N., B.M.P., K.M.R., J.I.R., N.L.S., N.S., B.H.Ch.S., J.C.M.W.
Conflicts
No potential conflicts of interest exist
References
- 1.Straus SM, et al. Prolonged QTc interval and risk of sudden cardiac death in a population of older adults. J Am Coll Cardiol. 2006;47:362–367. doi: 10.1016/j.jacc.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 2.Newton-Cheh C, et al. QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm. 2005;2:277–284. doi: 10.1016/j.hrthm.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Newton-Cheh C, Shah R. Genetic determinants of QT interval variation and sudden cardiac death. Curr Opin Genet Dev. 2007;17:213–221. doi: 10.1016/j.gde.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Splawski I, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 5.Arking DE, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 6.Bezzina CR, et al. A common polymorphism in KCNH2 (HERG) hastens cardiac repolarization. Cardiovasc Res. 2003;59:27–36. doi: 10.1016/s0008-6363(03)00342-0. [DOI] [PubMed] [Google Scholar]
- 7.Pfeufer A, et al. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ Res. 2005;96:693–701. doi: 10.1161/01.RES.0000161077.53751.e6. [DOI] [PubMed] [Google Scholar]
- 8.Newton-Cheh C, et al. Common Genetic Variation in KCNH2 Is Associated With QT Interval Duration. The Framingham Heart Study. Circulation. 2007 doi: 10.1161/CIRCULATIONAHA.107.710780. [DOI] [PubMed] [Google Scholar]
- 9.Gouas L, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. EurJHumGenet. 2005;13:1213–1222. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- 10.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from five cohorts. Circulation Cardiovascular Genetics. 2008 doi: 10.1161/CIRCGENETICS.108.829747. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoSGenet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 13.Pfeufer A, et al. Common variants in ten loci modulate QT interval duration in individuals of European ancestry: the QTSCD consortium. submitted Nature Genetics. 2008 doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aarnoudse AJ, et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 15.Lehtinen AB, et al. Association of NOS1APgenetic variants with QT interval duration in families from the Diabetes Heart Study. Diabetes. 2008 doi: 10.2337/db07-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Post W, et al. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum Hered. 2007;64:214–219. doi: 10.1159/000103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobin MD, et al. Gender and effects of a common genetic variant in the NOS1 regulator NOS1AP on cardiac repolarization in 3761 individuals from two independent populations. Int J Epidemiol. 2008;37:1132–41. doi: 10.1093/ije/dyn091. [DOI] [PubMed] [Google Scholar]
- 18.Eijgelsheim M, et al. Identification of a Common Variant at the NOS1AP Locus Strongly Associated to QT -Interval Duration. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn341. [DOI] [PubMed] [Google Scholar]
- 19.Pietila E, et al. Association between HERG K897T polymorphism and QT interval in middle-aged Finnish women. Journal of the American College of Cardiology. 2002;40:511–514. doi: 10.1016/s0735-1097(02)01979-4. [DOI] [PubMed] [Google Scholar]
- 20.Sotoodehnia N, et al. KCNE1 Gene D85N Variant, QT Interval, and Risk Of Mortality. Circulation. 2005;111:e231. [Google Scholar]
- 21.Marjamaa A, et al. Common candidate gene variants are associated with QT interval duration in the general population. J Intern Med. 2008 doi: 10.1111/j.1365-2796.2008.02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei J, et al. KCNE1 polymorphism confers risk of drug-induced long QT syndrome by altering kinetic properties of I-Ks potassium channels. Circulation. 1999;100:495–495. [Google Scholar]
- 23.Paulussen AD, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med. 2004;82:182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 24.Salisbury BA, et al. The single nucleotide polymorphism D85N-KCNE1 is associated with both congenital and drug-induced long QT. Heart Rhythm. 3:S98–S98. (206/5/1) [Google Scholar]
- 25.Splawski I, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 26.Burke A, et al. Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation. 2005;112:798–802. doi: 10.1161/CIRCULATIONAHA.104.482760. [DOI] [PubMed] [Google Scholar]
- 27.Amsterdam A, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu X, et al. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev Biol. 2008;317:486–496. doi: 10.1016/j.ydbio.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milan DJ, et al. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107:1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 30.Luo W, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 32.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Splawski I, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Street VA, et al. Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology. 2003;60:22–6. doi: 10.1212/wnl.60.1.22. [DOI] [PubMed] [Google Scholar]
- 35.Marjamaa A, et al. Common candidate gene variants are associated with QT interval duration in the general population. Journal of Internal Medicine. doi: 10.1111/j.1365-2796.2008.02026.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawber T, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Heart Study. Annals of the New York Academy of Science. 1963;107:539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 37.Kannel WB, Feinleib M, McNamara PM. An investigation of coronary heart disease in families: The Framingham Offspring Study. American Journal of Epidemiology. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 38.Splansky GL, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. American Journal of Epidemiology. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 39.Hofman A, et al. The Rotterdam Study: objectives and design update. Eur JEpidemiol. 2007;22:819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur JEpidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- 41.Fried LP, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 42.van Bemmel JH, Kors JA, van HG. Methodology of the modular ECG analysis system MEANS. Methods Inf Med. 1990;29:346–353. [PubMed] [Google Scholar]
- 43.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J HumGenet. 2006;S79:2290. [Google Scholar]
- 45.R Developmental Core Team. A language and environment for statistical computing. R Foundationfor Statistical Computing. 2007 [Google Scholar]
- 46.Therneau T. kinship: mixed effects Cox models, sparse matrices, and modeling data from large pedigrees. R package version 1.1.0-19 edn. 2008 [Google Scholar]
- 47.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of themultiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.