Abstract
Transcriptional activity of nuclear receptors (NRs) is influenced by a large number of coregulators that exert their actions predominantly at the transcription initiation step. Unlike most well-characterized NR coregulators, Cofactor of BRCA1 (COBRA1), a subunit of the negative elongation factor (NELF), binds to estrogen receptor α (ERα) and modulates estrogen-dependent transcription by impeding the movement of RNA polymerase II (RNAPII) during the transcription elongation stage. Here we show that, in addition to ERα, COBRA1 also displays various degrees of affinity for several other NRs. In particular, COBRA1 binds strongly to androgen receptor (AR) via its ligand-binding domain (LBD). Small hairpin RNA (shRNA)-mediated reduction of endogenous COBRA1 enhances androgen-mediated transcription. The effect of COBRA1 knockdown can be rescued by a silent mutant COBRA1 that is refractory to the shRNA action. Using a reporter assay for alternative splicing, we also provide evidence for a role of COBRA1 in influencing the exon skipping/inclusion of nascent transcripts produced from an androgen-dependent promoter. These findings suggest that COBRA1 may coordinate multiple steps in ligand-dependent gene expression, which in turn ensures both the quantity and quality of hormone-stimulated gene products.
Keywords: COBRA1, androgen receptor, transcriptional repression, alternative splicing, NELF
Introduction
Steroid hormone receptors are a group of DNA-binding transcription factors that activate gene expression in a ligand-dependent and/or –independent manner [1]. Transcription potency of these site-specific transcription activators are largely dictated by the availability of the cognate hormones, as well as an array of transcription coregulators that are recruited to individual transcription promoters by the steroid hormone receptors [2, 3]. The vast majority of the coregulators exert their impact on the rate of transcription initiation by inducing chromatin modifications and mediating communications between steroid hormone receptors and the basal transcription machinery [2, 3]. However, mounting evidence indicates that transcription elongation represents an additional regulatory step in eukaryotic gene expression [4-6]. Furthermore, it has been increasingly appreciated that transcriptional regulation is physically and functionally linked with other aspects of gene regulation including RNA processing [7]. For example, it has been shown that steroid hormones including estrogen and androgen, as well as a subset of NR coactivators can influence the alternative splicing pattern in a ligand-dependent manner [8-12]. Control of distinct aspects of gene expression by the same set of the coregulators may ensure expression of hormone-regulated genes with the appropriate quantity and quality in response to various hormonal cues.
Cofactor of BRCA1 (COBRA1) was identified as a BRCA1-interacting protein [13] and subsequently found to be one of the four subunits of the negative elongation factor complex NELF (NELF-B) [14]. The NELF complex was biochemically purified based on its ability to inhibit transcription elongation by stalling RNAPII within the promoter-proximal region [15, 16]. Subsequent work indicates that COBRA1 binds to ERα in breast cancer cells and is recruited to several estrogen receptor (ER) target promoters in a ligand-stimulated fashion [17]. Consistent with the in vitro biochemical studies, COBRA1 in breast cancer cells represses transcription by impeding the movement of RNA polymerase II (RNAPII) at a subset of estrogen-responsive genes [17]. Thus, the COBRA1 action during transcription elongation represents a mode of hormone-dependent gene regulation that is distinct from the actions of most known NR coregulators.
In the current study, we compared the affinity of COBRA1 for several additional steroid hormone receptors as well as its impact on their hormone-dependent transcriptional activity. In particular, we found that COBRA1 binds strongly to the ligand-binding domain (LBD) of AR and serves as potent modulator of androgen-dependent transcription. Interestingly, COBRA1 also influences the alternative splicing pattern of a reporter gene driven by an androgen-stimulated promoter. Thus, our work suggests a role of COBRA1 in coordinating multiple aspects of ligand-dependent gene expression, which may have significant biological consequences in cancer development.
Materials and Methods
Antibodies and Plasmids
The following antibodies used in this study were purchased from Santa Cruz Biotechnology: α-AR (C-19 and N-20), α-PRB (H-190), and α-GR (E-20). The α-COBRA1 monoclonal anbibody used for immunoblotting was described previously [17]. The following plasmids have been previously reported: the expression vectors for various AR fragments [18], the COBRA1 expression vector and retroviral-based shRNA expression vectors [13, 17], and the MMTV-CD44 minigene cassette [10]. The internal control reporter vector phRL-SV40 was purchased from Promega (E6261). PCR-directed mutagenesis was used to construct the COBRA1 silent mutant that is resistant to the knockdown effect of COBRA1-specific shRNA 2A (silent mutant sequence: GATCTCCTCGAAAAAAGTT). The expression vectors for various Flag-tagged COBRA1 fragments were constructed by cloning the corresponding PCR fragments into pcDNA3 (Invitrogen).
Cell lines
T47D and HEK293T cells were purchased from ATCC and cultured in Dulbecco's modified minimum essential medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 μg/mL penicillin, and 100 μg/mL streptomycin. The EGFP control and COBRA1 stable knockdown cell lines in the T47D were generated as described previously [17].
Coimmunoprecipitation
Approximately 2 × 106 of HEK293T cells were transfected with 1 μg nuclear receptor expression vectors and 1 μg control or Flag-COBRA1 expression vectors using Lipofectamine 2000 (Invitrogen). Cells were harvested 24 hours after transfection, washed twice with ice-cold PBS, and lysed in 0.5 mL of lysis buffer (20 mM Hepes, pH 8.0, 500 mM NaCl, 1% NP-40, 5 mM EDTA, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 μg/mL pepstatin, and 1 mM PMSF). Genomic DNA was sheared by passing the lysates through a 21G needle for five times. The lysates were then centrifuged at 16,000 g for 10 min at 4°C. 20 μL of the supernatant was saved as input control. The rest of the supernatant was pre-cleared with 20 μL of 50% slurry of protein-A agarose beads (Roche) for 1 hour at 4°C, and immunoprecipitated overnight with 20 μL of 50% slurry of the anti-Flag agarose beads (Sigma). The beads were washed four times with the lysis buffer, each lasting for at least 30 min. The immunoprecipitates were eluted in 15 μL of 2 × Laemmli buffer. Gel electrophoresis and subsequent immunoblotting were carried out according to standard procedures.
GST-pull down assay
GST-pull down assay was performed as previously described [17].
Luciferase Reporter Assay
Luciferase reporter assay was performed as previously described [17] with several minor modifications. Briefly, cells were starved for three days in phenol red-free DMEM supplemented with 5% charcoal/dextran-treated fetal calf serum. The starved cells were then seeded in Nunclon™ Δ 12-well plates (Fisher). T47D-derived cell lines were transfected with 250 ng of MMTV-luciferase or PSA-luciferase reporter plasmid, using the Lipofectamine Plus transfection reagent (Invitrogen) according to the manufacture's instructions. Vehicle or ligands were added to the cell culture 24 hours after transfection and cells were incubated for another 24 hours before harvesting for the luciferase assay. The following steroid hormones were used at the indicated final concentrations: dihydrotestosterone (DHT) 10 nM; synthetic progesterone promegestone (R5020) 10 nM; synthetic glucocorticoid dexamethasone (Dex) 100 nM; synthetic non-aromatizable androgen (R1881) 1 nM; and AR antagonist flutamide 10 μM. 50 ng of the Renilla luciferase reporter vector phRL-SV40 (Promega) was co-transfected as an internal control. Triplicate transfections were carried out for each condition, and the data shown were representative of at least three independent experiments.
Alternative splicing assay
Alternative splicing assay was performed as previously described [10] with minor modifications. The T47D-derived cells were transfected with 300 ng of splicing minigene plasmid as mentioned above. HEK293T cells were transfected with the splicing minigene plasmid plus 50 ng of the nuclear receptor expression vectors as indicated. Following incubation at 37°C for 24 hrs, the transfected cells were treated with vehicle or ligand for additional 24 hrs. Total RNA was isolated with the Trizol reagent (Invitrogen). 1 μg of DNase-treated RNA was used in one-step RT-PCR with the Access RT-PCR system (Promega), as previously described [10]. 25 cycles of PCR were performed with 32P-labeled CD44 sense/antisense primers. Radioactive RT-PCR products were resolved by 8% nondenaturing polyacrylamide gel electrophoresis. Data were quantitated with a phosphoimager from Molecular Dynamics and Imagequant software. The average values from duplicated transfections were presented.
Results
Interactions between COBRA1 and multiple steroid hormone receptors
COBRA1 was previously identified in our laboratory as a corepressor of estrogen receptor α (ERα). Given the structural and sequence homology between ERα and other members of the steroid hormone receptor subfamily, we sought to examine potential physical and functional interactions between COBRA1 and androgen receptor (AR), progesterone receptor B (PRB), and glucocorticoid receptor (GR). In a co-immunoprecipitation (co-IP) assay as shown in Fig. 1A, both AR and GR were readily co-immunoprecipitated with Flag-COBRA1, whereas the association between PRB and COBRA1 was very weak but consistently detectable. When compared at the equivalent intensity of ectopically expressed nuclear receptors (lane 4, Fig.1A) and Flag-COBRA1 (Fig. 1B), a significantly larger proportion of AR was co-immunoprecipitated with COBRA1 than the other two nuclear receptors (lane 2 in Fig. 1A), suggesting a higher affinity of COBRA1 for AR than PRB and GR.
FIGURE 1. Interaction between COBRA1 and multiple steroid hormone receptors.
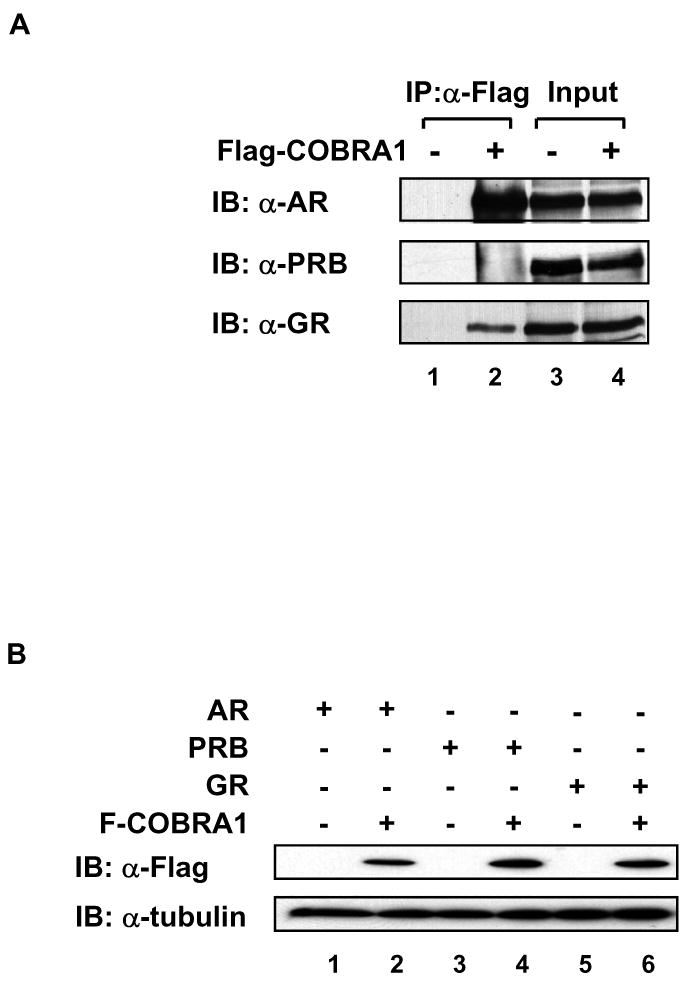
A. Empty or Flag-COBRA1-expressing vector was co-transfected with AR, PRB, or GR expression vectors into HEK293T cells. 24 hours after transfection, whole cell lysate was prepared and immunoprecipitated with an α-Flag antibody. Immunoprecipitates were resolved by SDS-PAGE and blotted with α-AR, α-PR, or α-GR antibodies. 2% of the whole cell lysate was loaded as input control. B. The protein levels of Flag-COBRA1 in the whole cell lysate were determined by immunoblotting with the α-Flag antibody. α-Tubulin was used as a loading control.
COBRA1 interacts with AR via the ligand-binding domain (LBD) of AR
In light of the strong association between COBRA1 and AR, we sought to further characterize the protein-protein interaction. First, the COBRA1-AR interaction was verified in a GST-pulldown assay, where in vitro translated, 35S-labeled AR was specifically pulled down by recombinant GST-COBRA1 protein (Fig. 2A). Second, we carried out a domain-mapping study of AR using the co-IP assay. As summarized in Fig. 2B, the ligand-binding domain (LBD) of AR bound to Flag-COBRA1 with a similar affinity as the full-length AR protein. In contrast, neither the amino terminus of AR (AF-1) nor the DNA binding domain and hinge region (D, H) exhibited any appreciable affinity for COBRA1. Thus, as has previously been shown with ERα [17], AR interacted with COBRA1 primarily via its LBD.
FIGURE 2. COBRA1 binds to the ligand-binding domain of AR.
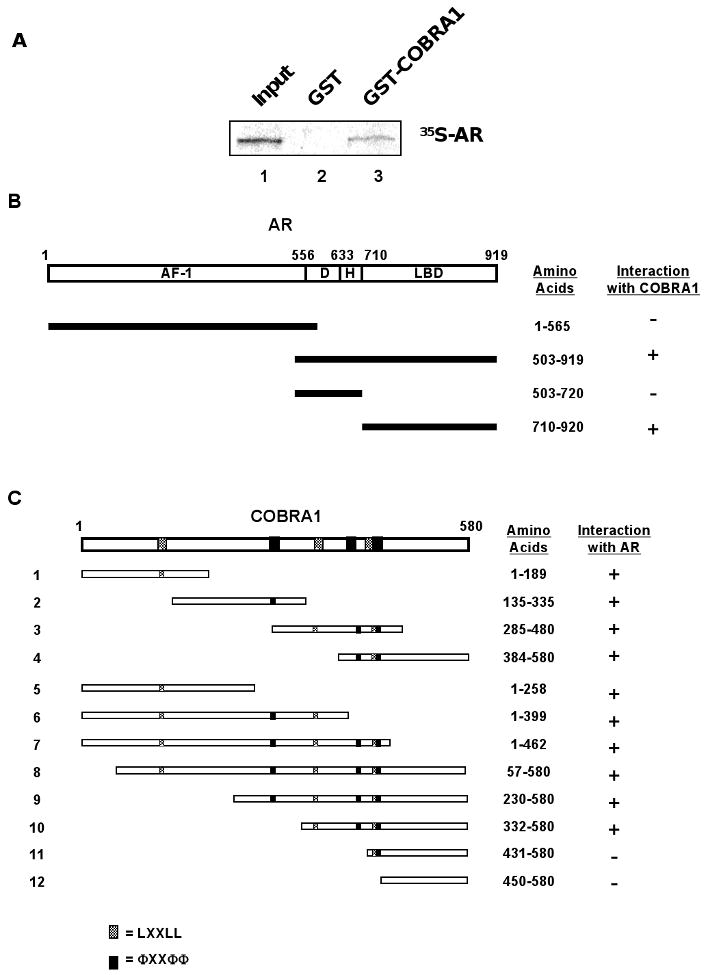
A. GST-pulldown assay was performed to detect the interaction between in vitro translated 35S-labeled AR and bacterially expressed GST-COBRA1. 1/15 of the in vitro translated AR used for GST-pull down was loaded as an input control. B. Full-length COBRA1 protein and various AR fragments were co-expressed in HEK293T cells, and the binding affinity was compared by co-immunoprecipitation. The diagram summarizes the interaction of COBRA1 with various AR fragments. AF-1: activation function 1; D: DNA binding domain; H: Hinge region; LBD: Ligand binding domain. C. Full-length AR was co-expressed with various Flag-COBRA1 fragments in HEK293T cells, and their interactions were examined with co-immunoprecipitation. The diagram is a summary of the binding affinity of COBRA1 fragments for the full-length AR protein. The shaded and solid boxes represent the NR-binding signature motifs LXXLL and ΦXXΦΦ, respectively (L for leucine; X for any amino acid; and Φ for hydrophobic residue).
To further characterize the COBRA1-AR interaction, we also tested the AR-binding ability for a series of COBRA1 deletion mutants (Fig. 2C). Surprisingly, all four COBRA1 fragments that collectively encompass the entire protein bound to AR as efficiently as the full-length COBRA1 protein, suggesting the presence of multiple AR-binding regions in COBRA1. As indicated in the diagram in Fig. 2C, COBRA1 protein contains multiple copies of the NR-binding motifs (“LXXLL” where L is leucine and X any amino acid; “ΦXXΦΦ” where Φ is hydrophobic residue) [2, 3, 19]. Conceivably, more than one of these motifs could mediate the interactions between AR and various COBRA1 fragments. Consistent with this possibility, removal of sequence between amino acid 384 and 431, which includes one of the “ΦXXΦΦ” sequences in COBRA1, rendered the remaining C-terminal fragment incapable of AR-binding (compare Fragment #4 with #11 in Fig. 2C).
Effect of COBRA1 on steroid hormone-dependent transcription
To determine the functional consequence of the physical interactions between COBRA1 and the three steroid hormone receptors, we used retroviral vectors that express COBRA1-specific shRNA sequences to stably knock down endogenous COBRA1 expression in T47D breast cancer cells. T47D cells express functional AR, PR, and GR, thus allowing for a direct comparison of the impact of COBRA1 on the transcriptional activity of all three nuclear receptors in the same cellular context. As compared with the control cell line (shEGFP), COBRA1 protein levels in the two shCOBRA1-expressing cell lines (2A and GA) were substantially reduced (inlet in Fig. 3A). Reduction of COBRA1 expression did not affect the levels of AR, PR, or GR (data not shown). We then conducted an MMTV-driven luciferase reporter assay in the control and COBRA1-knockdown cells. The MMTV-LTR promoter can be stimulated by all three nuclear hormone receptors of interest, thus providing the same promoter context for the comparative study. Consistent with a previous report [20], the stimulatory effects of progesterone and glucocorticoid on the MMTV-LTR promoter in the control T47D cells were much more robust than that of androgen (compare lane 2 with 3 and 4 in Fig. 3A; 41, 626, and 331 fold for AR, PR, and GR, respectively). Remarkably, ligand-dependent transcriptional activation by AR was significantly elevated in two independent COBRA1 knockdown cell lines (compare 41 fold in lane 2 with 667 fold in lane 6 and 472 fold in lane 10), whereas GR and PR-dependent transcription was only moderately enhanced by COBRA1 knockdown (for PR, 626 fold in lane 3,1081 fold in lane 7, and 692 fold in lane 11; for GR, 331 fold in lane 4, 1120 fold in lane 8, and 752 fold in lane 12). As a consequence, the ligand-stimulated transcription by the three steroid hormones reached comparable levels in the COBRA1 knockdown cells (compare lane 6-8; 10-12 in Fig. 3A). Notably, the extent of the impact of COBRA1 knockdown on the ligand-dependent transcription correlates quite well with the differential affinity of COBRA1 for the three steroid hormone receptors (AR>GR>PRB; Fig. 1A).
FIGURE 3. Reduction of COBRA1 in breast cancer cells results in elevated transcriptional activation by AR.
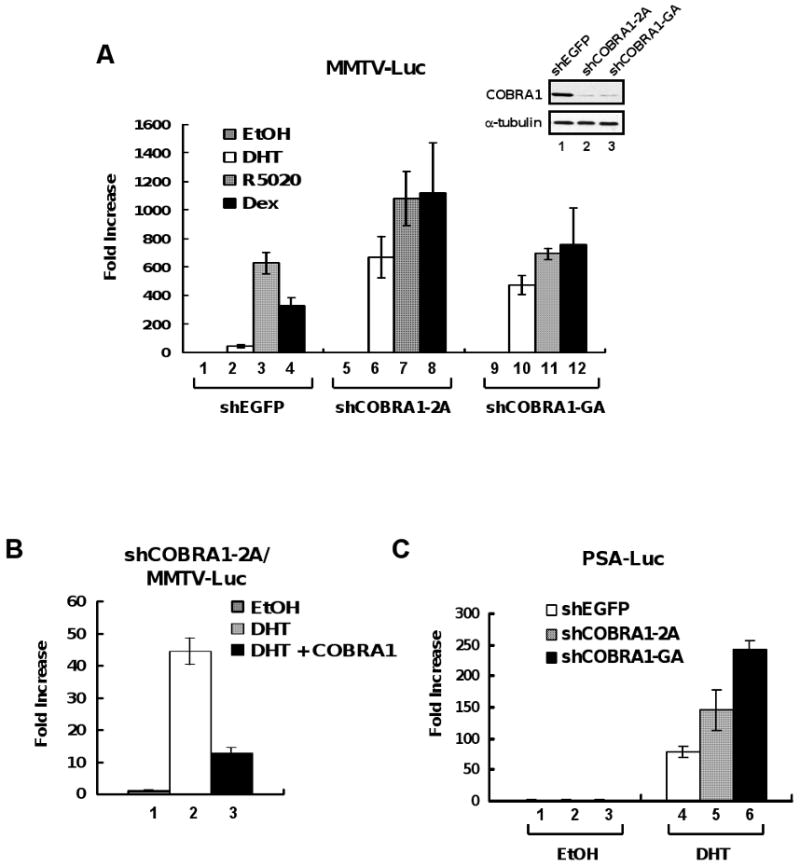
A. The inlet shows an α-COBRA1 immunoblot in T47D-derived cell lines that stably expressed short hairpin RNAs against EGFP control (shEGFP) or COBRA1 (shCOBRA1-2A and shCOBRA1-GA). α-Tubulin was used as a loading control. MMTV-luciferase reporter plasmid was transiently transfected in the control or two COBRA1 knockdown cell lines. The cells were treated with DHT (10 nM; open bars), R5020 (10 nM; shaded bars), or dexamethasone (Dex, 100 nM; solid bars) for 24 hours before harvesting for luciferase assay. Ethanol was used as vehicle. Steroid hormone-dependent transcription was expressed as fold increase over vehicle. B. A COBRA1 silent mutant (COBRA1) that is refractory to shCOBRA1-2A was co-expressed with the MMTV-luciferase reporter plasmid in stable knockdown cells expressing shCOBRA-2A. Transfected cells were treated with DHT (10 nM) for 24 hours before harvesting for luciferase assay. C. DHT (10 nM)–dependent transcription from the prostate-specific antigen (PSA) promoter in the control and two COBRA1 knockdown cells as measured by luciferase assay.
Despite the superior power of the shRNA technique, interpretation of the knockdown result can be complicated by spurious “off-targeting” effects. The fact that two independent COBRA1 shRNA sequences give rise to similar effects on the androgen-dependent transcription strongly suggests a high specificity of the COBRA1-targeted shRNA. Furthermore, ectopic expression of a COBRA1 silent mutant that was refractory to the shRNA-2A-mediated degradation blunted the shRNA effect on the androgen-dependent transcription (Fig. 3B), thus unambiguously demonstrating the specificity of the shRNA-mediated effect. In addition to the MMTV-LTR promoter, ligand-dependent transcription of the native prostate specific antigen (PSA) promoter was also elevated in the COBRA1 knockdown cells, albeit to a lesser extent (compare lane 4 with 5 and 6 in Figure 3C).
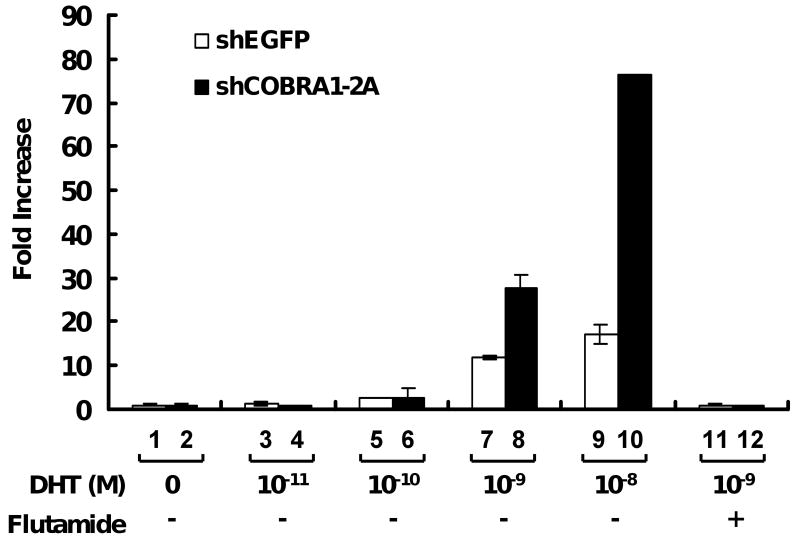
Several known AR coactivators have been shown to alter the ligand sensitivity of the receptor in transcriptional activation [21-24]. To test whether COBRA1 knockdown would alter ligand sensitivity of AR, we compared ligand-dependent transcription from the MMTV-LTR in the control and COBRA1-knockdown T47D cells using a range of final concentrations of dehydrotestosterone (DHT; 10-11 to 10-8 M). As shown in Fig. 4, the same minimal concentration of DHT (10-10 M) was required to stimulate the ligand-dependent transcription in the control and COBRA1 knockdown cells. In fact, the effect of COBRA1 knockdown only became obvious at higher DHT concentrations (10-9 and 10-8 M). In addition, the ligand-dependent transcriptional activation in both cell lines can be completely abolished by flutamide, a known AR antagonist. Thus, while COBRA1 reduction significantly increases the magnitude of androgen-dependent transcription, it does not change the AR sensitivity to either agonists or antagonists.
FIGURE 4. COBRA1 does not affect the ligand sensitivity of AR-mediated transcriptional activation.
The control and COBRA1 knockdown cells were transfected with the MMTV-luciferase reporter plasmid, and subsequently treated with a serial 10-fold dilution of DHT, ranging from 10-11 M to 10-8 M. Transfected cells were also treated with AR antagonist flutamide (10-6 M) in the presence of 10-9 M DHT to ascertain the androgen-dependency of the observed luciferase expression.
A novel role of COBRA1 in modulating hormone-dependent alternative splicing
While the majority of transcriptional coactivators exert their actions at the step of transcriptional initiation, emerging evidence indicates that a subset of coregulators for steroid hormone receptors share structural and functional similarity with factors involved in RNA splicing [25]. Indeed, various steroid hormones and a number of coactivators can simultaneously activate ligand-dependent transcription and influence the alternative splicing pattern from the same genes [8-12]. Compared to these coactivators, a role of corepressors in alternative splicing is largely un-explored. In light of the negative impact of COBRA1 on androgen-dependent transcription and its established function in transcription elongation, we sought to test whether COBRA1 could affect alternative splicing from the same androgen-dependent promoter.
We took advantage of the MMTV-CD44 mini-gene reporter system that has been previously used to demonstrate the effect of steroid hormones on alternative splicing [10]. In this mini-gene cassette, the variable exons v4 and v5 of the human CD44 gene, along with their surrounding intron sequences, were inserted into an intron of the β-globin gene (diagram in Fig. 5) [10, 26]. The reporter gene is expected to yield two major spliced RNA products that have either both variable exons (inclusion) or neither exon (skipping), and a minor product that only has one variable exon. Administration of synthetic androgen (R1881) indeed promoted the production of the skipping form of the transcript and concomitantly reduced the inclusion form, resulting in a significant increase in the ratio of the skipping over inclusion species (compare lane/column 1 and 2 in Fig. 5). Ectopic expression of COBRA1 gave rise to a three-fold reduction in the skipping/inclusion ratio (lane/column 3 in Fig. 5).
FIGURE 5. Over-expression of COBRA1 reduces the skipping/inclusion ratio of androgen-dependent alternative splicing.
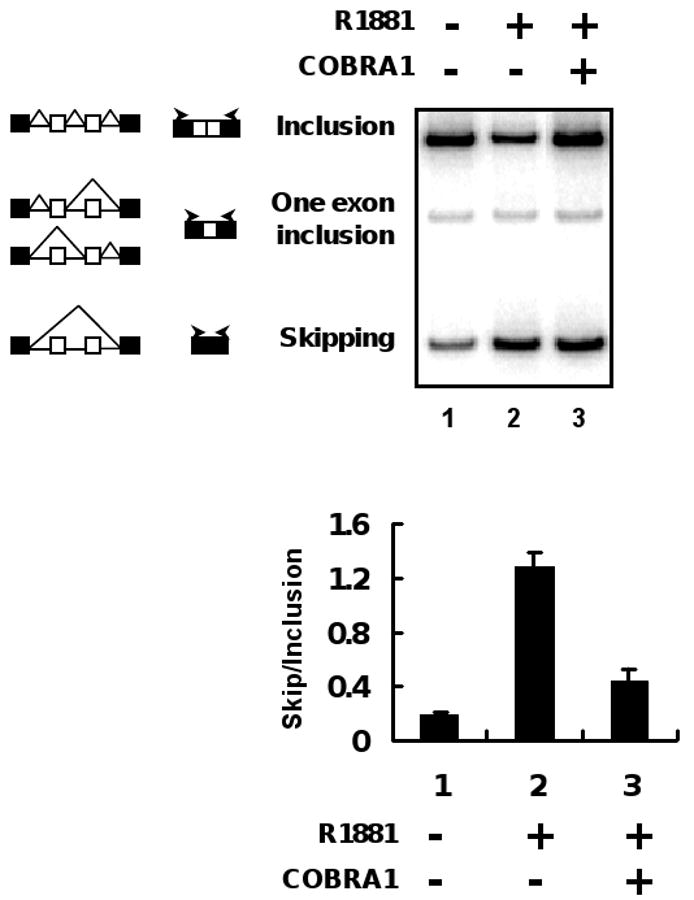
COBRA1 and AR were co-expressed with the MMTV-CD44 mini-gene cassette in HEK293T cells. The transfected cells were treated with 1 nM R1881 for 24 hours before harvesting for alternative splicing assay. Two major products of CD44 mini-gene resulted from alternative splicing were designated as inclusion and skipping, respectively. The minor splicing product with one exon inclusion is also indicated in the figure, but not considered in the calculation of the skipping/inclusion ratio. The skipping/inclusion ratio was calculated by averaging two independent transfections in each assay. The diagram indicates various splicing events. The open and solid boxes in the diagram refer to the variable and invariable exons, respecitively. The arrows indicate the positions of the primers used in the PCR reactions.
To corroborate the effect of COBRA1 over-expression, we also conducted the same alternative splicing experiment in control T47D cells and the two independent COBRA1 knockdown cell lines (shCOBRA1-2A and -GA). As shown in Fig. 6, COBRA1 knockdown resulted in increased skipping/inclusion ratios (compare lane/column 4 with 5 and 6), opposite to the effect of COBRA1 over-expression. Taken together with findings from the transcription assay, these results strongly suggest a dual function of COBRA1 in regulating androgen-dependent transcription and RNA processing.
FIGURE 6. COBRA1 knockdown increases the skipping/inclusion ratio of the alternative splicing.
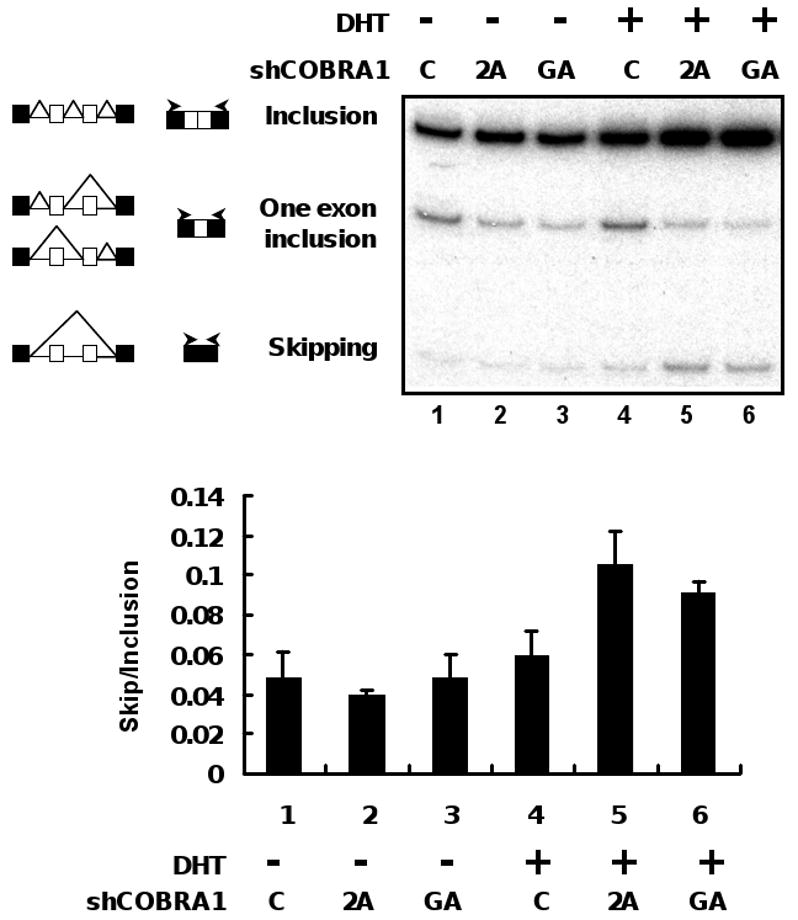
The MMTV-CD44 mini-gene were transfected into T47D derived stable cell lines expressing EGFP (control) or COBRA1-specific shRNAs (2A and GA). Cells were treated with DHT (10 nM) for 24 hours before harvesting for alternative splicing assay. Quantitation of the results as shown in the graph was performed in the same way as shown in Fig. 5.
Discussion
Extensive in vitro biochemical investigation has clearly established that the human NELF complex, of which COBRA1 is an integral subunit, acts at an early stage of transcription elongation to stall the movement of the transcription complex [14, 16, 27]. Consistent with the in vitro biochemical observation, in vivo studies indicate that COBRA1 together with the other NELF subunits inhibits ligand-dependent transcriptional activation by ERα in breast cancer cells [17]. The current work extends the previous findings by demonstrating physical and functional interactions between COBRA1 and other members of the steroid hormone receptor family. Importantly, we provide evidence for a dual role of COBRA1 in modulating androgen-dependent transcription and alternative splicing, which is consistent with the emerging theme of unified eukaryotic gene expression.
Most known NR coregulators are capable of binding to multiple receptors and mediating transcriptional regulation in response to a variety of ligands. The functional promiscuity of these coregulators has been attributed to the presence of the multiple NR-binding motifs LXXLL or ΦXXΦΦ [2, 3, 19]. These structural and functional features are obviously shared by COBRA1 and other NR coregulators. In addition, COBRA1 also exhibits some distinct properties in modulating hormone-dependent gene expression. For example, COBRA1 displays a differential degree of affinity for various steroid hormone receptors, which correlates with the magnitude of COBRA1-mediated repression of the ligand-dependent transcription. Of the several nuclear receptors examined in the current study, AR displays the strongest affinity for COBRA1 and most prominent susceptibility to COBRA1-mediated repression. Second, unlike most coregulators that act at transcription initiation, COBRA1 is known to exert its effect during the early stage of transcription elongation. Conceivably, once recruited to the ligand-responsive promoters by the corresponding site-specific transcription factor, COBRA1 and the rest of the NELF complex may no longer depend on the promoter-bound factor to modulate the transcription rate during the elongation stage. This could explain the lack of impact of COBRA1 on the ligand-sensitivity and specificity of AR. Lastly, results from the current study put COBRA1 in a special group of coregulators that are capable of simultaneously influencing ligand-dependent transcription and alternative splicing [25]. Most coregulators with dual functions in transcription and RNA processing contain the RNA recognition motif (RRM) [25]. In this regard, it is worth noting that, while COBRA1 itself does not have such a motif, another subunit of the NELF complex (NELF-E) is an RRM-containing protein [16].
Mounting evidence from both genetic and biochemical studies strongly suggests that pre-mRNA processing of eukaryotic gene expression occurs in a “co-transcriptional” fashion [28]. To our knowledge, COBRA1 is the first known corepressor of steroid hormone receptors that promotes inclusion and/or attenuates skipping. We can envision several possible mechanisms that could be used by COBRA1 to alter the alternative splicing pattern. For example, upon its recruitment to an androgen-dependent promoter, COBRA1 may directly counteract those coactivators that facilitate the skipping event [10]. Alternatively, COBRA1 may promote exon inclusion by slowing down the movement of RNAPII, a key factor in determining the alternative splicing fate of pre-mRNA [7]. A slower-moving transcription elongation complex could in turn have ample time to recruit splicing factors and/or select cis-acting elements for alternative splicing [29]. It is also possible that COBRA1 may directly interact with components of the splicing apparatus that are required for alternative splicing. Consistent with this possibility, NELF complex has been shown to functionally interact with the RNA-capping enzyme [30]. Regardless of the underlying mechanisms, a dual function of COBRA1 in transcription and RNA processing may ensure efficient control of ligand-dependent gene expression in terms of the quantity as well as nature of the mature gene products. In addition, this coupling role of COBRA1 may allow a prompt and adequate response of the gene expression machinery to the rapid change of hormone levels in vivo.
It is estimated that at least 30% of human genes are alternatively spliced [31, 32]. Because the splice variant products could have very distinct biological activity as compared to the wild-type counterpart, the contribution of alternative splicing to the functional complexity and diversity of human proteins has been increasingly appreciated. A large number of cancer-associated genes are known to express various alternatively spliced transcripts [31], and cancer-specific mRNA isoform signatures have been identified in tumor specimens [33]. It is conceivable that cancer-specific splice variants could contribute to the initiation and/or progression of disease, or simply act as surrogate markers. In this regard, active research is underway to identify cancer-specific alternative splicing patterns of endogenous genes that are subject to regulation by COBRA1. Furthermore, given the paramount importance of androgen and AR in the development of prostate cancer, it will be of great interest to explore a role of COBRA1 in androgen-dependent gene regulation and tumorigenesis in prostate tissue.
Acknowledgments
We thank Drs. Bryce Paschal, Daniel Gioeli, Bert O'Malley, and Rosalie Uht for providing various plasmids used in the study. We also thank Dr. Yanfen Hu for critical reading of the manuscript. The work was supported by a predoctoral fellowship from D.O.D. Breast Cancer Research Program to J.S. and an NIH grant to R.L. (DK064604).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jianlong Sun, Department of Molecular Medicine, Institute of Biotechnology, 15355 Lambda Drive, University of Texas Health Science Center at San Antonio, San Antonio, TX 78245.
Ashley L. Blair, Department of Biochemistry and Molecular Genetics, School of Medicine, P.O. Box 800733, University of Virginia, Charlottesville, VA 22908-0733
Sarah E. Aiyar, Department of Biochemistry and Molecular Genetics, School of Medicine, P.O. Box 800733, University of Virginia, Charlottesville, VA 22908-0733
Rong Li, Department of Biochemistry and Molecular Genetics, School of Medicine, P.O. Box 800733, University of Virginia, Charlottesville, VA 22908-0733.
References
- 1.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pttersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 2.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 3.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 4.Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 5.Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2:1006.1–1006.3. doi: 10.1186/gb-2001-2-2-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zorio DAR, Bentley DL. Transcription elongation: the “Foggy” is lifting. Curr Biol. 2001;11:R114–R146. doi: 10.1016/s0960-9822(01)00063-x. [DOI] [PubMed] [Google Scholar]
- 7.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 8.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, O'Malley BW. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc Natl Acad Sci U S A. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'Malley BW. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auboeuf D, Honig A, Berget SM, O'Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 11.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 13.Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001;155:911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose H, Sugimoto S, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymeerase II processitivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 17.Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lv Y, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, Li R. Attenuation of Estrogen Receptor alpha-Mediated Transcription through Estrogen-Stimulated Recruitment of a Negative Elongation Factor. Genes & Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang CS, Vitto MJ, Busby SA, Garcia BA, Kesler CT, Gioeli D, Shabanowitz J, Hunt DF, Rundell K, Brautigan DL, Paschal BM. Simian virus 40 small t antigen mediates conformation-dependent transfer of protein phosphatase 2A onto the androgen receptor. Mol Cell Biol. 2005;25:1298–1308. doi: 10.1128/MCB.25.4.1298-1308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 20.Gowland PL, Buetti E. Mutations in the hormone regulatory element of mouse mammary tumor virus differentially affect the response to progestins, androgens, and glucocorticoids. Mol Cell Biol. 1989;9:3999–4008. doi: 10.1128/mcb.9.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto H, Yeh S, Wilding G, Chang C. Promotion of agonist activity of antiandrogens by the androgen receptor coactivator, ARA70, in human prostate cancer DU145 cells. Proc Natl Acad Sci USA. 1998;95:7379–7384. doi: 10.1073/pnas.95.13.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 2000;60:4709–4713. [PubMed] [Google Scholar]
- 23.Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
- 24.Agoulnik IU, Vaid A, Bingman WEr, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 25.Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O'Malley BW. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol Cell Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Handa H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differnces between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- 28.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Op Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 29.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkman BMN. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 33.Li HR, Wang-Rodriguez J, Nair TM, Yeakley JM, Kwon YS, Bibikova M, Zheng C, Zhou L, Zhang K, Downs T, Fu XD, Fan JB. Two-dimensional transcriptome profiling: identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–88. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]