Abstract
Human papilloma (HPV) virus-like particle (VLP) vaccines were recently licensed. Though neutralizing antibody titers are thought to be the main effectors of protection against infection, early predictors of long-term efficacy are not yet defined and a comprehensive understanding of innate and adaptive immune responses to vaccination is still lacking. Here, microarrays were used to compare the gene expression signature in HPV-16 L1 VLP-stimulated PBMC from 17 vaccine and 4 placebo recipients before vaccination, and 1 month after receiving the second immunization. Vaccination with a monovalent HPV-16 L1 VLP vaccine was associated with modulation of genes involved in the inflammatory/defense response, cytokine, interferon and cell cycle pathways in VLP-stimulated PBMC. Additionally, there was up-regulation of probesets associated with cytotoxic (GZMB, TNFSF10) and regulatory (INDO, CTLA4) activities. The strongest correlations with neutralizing antibody titers were found for cyclin d2 (CCND2) and galectin (LGALS2). Twenty-two differentially expressed probesets were selected for confirmation by RT-PCR in an independent sample set. Agreement with microarray data was seen for over two-thirds of these probesets. Up-regulation of immune/defense response genes by HPV-16 L1 VLP, in particular interferon-induced genes was observed in PBMC collected prior to vaccination, with many of these genes being further induced following vaccination. In conclusion, we identified important innate and adaptive response related- genes induced by vaccination with HPV-16 L1 VLP. Further studies are needed to identify gene expression signatures of immunogenicity and long-term protection with potential utility in prediction of long-term HPV vaccination outcomes in clinical trials.
Keywords: Human, Vaccination, Microarray
Introduction
Cervical cancer is the second most common cancer among women worldwide (1, 2). The recent development of prophylactic HPV vaccines provides an important new avenue for the prevention of this cancer and others linked to HPV infection.
The current HPV prophylactic vaccines consist of non-infectious, L1 recombinant HPV-like particles (VLP) (3, 4). Vaccine clinical trials have shown near complete protection against cervical intraepithelial neoplasia (CIN) 2 or greater lesions (5–7). Neutralizing antibodies are believed to be the main effectors of protection. A robust humoral response has been observed after vaccination (8–10), and antibody levels against HPV-16 remain relatively high in the first few years after vaccination (11–13). However, length of protection afforded by vaccination is not currently known.
Studies by our group and others have shown that in addition to antibody responses, strong cell-mediated immune responses are observed after vaccination (14–17). It is known that cell-mediated immune responses are required for antibody production and for maintenance of antibody levels over time. Thus, it could be postulated that the patterns of innate and acquired cellular immune responses following initial vaccination might influence antibody responses and duration of protection afforded by vaccination (18, 19). A better understanding of these early responses to vaccination might help elucidate mechanisms of vaccine protection via neutralizing antibodies, and assist in the identification of early markers of long-lasting vaccine responses.
Microarray analysis has been applied to determine gene expression patterns in studies of disease pathogenesis (20–23), immune response to infection (24–30), and immune response after immunization (31, 32). However, few studies have looked at vaccine-induced responses in humans (33–35).
To characterize the immune response to HPV-16 L1VLP vaccination, we used microarray chips that cover 8638 verified sequences to investigate the gene profile associated with a recall response to the vaccine in leukocytes from vaccine recipients. We identified probesets that were differentially expressed in PBMC from vaccine recipients after in vitro stimulation with HPV-16 L1 VLP compared to pre-vaccination samples. We also analyzed the “primary” response to HPV-16 L1 VLP, independent of vaccination, by comparing the gene expression pattern of cells incubated with HPV-16 L1 VLP and unstimulated cells, using pre-vaccination samples.
In addition, the correlation of gene expression to neutralizing antibody titers developed after vaccination was examined to evaluate potential determinants of strong antibody responses. Key results were confirmed by RT-PCR in an independent set of vaccinated individuals.
This is the first study that evaluates the immune response to an HPV VLP vaccine using microarray technology. Our results contribute to a broader understanding of the effects of vaccination with a monovalent HPV-16 L1 VLP vaccine. The approach used here and the gene expression profile defined in this study may prove to be useful for future prediction of long-term HPV vaccination outcomes in ongoing clinical trials.
Materials and Methods
Study Design
Participants were selected from a double-blind, randomized, placebo-controlled phase II trial of a monovalent HPV-16 L1 VLP vaccine without adjuvant, which was conducted in a sample of 220 healthy, HIV seronegative adult female volunteers 18–25 years of age, as described previously (17). Briefly, subjects were enrolled at The Johns Hopkins University Center for Immunization Research (Baltimore). Pre-vaccination HPV-16 antibody or DNA status was not a criterion for eligibility into the trial. Subjects were determined by history to be at low risk for HPV16 exposure. Individuals were not eligible to participate if they had a history of more than four lifetime sexual partners or more than two sexual partners within the preceding 6 months. Additional exclusion criteria included history of abnormal cervical cytology, immunodeficiency, anaphylaxis to medicines or vaccines, receipt of blood products within 3 months of enrollment, current pregnancy or lactation, and any other condition that might interfere with the study objectives. Women were randomly assigned to receive three intramuscular doses of either 50 µg of HPV-16 L1 VLP vaccine without adjuvant, or placebo (0.5 mL saline). Blood specimens were collected before the initial dose (month 0), and 1 month after each of the subsequent vaccinations (months 2 and 7). The Johns Hopkins University Institutional Review Board approved the protocol for this study. Blood specimens were shipped to the HPV Immunology Laboratory (Frederick, MD), where PBMCs were cryopreserved.
Twenty-seven subjects (20 vaccine and 7 placebo recipients) were randomly selected for microarray measurements. Only month 0 and month 2 samples were selected for this study because earlier findings indicated that the largest increases in cytokine responses were typically observed at month 2 (17). Month 0 and 2 samples from 10 additional vaccine recipients were selected for a second, confirmatory phase of our study by RT-PCR.
HPV-16 L1 VLP vaccine
HPV-16 L1 VLPs were expressed in baculovirus-infected Sf9 insect cells (Novavax, Rockville, MD). Production of clinical lots of recombinant HPV-16 L1 VLP vaccines was performed in accordance with GMP guidelines as previously reported (36). The VLPs used to in vitro to stimulate PBMCs were similar to the ones used to vaccinate the subjects included in this study. VLPs were provided at 1 mg/mL, stored at −80°C and thawed immediately preceding in vitro stimulation, as previously described (16).
PBMC incubation and Microarray Analysis
Cryopreserved PBMCs were thawed and cultured (2.0×106 cells/mL) as previously described (17). 10×106 PBMCs were plated in each well of a 6-well plate (Costar) in RPMI-1640 supplemented with penicillin-streptomycin (100 µg/mL-100 U/mL; Gibco), L-glutamine (2mM), HEPES buffer (10 mM) and 10% heat-inactivated fetal calf serum (HyClone). Cells were cultured for 72 hours at 37°C with: media; HPV-16 L1 VLP (2.5 µg/mL); or Sf9/baculovirus insect cell lysate (0.1 µg/mL, Novavax) all diluted in cell culture media. Media was used as a background measurement for untreated cells. The Sf9/baculovirus insect cell lysate was used as a control antigen for the L1 VLP production system.
A total of eight incubations were setup on the same day for each subject. Incubations for two subjects were setup on the same day. The order of sample preparation was randomly defined from the list of selected subjects.
When harvesting, cultures were centrifuged and cell-free supernatants were aliquoted and frozen at −80°C. Total RNA extracts were performed using an RNeasy Total RNA isolation Kit (Qiagen). RLT lysis buffer was quickly added to the culture well and then to the cell pellet, in order to include both adherent and suspension cells. RNA purity and integrity were tested by microcapillary electrophoresis using the Agilent 2100 bioanalyzer (Agilent Technologies).
Microarray gene expression analysis was performed using the using the Human Genome Focus Array from the Affymetrix GeneChip system (Affymetrix, Inc.) that contains about 8700 probesets to 8638 characterized human genes. Total RNA preparation and labeled cRNA synthesis and hybridization were performed according to the manufacturer's recommended protocol (Affymetrix, Inc.). In short, 10 µg of total RNA were used for double-stranded cDNA (ds-cDNA) synthesis with the SuperScript™ II Reverse Transcriptase Kit (Invitrogen) and an oligo (dT) primer containing a T7 RNA polymerase promoter to prime the first-strand synthesis. Biotin-labeled cRNA was obtained by in vitro transcription after addition of T7 RNA polymerase and biotinylated nucleotides (Enzo Biochem) to the ds-cDNA. Labeled cRNA was fragmented and hybridized to the GeneChips, which were then washed and stained with streptavidin-conjugated PE by using the Affymetrix GeneChip Fluid Station 400 (Affymetrix, Inc.). To assess the quality of hybridization, the following quality control filters were used: scaling factor, background, percentage of present calls, noise, housekeeping genes (3'/5' ratios and intensity for GAPDH and Actin), RNA degradation slope and presence or absence of internal spike controls.
Gene expression levels were determined by laser scanning of the GeneChip at 570 nm and were log2 transformed following MAS5 probeset summarization. All results from a donor were excluded if one of the conditions (media and VLP cultures for the pre- and post-vaccination specimens) did not fulfill one of the previously mentioned quality control filters used to ensure that all chips were of comparable quality. These quality control procedures resulted in exclusion of two of the vaccine recipients and one placebo. One additional vaccine recipient was excluded due to a high degree of granulocyte contamination (61% versus 0.2–13.6% granulocytes for remaining samples), possibly due to poor PBMC Ficoll isolation, and because neutralizing antibodies against HPV-16 were detectable before vaccination. Two additional placebos were excluded because they had detectable antibodies in the course of our study. After all exclusions, 17 vaccine and 4 placebo recipients were used in subsequent analysis.
Data from probesets that were not reliably detected with a mean intensity> log2 = 5 in at least one experimental condition (media and VLP cultures for the pre- and post-vaccination specimens) were excluded before further analysis. A total of 7145 probesets remained after applying this criterion.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (37), and are accessible through GEO Series accession number GSE13587 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13587).
Flow Cytometry
Major lymphocyte subsets for all samples were determined by flow cytometry (FC-500, Beckman-Coulter), as previously described (38). Flow cytometric analysis of main lymphocyte subsets indicated that overall levels of major lymphocyte subsets did not change considerably after vaccination (mean + SD pre versus post vaccination, were CD3+ cells: 78±7 versus 77±9, CD19+ cells: 10±6 versus 10±5, CD16+/56+ cells: 11±6 versus 11±6). In addition, placebo recipients had also similar overall levels of leukocyte subsets (mean ± SD month 0 versus month 2, were CD3: 77±4 versus 79±5, CD19: 11±3 versus 11±6, CD16+/CD56+ cells 9±1 versus 8±3).
Neutralizing antibodies
HPV-16 neutralizing antibody titers were determined using a pseudovirus-based neutralization assay performed as previously described (39). Serum neutralization titers were defined as the reciprocal of the highest dilution that caused at least 50% reduction in SEAP activity. A negative control using the Bovine Papillomavirus (BPV) pseudovirus was performed for all samples. Undetectable antibody levels were considered as 0. All individuals selected for the microarray analysis had no detectable antibodies prior to vaccination. After vaccination, all individuals developed antibody titers against HPV-16 (month 2 mean titer of 843±748, range of 78 to 2602; month 7 mean titer of 5209±7383, range of 111 to 25571; month 12 mean titer of 2366±4184, range of 148 to 15317).
Quantitative real-time PCR and confirmation of microarray results
Real-time PCR measurements were performed with samples from 10 independent vaccine recipients collected at month 0 and 2 post-vaccination within this same clinical trial. These individuals were selected randomly based on availability of cryopreserved PBMC at both time points and were not tested by microarray. One subject was excluded from analysis because anti-HPV-16 neutralizing antibodies were detectable before vaccination. All remaining nine donors, had no detectable neutralizing antibody titers before vaccination, and thus, a similar prevaccination antibody status as the individuals used for the microarray experiments. In addition, an induction in anti-HPV-16 titers was observed at month two after the first dose (mean titer at month 2 of 2675±2644, neutralizing titer range: 333–8508).
A total of 22 probesets (Table IV) were selected for confirmation. All selected probesets had an average fold induction of 2.0 or higher by microarray (range: 2.0–5.01), which was observed in at least 50% of subjects (range: 50–83%). Ten selected probesets had statistically significant differences before and after vaccination with significance levels of p≤0.001. These included GZMB, IFNG, HSD11B1, INDO, GGTLA1, KIF20A, GOLPH2, IL6, IL3 and AICDA. The remaining 12 probesets were selected either because their expression levels correlated with antibody titers at month 7 or 12 (r>0.52; 5 probesets: CYP27B1, INHBA, MMP12, CD1B and CCL7) or they were cytokines/chemokines of potential interest (7 probesets: CSF2, IL2, IL5, CXCL13, LTA, LIF, CCL13).
Table IV.
Spearman Correlation of Gene Expression with Neutralizing Antibody Levels*
| Affy ID | Gene Symbol | Gene name | Fold Change# | p-value-Microarray | Spearman's r | ||
|---|---|---|---|---|---|---|---|
| Month 2 | Month 7** | Month 12** | |||||
| 200953_s_at | CCND2 | cyclin d2 | 1.42 | 0.000378 | ** | 0.88 | 0.82 |
| 208450_at | LGALS2 | lectin, galactoside-binding, soluble, 2 (galectin 2) | 3.03 | 0.0040252 | ** | 0.86 | 0.74 |
| 212657_s_at | IL1RN | interleukin 1 receptor antagonist | 1.61 | 0.035551 | ** | 0.75 | 0.73 |
| 206749_at | CD1B | cd1b antigen | 2.91 | 0.0049094 | ** | 0.68 | 0.73 |
| 203564_at | FANCG | fanconi anemia, complementation group g | 1.52 | 0.006110 | ** | 0.68 | 0.65 |
| 221287_at | RNASEL | ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent) | −1.84 | 0.0158257 | ** | 0.64 | 0.84 |
| 206181_at | SLAMF1 | signaling lymphocytic activation molecule family member 1 | 1.42 | 0.0004684 | ** | 0.64 | 0.60 |
| 205638_at | BAI3 | brain-specific angiogenesis inhibitor 3 | −1.71 | 0.037898 | ** | 0.63 | 0.71 |
| 205633_s_at | ALAS1 | aminolevulinate, delta-, synthase 1 | 1.60 | 0.0005912 | ** | 0.61 | 0.70 |
| 204092_s_at | AURKA | aurora kinase a | 1.49 | 0.037044 | ** | 0.61 | 0.63 |
| 204504_s_at | HIRIP3 | hira interacting protein 3 | 1.55 | 0.031187 | ** | 0.60 | 0.81 |
| 206206_at | CD180 | cd180 antigen | −1.39 | 0.0357298 | ** | −0.61 | −0.66 |
| 203126_at | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 | −1.34 | 0.013930 | ** | −0.62 | −0.62 |
| 34187_at | RBMS2 | rna binding motif, single stranded interacting protein 2 | 1.89 | 0.0318612 | ** | −0.62 | −0.65 |
| 202838_at | FUCA1 | fucosidase, alpha-l- 1, tissue | −2.05 | 0.010039 | ** | −0.64 | −0.76 |
| 201753_s_at | ADD3 | adducin 3 (gamma) | −1.35 | 0.026385 | ** | −0.64 | −0.64 |
| 209774_x_at | CXCL2 | chemokine (c-x-c motif) ligand 2 | −1.76 | 0.0020535 | ** | −0.69 | −0.61 |
| 219697_at | HS3ST2 | heparan sulfate (glucosamine) 3-o-sulfotransferase 2 | −2.64 | 0.0002601 | ** | −0.69 | −0.62 |
| 204394_at | SLC43A1 | solute carrier family 43, member 1 | −1.37 | 0.004684 | ** | −0.69 | −0.64 |
| 221558_s_at | LEF1 | lymphoid enhancer-binding factor 1 | −1.36 | 0.0000261 | ** | −0.76 | −0.69 |
| 212552_at | HPCAL1 | hippocalcin-like 1 | −1.32 | 0.0029172 | ** | −0.76 | −0.75 |
| 201820_at | KRT5 | keratin 4 | −2.00 | 0.012345 | ** | −0.90 | −0.73 |
Neutralizing antibody titers [Median (range)]: Month 0=0 (0–0); Month 2=664 (78–4949); Month 7=2401 (111–25571); Month 12=986 (148–15316)
Only probe sets with correlation coefficient values of r≥0.60 or r≤−0.60 at at both Month 7 and Month 12 are shown (p≤0.05 in all cases).
(VLP-Media) expression levels as defined in the Materials and Methods Section
PBMC incubation and total RNA extraction were performed as described above. One µg of total RNA extract was reverse-transcribed with random hexamer primers, using Super Script™ Reverse Transcriptase (Invitrogen). 1/20 of the first-strand cDNA synthesis reaction was used as template. Pre-designed primer and probe kits (TaqMan® Gene Expression Assays, Applied Biosystems) and the TaqMan® Universal PCR Master Mix (Applied Biosystems) were used according to manufacturer’s recommendations. Briefly, stock reaction solutions were prepared by mixing 10 µL of PCR Master Mix (2X), 1 µL of Primer kit (20X) and 7.1 µL of DEPC-treated water per reaction. 18.1 µL of stock solution were dispensed to each well, and 1.9 µL of cDNA was then added (total volume per reaction: 20 µL). The following temperature program was used for all genes: 2 minutes at 52°C, 10 minutes at 95 °C and 50 cycles of 15 seconds at 95°C and 1 minute at 60°C. Fluorescence intensity was measured in real time during the extension step, using the iCycler IQ multicolor real time PCR detection system (Bio-Rad). The following formula was used to determine fold induction for each gene:
Where CtXVpost = threshold cycle of the analyzed gene in month 2 cells treated with VLP; CtRVpost = threshold cycle of RPLPO (acidic ribosomal phosphoprotein P0 subunit) in month 2 cells treated with VLP; CtXMpost = threshold cycle of the analyzed gene in month 2 cells treated with media; CtRMpost = threshold cycle of RPLPO in month 2 cells treated with media; CtXVpre = threshold cycle of the analyzed gene in month 0 cells treated with VLP; CtRVpre = threshold cycle of RPLPO in month 0 cells treated with VLP; CtXMpre = threshold cycle of the analyzed gene in month 0 cells treated with media; CtRMpre = threshold cycle of RPLPO in month 0 cells treated with media.
To determine the cutoff value for PCR results, the fold change for RPLPO was calculated as follows:
This was used to determine variability of RPLPO for all subjects, and define the lowest fold change to consider a gene as up-regulated. Based on this, only genes with a fold change higher than the average plus two standard deviations of the value obtained for RPLPO were considered positive (i.e. 2.14-fold change).
Data analysis
To determine vaccination-induced changes, gene expression levels (presented as log2 of intensity) for each probeset were compared before and after vaccination. Media-incubated expression levels were subtracted from VLP-incubated samples to adjust for background noise. Paired t-tests comparing the mean before and after vaccination were used to determine statistical significance. To account for the evaluation of the large number of genes, p-values < 0.001 were considered significant. Further, only probesets with log2 expression change (n-fold) <–1.3 (downregulated) or >1.3 (upregulated) were considered. For the a priori analysis, changes in gene expression for a predefined set of probesets within pre-selected pathways (Defense, Inflammation, Cytokine, Interferon, Cell cycle, Signal transduction) were evaluated. The selection of probesets was based on the list of Gene Ontology pathways obtained for the probesets present in the Human Genome Focus Array. A p-value of less than 0.001 (and fold change>1.3) was used to define a significantly differentially expressed probeset. The probesets were then mapped to our a priori list of probesets in pathways of interest. The rates of differential expression (DE) in these pathways were compared to the rates of DE amongst non-pathway probesets. Gene-set analysis, a permutation-based approach was used to compute p-values for the pre-defined gene sets (25).
We compared the direction and magnitude of expression values from the vaccine recipients to that from four placebos to ensure that effects seen in vaccinated women were specific and not observed among placebo controls. Since the number of placebo recipients was small, we relied on the magnitude of effect rather than statistical significance to determine the specificity of findings among vaccinated women.
For the exploratory analyses, functional annotation and biological term enrichment analysis of regulated probesets were performed by using the DAVID knowledge base (40). This approach assumes independence under the null hypothesis.
Expression values for PBMCs incubated in culture media were evaluated before and after vaccination for background measurements. Gene expression values of VLP-treated cells were corrected for background by subtracting results from unstimulated cells. Paired t-tests were conducted to compare expression levels for each probeset before and after vaccination. All probesets that were differentially expressed at a significance level of p<0.05 (and fold change >1.3) were considered, including the 113 probesets that were significant at p<0.001 (and fold change >1.3). Since the purpose of this post-hoc evaluation was of exploratory nature, we used a high p-value threshold to define probesets of interest and did not adjust for multiple comparisons. Functional annotation of the differentially expressed probesets was performed using DAVID version 2.0 (41).
In order to determine if there is an association between gene expression and antibody levels for differentially expressed probesets, we calculated the Spearman correlation of the difference in expression levels post-pre vaccination (corrected for background, as summarized above) with neutralizing antibody titers at months 2, 7, and 12.
Results
Evaluation of Differential Response to HPV-16 L1 VLP Stimulation Induced by Vaccination
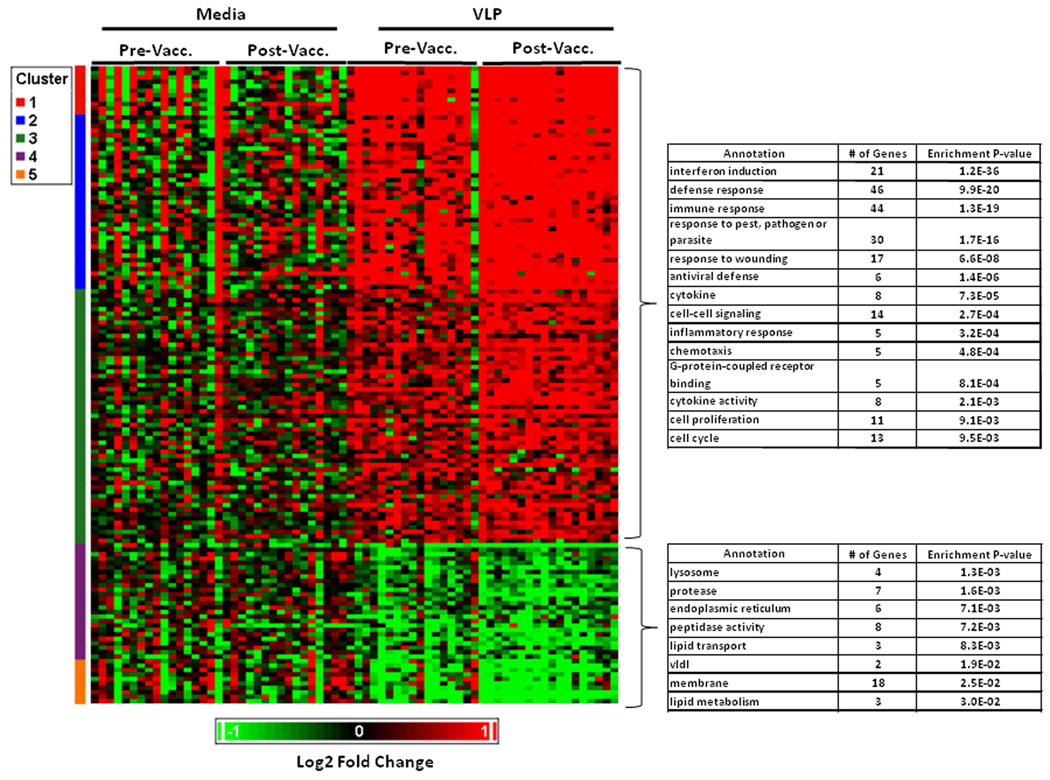
The main aim of our study was to determine changes in gene expression that are associated with HPV vaccination. To this end, we compared gene expression levels in PBMC from vaccine recipients obtained before vaccination and one month after the second vaccine dose (month 2). After filtering, we observed that overall 128 of 7145 probesets (1.8%) were found to be significantly differentially expressed after vaccination (p<0.001). Among these, 113 probesets met the fold-change criteria (fold-change >1.3). Of these, 63 (56%) were up-regulated and 50 (44%) were down-regulated. None of these probesets were significantly differentially expressed in baculovirus-treated cultures following vaccination, when compared with levels in baculovirus-treated pre-vaccination samples (all p-values were > 0.001, data not shown). Also, with the exception of probeset PLA2G4B, none of these probesets demonstrated evidence of differential expression of comparable magnitude among the placebo subjects evaluated (data not shown).
Forty-eight of the 113 significantly differentiated probesets mapped within the 2345 probesets identified within the six pathways targeted for our a priori analysis (cell cycle, cytokines, defense, inflammation, interferon response and cell signaling). As shown in Table I, the percentage of differentially expressed probesets was significantly increased in the cell cycle (1.7 fold, p<0.001), cytokine (2.3-fold, p=0.002), defense (2.7-fold, p=0.002), inflammation (3.0-fold, p=0.004) and interferon (4.6-fold, p=0.037) pathways, when compared to rates of differential expression among the 4800 probesets that did not map to probesets within a priori pathways (1.4%). The lists of significantly differentially expressed probesets (p < 0.001) within these a priori pathways are presented in Table II A–E. The most significant upregulated probesets were RRM2 (cell cycle pathway), CTSC (defense pathway), IFNG (cytokine and defense pathways), RFC3 (cell cycle pathway) and INDO (defense and interferon pathways), and the most significant downregulated probesets were TSC1 (cell cycle pathway), CD14 (defense and inflammation pathways), ADORA2B and DPP4 (defense pathway), as well as GRN (cytokine pathway) and DPP4 (defense pathway).
Table I.
Frequency of Differentially Expressed (DE) Probe-Sets within A-Priori Defined Biological Pathways and Enrichment Ratio Compared to Probe-Sets not within A-Priori Defined Pathways
| A-Priori Pathway | % DE probe-sets in pathway (# DE probe-sets/# total probe-sets in pathway) | DE enrichment ratio (a-priori pathway/not in a priori pathways**) | p-value* | # Up-/down-regulated probe sets |
|---|---|---|---|---|
| Complete Array** | 1.35 (65/4800) | 34 / 30 | ||
| Cell Cycle | 2.29 (12/525) | 1.69 | <0.001 | 8 / 4 |
| Cytokine | 3.11 (9/289) | 2.30 | 0.002 | 7 / 2 |
| Defense | 3.63 (22/606) | 2.68 | 0.002 | 15 / 7 |
| Inflammation | 4.00 (6/150) | 2.95 | 0.004 | 3 / 3 |
| Interferon | 6.25 (2/32) | 4.62 | 0.037 | 2 / 0 |
| Signal transduction | 1.78 (28/1577) | 1.31 | 0.920 | 11 / 17 |
multiple comparison p-value computed using a bootstrap method as described in the Materials and Methods Section. p<0.05 is considered significant
Excluding probe-sets mapping to a-priori pathways listed in the table. Note there is overlap of probe-sets across pathways.
Table II.
| Table II A. List of Genes Demonstrating Significant DE within the Cell Cycle Pathway | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | AffyID | Fold-Change | p-value |
| Up-regulated genes (expression higher post-vaccination than pre-vaccination) | ||||
| RRM2 | ribonucleotide reductase M2 polypeptide | 209773_s_at | 2.06 | 0.000002 |
| RFC3 | replication factor C (activator 1) 3, 38kDa | 204127_at | 1.78 | 0.000022 |
| PMS2 /// PMS2CL | PMS2 postmeiotic segregation increased 2 (S. cerevisiae) | 209805_at | 1.93 | 0.000257 |
| PRC1 | protein regulator of cytokinesis 1 | 218009_s_at | 1.92 | 0.000321 |
| CCNB2 | cyclin B2 | 202705_at | 1.66 | 0.000377 |
| CCND2 | cyclin D2 | 200953_s_at | 1.42 | 0.000378 |
| GINS2 | DNA replication complex GINS protein PSF2 | 221521_s_at | 3.50 | 0.000599 |
| BUB1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | 209642_at | 2.16 | 0.000754 |
| Down-regulated genes (expression lower post-vaccination than pre-vaccination) | ||||
| TSC1 | tuberous sclerosis 1 | 209390_at | −1.36 | 0.000023 |
| LCK | lymphocyte-specific protein tyrosine kinase | 204891_s_at | −1.31 | 0.000244 |
| CDC25B | cell division cycle 25B | 201853_s_at | −1.36 | 0.000428 |
| ARHGEF1 | Rho guanine nucleotide exchange factor (GEF) 1 | 203055_s_at | −1.36 | 0.000724 |
| Table II B. List of Genes Demonstrating Significant DE within the Cytokine Pathway | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | AffyID | Fold-Change | p-value |
| Up-regulated genes (expression higher post-vaccination than pre-vaccination) | ||||
| IFNG | interferon, gamma | 210354_at | 2.57 | 0.000022 |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 210163_at | 2.39 | 0.000086 |
| IL6 | interleukin 6 (interferon, beta 2) | 205207_at | 2.18 | 0.000295 |
| IL3 | interleukin 3 (colony-stimulating factor, multiple) | 207906_at | 4.03 | 0.000303 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 203915_at | 1.91 | 0.000507 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | 207113_s_at | 1.81 | 0.000638 |
| IL15 | interleukin 15 | 205992_s_at | 1.32 | 0.000979 |
| Down-regulated genes (expression lower post-vaccination than pre-vaccination) | ||||
| GRN | granulin | 211284_s_at | −1.52 | 0.000082 |
| CSF3R | colony stimulating factor 3 receptor (granulocyte) | 203591_s_at | −1.44 | 0.000355 |
| Table II C. List of Genes Demonstrating Significant DE within the Defense Pathway | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | AffyID | Fold-Change | p-value |
| Up-regulated genes (expression higher post-vaccination than pre-vaccination) | ||||
| CTSC | cathepsin C | 201487_at | 1.52 | 0.000006 |
| IFNG | interferon, gamma | 210354_at | 2.57 | 0.000022 |
| INDO | indoleamine-pyrrole 2,3 dioxygenase | 210029_at | 2.47 | 0.000058 |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 210163_at | 2.39 | 0.000086 |
| KIR2DS5 | killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 5 | 208203_x_at | 2.11 | 0.000110 |
| ADAR | adenosine deaminase, RNA-specific | 201786_s_at | 1.37 | 0.000249 |
| IL6 | interleukin 6 (interferon, beta 2) | 205207_at | 2.18 | 0.000295 |
| IL3 | interleukin 3 (colony-stimulating factor, multiple) | 207906_at | 4.03 | 0.000303 |
| AICDA | activation-induced cytidine deaminase | 219841_at | 2.91 | 0.000333 |
| SLAMF1 | signaling lymphocytic activation molecule family member 1 | 206181_at | 1.42 | 0.000468 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 203915_at | 1.91 | 0.000507 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | 207113_s_at | 1.81 | 0.000638 |
| GZMA | granzyme A (granzyme 1, CTL-associated serine esterase 3) | 205488_at | 1.59 | 0.000755 |
| C1QB | complement component 1, q subcomponent, beta polypeptide | 202953_at | 1.59 | 0.000775 |
| IL15 | interleukin 15 | 205992_s_at | 1.32 | 0.000979 |
| Down-regulated genes (expression lower post-vaccination than pre-vaccination) | ||||
| CD14 | CD14 antigen | 201743_at | −2.46 | 0.000031 |
| ADORA2B | adenosine A2b receptor | 205891_at | −1.69 | 0.000077 |
| DPP4 | dipeptidylpeptidase 4 (CD26, adenosine deaminase complexing protein 2) | 203717_at | −1.35 | 0.000241 |
| LY86 | lymphocyte antigen 86 | 205859_at | −1.45 | 0.000340 |
| CSF3R | colony stimulating factor 3 receptor (granulocyte) | 203591_s_at | −1.44 | 0.000355 |
| PLA2G4B | phospholipase A2, group IVB (cytosolic) | 60528_at | −1.34 | 0.000546 |
| FCGRT | Fc fragment of IgG, receptor, transporter, alpha | 218831_s_at | −1.68 | 0.000678 |
| Table II D. List of Genes Demonstrating Significant DE within the Inflammation Pathway | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | AffyID | Fold-Change | p-value |
| Up-regulated genes (expression higher post-vaccination than pre-vaccination) | ||||
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 210163_at | 2.39 | 0.000086 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 203915_at | 1.91 | 0.000507 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | 207113_s_at | 1.81 | 0.000638 |
| Down-regulated genes (expression lower post-vaccination than pre-vaccination) | ||||
| CD14 | CD14 antigen | 201743_at | −2.46 | 0.000031 |
| LY86 | lymphocyte antigen 86 | 205859_at | −1.45 | 0.000340 |
| PLA2G4B | phospholipase A2, group IVB (cytosolic) | 60528_at | −1.34 | 0.000546 |
| Table 2E. List of Genes Demonstrating Significant DE within the Interferon Pathway | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | AffyID | Fold-Change | p-value |
| Up-regulated genes (expression higher post-vaccination than pre-vaccination) | ||||
| INDO | indoleamine-pyrrole 2,3 dioxygenase | 210029_at | 2.47 | 0.0000584 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 203915_at | 1.91 | 0.0005074 |
(VLP-Media) expression levels as defined in the Materials and Methods Section
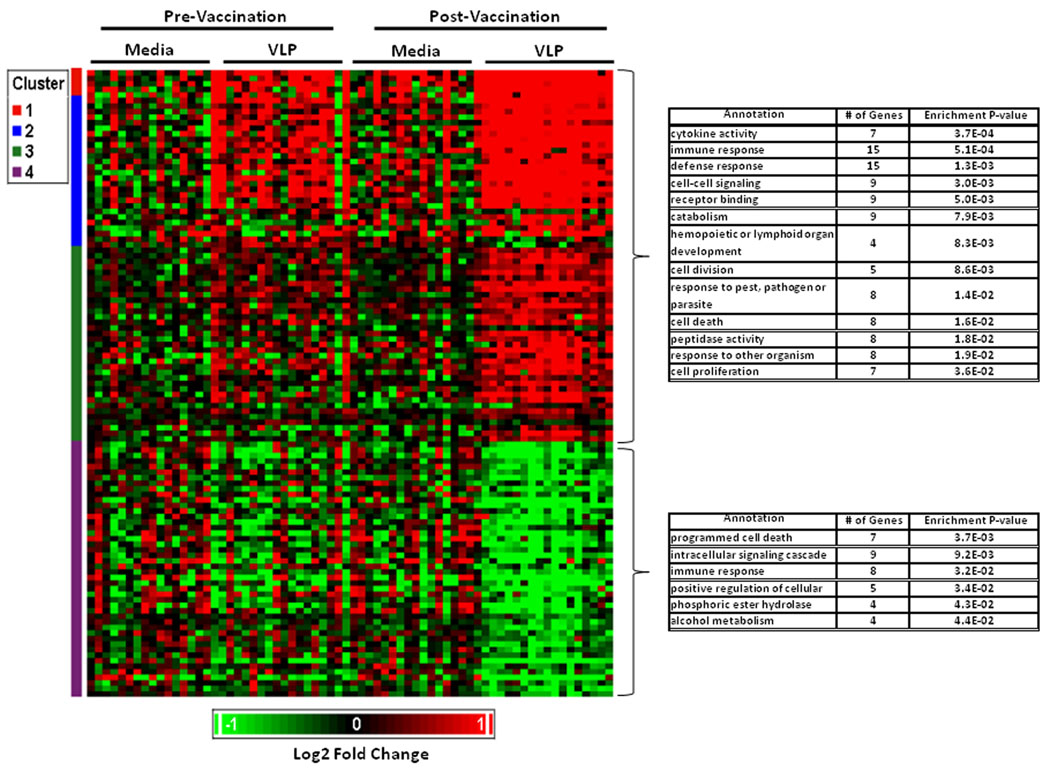
A similar list for the pathway that did not have significant evidence for enrichment in our a priori evaluation (cell signaling) is presented in Supplementary Table I. The list of significantly differentially expressed probesets not within one of our a priori pathways is shown in Supplementary Table II A–B. A summary of overall gene expression profiles induced by vaccination (p<0.001) is summarized in Figure 1. Biological term enrichment analysis was performed using DAVID. In addition to the above a priori pathways, the following were found to have evidence for enrichment. Upregulated pathways: Receptor binding (9 probesets), catabolism (9 probesets), hemopoietic or lymphoid cell development (4 probesets), cell death (8 probesets) and peptidase activity (8 probesets). Downregulated pathways: programmed cell death (7 probesets), phosphoric ester hydrolase (4 probesets) and alcohol metabolism (4 probesets).
Figure 1. Expression of profiles of probesets significantly modulated by VLP in PBMC from vaccine recipients before and after vaccination with HPV-16 L1 VLP.
PBMC from vaccine recipients were incubated with HPV-16 L1 VLP as indicated in Materials and Methods. Functional annotation of probesets differentially expressed (p<0.001) was done using DAVID. Upregulated genes are shown in red and downregulated genes in green. Each row represents a probset and each column represents an individual.
For our exploratory analysis, we expanded the evaluation to probesets differentially expressed at a p<0.05 cutoff level, 624 of 7145 probesets (8.7%) were found to be differentially expressed (330 up-regulated, and 294 down-regulated; Table III).
Table III.
List of DE Genes After Vaccination Using p<0.05 as Cutoff
| Affy ID | F.C. post vs. pre |
p value | Gene Symbol | Gene Name | Affy ID | F.C. post vs. pre |
p value | Gene Symbol |
Gene Name |
|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated Genes | Down-Regulated Genes | ||||||||
| 209773_s_at | 2.06 | 0.000002 | RRM2 | ribonucleotide reductase m2 polypeptide | 201005_at | −1.82 | 0.00000007 | CD9 | cd9 antigen (p24) |
| 208921_s_at | 1.48 | 0.000004 | SRI | sorcin | 201029_s_at | −1.30 | 0.000002 | CD99 | cd99 antigen |
| 201487_at | 1.52 | 0.000006 | CTSC | cathepsin c | 205695_at | −2.51 | 0.000009 | SDS | serine dehydratase |
| 200039_s_at | 1.34 | 0.000010 | PSMB2 | proteasome (prosome, macropain) subunit, beta type, 2 | 203509_at | −1.71 | 0.000012 | SORL1 | sortilin-related receptor, l(dlr class) a repeats-containing |
| 210164_at | 2.33 | 0.000015 | GZMB | granzyme b (granzyme 2, cytotoxic t-lymphocyte-associated serine esterase 1) | 201591_s_at | −1.51 | 0.000020 | NISCH | nischarin |
| 210354_at | 2.57 | 0.000022 | IFNG | interferon, gamma | 209390_at | −1.36 | 0.000023 | TSC1 | tuberous sclerosis 1 |
| 204127_at | 1.78 | 0.000022 | RFC3 | replication factor c (activator 1) 3, 38kda | 221558_s_at | −1.36 | 0.000026 | LEF1 | lymphoid enhancer-binding factor 1 |
| 204777_s_at | 1.65 | 0.000030 | MAL | mal, t-cell differentiation protein | 201743_at | −2.46 | 0.000031 | CD14 | cd14 antigen |
| 217848_s_at | 1.86 | 0.000048 | PPA1 | pyrophosphatase (inorganic) 1 | 203979_at | −2.14 | 0.000032 | CYP27A1 | cytochrome p450, family 27, subfamily a, polypeptide 1 |
| 201274_at | 1.39 | 0.000049 | PSMA5 | proteasome (prosome, macropain) subunit, alpha type, 5 | 204647_at | −1.99 | 0.000056 | HOMER3 | homer homolog 3 (drosophila) |
| 205404_at | 3.49 | 0.000052 | HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 | 205891_at | −1.69 | 0.000077 | ADORA2B | adenosine a2b receptor |
| 210029_at | 2.47 | 0.000058 | INDO | indoleamine-pyrrole 2,3 dioxygenase | 205960_at | −2.80 | 0.000082 | PDK4 | pyruvate dehydrogenase kinase, isozyme 4 |
| 219210_s_at | 1.40 | 0.000063 | RAB8B | rab8b, member ras oncogene family | 211284_s_at | −1.52 | 0.000082 | GRN | granulin |
| 205582_s_at | 3.90 | 0.000065 | GGTLA1 | gamma-glutamyltransferase-like activity 1 | 203695_s_at | −1.82 | 0.000089 | DFNA5 | deafness, autosomal dominant 5 |
| 216268_s_at | 1.51 | 0.000077 | JAG1 | jagged 1 (alagille syndrome) | 214198_s_at | −1.34 | 0.000101 | DGCR2 | kiaa0163 gene product |
| 202760_s_at | 1.58 | 0.000082 | AKAP2 /// PALM2-AKAP2 | a kinase (prka) anchor protein 2 | 204134_at | −2.87 | 0.000153 | PDE2A | phosphodiesterase 2a, cgmp-stimulated |
| 210163_at | 2.39 | 0.000086 | CXCL11 | chemokine (c-x-c motif) ligand 11 | 222240_s_at | −1.36 | 0.000158 | ISYNA1 | myo-inositol 1-phosphate synthase a1 |
| 203471_s_at | 1.40 | 0.000087 | PLEK | pleckstrin | 203382_s_at | −2.95 | 0.000183 | APOE | apolipoprotein e |
| 202941_at | 1.34 | 0.000092 | NDUFV2 | nadh dehydrogenase (ubiquinone) flavoprotein 2, 24kda | 204906_at | −1.36 | 0.000190 | RPS6KA2 | ribosomal protein s6 kinase, 90kda, polypeptide 2 |
| 209765_at | 1.77 | 0.000096 | ADAM19 | adam metallopeptidase domain 19 (meltrin beta) | 52940_at | −1.52 | 0.000204 | SIGIRR | single immunoglobulin and toll-interleukin 1 receptor (tir) domain |
| 208203_x_at | 2.11 | 0.000110 | KIR2DS5 | killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 2 | 219889_at | −1.41 | 0.000207 | FRAT1 | frequently rearranged in advanced t-cell lymphomas |
| 212581_x_at | 1.34 | 0.000120 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 209230_s_at | −2.44 | 0.000232 | P8 | p8 protein (candidate of metastasis 1) |
| 202638_s_at | 1.50 | 0.000145 | ICAM1 | intercellular adhesion molecule 1 (cd54), human rhinovirus receptor | 209831_x_at | −1.35 | 0.000235 | DNASE2 | deoxyribonuclease ii, lysosomal |
| 208799_at | 1.34 | 0.000145 | PSMB5 | proteasome (prosome, macropain) subunit, beta type, 5 | 203717_at | −1.35 | 0.000241 | DPP4 | dipeptidyl-peptidase 4 (cd26, adenosine deaminase complexing protein 2) |
| 218755_at | 4.62 | 0.000152 | KIF20A | kinesin family member 20a | 204891_s_at | −1.31 | 0.000244 | LCK | lymphocyte-specific protein tyrosine kinase |
| 203217_s_at | 1.35 | 0.000152 | ST3GAL5 | st3 beta-galactoside alpha-2,3-sialyltransferase 5 | 202054_s_at | −1.48 | 0.000256 | ALDH3A2 | aldehyde dehydrogenase 3 family, member a2 |
| 201037_at | 1.52 | 0.000167 | PFKP | phosphofructokinase, platelet | 219697_at | −2.64 | 0.000260 | HS3ST2 | heparan sulfate (glucosamine) 3-o-sulfotransferase 2 |
| 205692_s_at | 1.92 | 0.000181 | CD38 | cd38 antigen (p45) | 208626_s_at | −1.30 | 0.000269 | VAT1 | vesicle amine transport protein 1 homolog (t californica) |
| 217771_at | 3.13 | 0.000200 | GOLPH2 | golgi phosphoprotein 2 | 204466_s_at | −1.58 | 0.000286 | SNCA | synuclein, alpha (non a4 component of amyloid precursor) |
| 213011_s_at | 1.32 | 0.000211 | TPI1 | triosephosphate isomerase 1 | 205859_at | −1.45 | 0.000340 | LY86 | lymphocyte antigen 86 |
| 201761_at | 1.42 | 0.000226 | MTHFD2 | methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | 203591_s_at | −1.44 | 0.000355 | CSF3R | colony stimulating factor 3 receptor (granulocyte) |
| 202589_at | 1.75 | 0.000226 | TYMS | thymidylate synthetase | 221872_at | −2.32 | 0.000403 | RARRES1 | retinoic acid receptor responder (tazarotene induced) 1 |
| 202352_s_at | 1.35 | 0.000231 | PSMD12 | proteasome (prosome, macropain) 26s subunit, non-atpase, 12 | 219445_at | −1.58 | 0.000404 | GLTSCR1 | glioma tumor suppressor candidate region gene 1 |
| 201323_at | 1.43 | 0.000244 | EBNA1BP2 | ebna1 binding protein 2 | 204867_at | −1.75 | 0.000408 | GCHFR | gtp cyclohydrolase i feedback regulator |
| 209146_at | 1.39 | 0.000247 | SC4MOL | sterol-c4-methyl oxidase-like | 201853_s_at | −1.36 | 0.000428 | CDC25B | cell division cycle 25b |
| 201786_s_at | 1.37 | 0.000249 | ADAR | adenosine deaminase, rna-specific | 200878_at | −1.59 | 0.000443 | EPAS1 | endothelial pas domain protein 1 |
| 209805_at | 1.93 | 0.000257 | PMS2 /// PMS2CL | pms2 postmeiotic segregation increased 2 (s. cerevisiae), pms2-c terminal -like | 220776_at | −1.89 | 0.000452 | KCNJ14 | potassium inwardly-rectifying channel, subfamily j, member 14 |
| 205569_at | 1.66 | 0.000283 | LAMP3 | lysosomal-associated membrane protein 3 | 201278_at | −1.52 | 0.000496 | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (drosophila) |
| 205207_at | 2.18 | 0.000295 | IL6 | interleukin 6 (interferon, beta 2) | 209122_at | −1.60 | 0.000511 | ADFP | adipose differentiation-related protein |
| 207906_at | 4.03 | 0.000303 | IL3 | interleukin 3 (colony-stimulating factor, multiple) | 200766_at | −1.54 | 0.000544 | CTSD | cathepsin d (lysosomal aspartyl peptidase) |
| 218009_s_at | 1.92 | 0.000321 | PRC1 | protein regulator of cytokinesis 1 | 60528_at | −1.34 | 0.000546 | PLA2G4B | phospholipase a2, group ivb (cytosolic) |
| 219841_at | 2.91 | 0.000333 | AICDA | activation-induced cytidine deaminase | 203665_at | −1.61 | 0.000564 | HMOX1 | heme oxygenase (decycling) 1 |
| 214210_at | 1.79 | 0.000349 | SLC25A17 | solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kda), member 17 | 218831_s_at | −1.68 | 0.000678 | FCGRT | fc fragment of igg, receptor, transporter, alpha |
| 202705_at | 1.66 | 0.000377 | CCNB2 | cyclin b2 | 212774_at | −1.54 | 0.000688 | ZNF238 | zinc finger protein 238 |
| 200953_s_at | 1.42 | 0.000378 | CCND2 | cyclin d2 | 202500_at | −1.50 | 0.000695 | DNAJB2 | dnaj (hsp40) homolog, subfamily b, member 2 |
| 201317_s_at | 1.41 | 0.000395 | PSMA2 | proteasome (prosome, macropain) subunit, alpha type, 2 | 203055_s_at | −1.36 | 0.000724 | ARHGEF1 | rho guanine nucleotide exchange factor (gef) 1 |
| 203376_at | 1.34 | 0.000400 | CDC40 | cell division cycle 40 homolog (yeast) | 214780_s_at | −1.42 | 0.000801 | MYO9B | myosin ixb |
| 212185_x_at | 1.72 | 0.000417 | MT2A | metallothionein 2a | 219113_x_at | −1.51 | 0.000846 | DHRS10 | dehydrogenase/reductase (sdr family) member 10 |
| 206181_at | 1.42 | 0.000468 | SLAMF1 | signaling lymphocytic activation molecule family member 1 | 204638_at | −1.39 | 0.000861 | ACP5 | acid phosphatase 5, tartrate resistant |
| 203915_at | 1.91 | 0.000507 | CXCL9 | chemokine (c-x-c motif) ligand 9 | 218855_at | −1.51 | 0.000959 | GPR175 | seven transmembrane domain orphan receptor |
| 208581_x_at | 1.94 | 0.000511 | MT1X | metallothionein 1x | 204046_at | −1.42 | 0.001080 | PLCB2 | phospholipase c, beta 2 |
| 209803_s_at | 1.84 | 0.000566 | PHLDA2 | pleckstrin homology-like domain, family a, member 2 | 221601_s_at | −1.41 | 0.001082 | FAIM3 | fas apoptotic inhibitory molecule 3 |
| 205633_s_at | 1.60 | 0.000591 | ALAS1 | aminolevulinate, delta-, synthase 1 | 40225_at | −1.34 | 0.001110 | GAK | cyclin g associated kinase |
| 221521_s_at | 3.50 | 0.000599 | GINS2 | dna replication complex gins protein psf2 | 201185_at | −1.53 | 0.001140 | HTRA1 | htra serine peptidase 1 |
| 204962_s_at | 1.76 | 0.000638 | CENPA | centromere protein a, 17kda | 221579_s_at | −1.33 | 0.001161 | NUDT3 | diphosphoinositol polyphosphate phosphohydrolase |
| 207113_s_at | 1.81 | 0.000638 | TNF | tumor necrosis factor (tnf superfamily, member 2) | 221675_s_at | −1.48 | 0.001196 | CHPT1 | choline phosphotransferase 1 |
| 205126_at | 1.45 | 0.000693 | VRK2 | vaccinia related kinase 2 | 217963_s_at | −1.63 | 0.001204 | NGFRAP1 | nerve growth factor receptor (tnfrsf16) associated protein 1 |
| 209642_at | 2.16 | 0.000754 | BUB1 | bub1 budding uninhibited by benzimidazoles 1 homolog (yeast) | 218555_at | −1.47 | 0.001284 | ANAPC2 | anaphase promoting complex subunit 2 |
| 205488_at | 1.59 | 0.000755 | GZMA | granzyme a (granzyme 1, cytotoxic t-lymphocyte-associated serine esterase 3) | 219452_at | −2.48 | 0.001417 | DPEP2 | dipeptidase 2 |
| 202953_at | 1.59 | 0.000775 | C1QB | complement component 1, q subcomponent, b chain | 35626_at | −1.32 | 0.001587 | SGSH | n-sulfoglucosamine sulfohydrolase (sulfamidase) |
| 209267_s_at | 1.68 | 0.000802 | SLC39A8 | solute carrier family 39 (zinc transporter), member 8 | 201819_at | −1.35 | 0.001596 | SCARB1 | scavenger receptor class b, member 1 |
| 204326_x_at | 1.77 | 0.000952 | MT1X | metallothionein 1× | 211110_s_at | −2.00 | 0.001806 | AR | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; kennedy disease) |
| 205992_s_at | 1.32 | 0.000979 | IL15 | interleukin 15 | 209774_x_at | −1.76 | 0.002054 | CXCL2 | chemokine (c-x-c motif) ligand 2 |
| 205174_s_at | 2.29 | 0.001056 | QPCT | glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | 206608_s_at | −1.36 | 0.002175 | RPGRIP1 | retinitis pigmentosa gtpase regulator interacting protein 1 |
| 219960_s_at | 1.30 | 0.001160 | UCHL5 | ubiquitin carboxyl-terminal hydrolase 15 | 204949_at | −1.34 | 0.002253 | ICAM3 | intercellular adhesion molecule 3 |
| 208864_s_at | 1.34 | 0.001162 | TXN | thioredoxin | 214414_x_at | −1.59 | 0.002301 | HBA2 | hemoglobin, alpha 1 |
| 203213_at | 2.50 | 0.001193 | CDC2 | cell division cycle 2, g1 to s and g2 to m | 219549_s_at | −1.32 | 0.002316 | RTN3 | reticulon 3 |
| 210229_s_at | 3.34 | 0.001253 | CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | 205382_s_at | −1.84 | 0.002559 | CFD | complement factor d (adipsin) |
| 205676_at | 2.38 | 0.001331 | CYP27B1 | cytochrome p450, family 27, subfamily b, polypeptide 1 | 216092_s_at | −1.48 | 0.002565 | SLC7A8 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 |
| 217979_at | 1.51 | 0.001398 | TSPAN13 | tetraspanin 13 | 202449_s_at | −1.31 | 0.002571 | RXRA | retinoid x receptor, alpha |
| 202421_at | 1.59 | 0.001460 | IGSF3 | immunoglobulin superfamily, member 3 | 205090_s_at | −1.30 | 0.002597 | NAGPA | n-acetylglucosamine-1-phosphodiester alpha-n-acetylglucosaminidase |
| 207277_at | 1.65 | 0.001460 | CD209 | cd209 antigen | 218427_at | −1.50 | 0.002685 | SDCCAG3 | serologically defined colon cancer antigen 3 |
| 205505_at | 1.54 | 0.001515 | GCNT1 | glucosaminyl (n-acetyl) transferase 1, core 2 (beta-1,6-n-acetylglucosaminyltransferase) | 207105_s_at | −1.49 | 0.002882 | PIK3R2 | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) |
| 211269_s_at | 1.51 | 0.001562 | IL2RA | interleukin 2 receptor, alpha | 212346_s_at | −1.38 | 0.002913 | MXD4 | max dimerization protein 4 |
| 220358_at | 2.21 | 0.001584 | SNFT | jun dimerization protein p21snft | 212552_at | −1.32 | 0.002917 | HPCAL1 | hippocalcin-like 1 |
| 209825_s_at | 1.57 | 0.001612 | UCK2 | uridine-cytidine kinase 2 | 201427_s_at | −4.80 | 0.002990 | SEPP1 | selenoprotein p, plasma, 1 |
| 207900_at | 2.43 | 0.001637 | CCL17 | chemokine (c-c motif) ligand 17 | 209409_at | −1.50 | 0.002998 | GRB10 | growth factor receptor-bound protein 10 |
| 205890_s_at | 2.17 | 0.001649 | GABBR1 /// UBD | ubiquitin d | 207574_s_at | −1.31 | 0.003015 | GADD45B | growth arrest and dna-damage-inducible, beta |
| 201897_s_at | 1.45 | 0.001660 | CKS1B | cdc28 protein kinase regulatory subunit 1b | 200710_at | −1.38 | 0.003071 | ACADVL | acyl-coenzyme a dehydrogenase, very long chain |
| 203200_s_at | 1.36 | 0.001691 | MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 206471_s_at | −1.58 | 0.003112 | PLXNC1 | plexin c1 |
| 201013_s_at | 1.33 | 0.001713 | PAICS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 205466_s_at | −2.39 | 0.003130 | HS3ST1 | heparan sulfate (glucosamine) 3-o-sulfotransferase 1 |
| 221331_x_at | 2.49 | 0.001713 | CTLA4 | cytotoxic t-lymphocyte-associated protein 4 | 210980_s_at | −1.39 | 0.003134 | ASAH1 | n-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| 37145_at | 1.31 | 0.001870 | GNLY | granulysin | 200965_s_at | −1.37 | 0.003340 | ABLIM1 | actin binding lim protein 1 |
| 207849_at | 3.91 | 0.001871 | IL2 | interleukin 2 | 204955_at | −1.40 | 0.003354 | SRPX | sushi-repeat-containing protein, x-linked |
| 202675_at | 1.35 | 0.001876 | SDHB | succinate dehydrogenase complex, subunit b, iron sulfur (ip) | 202481_at | −1.31 | 0.003365 | DHRS3 | dehydrogenase/reductase (sdr family) member 3 |
| 202688_at | 1.55 | 0.001887 | TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | 205182_s_at | −1.86 | 0.003400 | ZNF324 | zinc finger protein 324 |
| 204924_at | 1.36 | 0.001944 | TLR2 | toll-like receptor 2 | 205683_x_at, 207741_x_at |
−2.07 | 0.003403 | TPSAB1 | tryptase, alpha |
| 201157_s_at | 1.34 | 0.002035 | NMT1 | n-myristoyltransferase 1 | 207826_s_at | −1.69 | 0.003407 | ID3 | inhibitor of dna binding 3, dominant negative helix-loop-helix protein |
| 209610_s_at | 1.49 | 0.002047 | SLC1A4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 221748_s_at | −1.41 | 0.003427 | TNS1 | tensin 1 |
| 210001_s_at | 1.60 | 0.002354 | SOCS1 | suppressor of cytokine signaling 1 | 221210_s_at | −1.36 | 0.003450 | NPL | n-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| 33304_at | 1.46 | 0.002364 | ISG20 | interferon stimulated exonuclease gene 20kda | 204446_s_at | −1.45 | 0.003510 | ALOX5 | arachidonate 5-lipoxygenase |
| 201625_s_at | 1.57 | 0.002371 | INSIG1 | insulin induced gene 1 | 206368_at | −3.15 | 0.003566 | CPLX2 | complexin 2 |
| 203097_s_at | 1.33 | 0.002475 | RAPGEF2 | rap guanine nucleotide exchange factor (gef) 2 | 206644_at | −2.17 | 0.003579 | NR0B1 | nuclear receptor subfamily 0, group b, member 1 |
| 204279_at | 1.39 | 0.002485 | PSMB9 | proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) | 209782_s_at | −1.66 | 0.003585 | DBP | d site of albumin promoter (albumin d-box) binding protein |
| 204929_s_at | 1.66 | 0.002546 | VAMP5 | vesicle-associated membrane protein 5 (myobrevin) | 203803_at | −3.43 | 0.003661 | PCYOX1 | prenylcysteine oxidase 1 |
| 205159_at | 1.40 | 0.002596 | CSF2RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | 205728_at | −2.76 | 0.003665 | ODZ1 | odz, odd oz/ten-m homolog 1(drosophila) |
| 204033_at | 1.61 | 0.002694 | TRIP13 | thyroid hormone receptor interactor 13 | 209355_s_at | −1.80 | 0.003724 | PPAP2B | phosphatidic acid phosphatase type 2b |
| 207500_at | 2.31 | 0.002995 | CASP5 | caspase 5, apoptosis-related cysteine peptidase | 202790_at | −2.17 | 0.003926 | CLDN7 | claudin 7 |
| 204103_at | 1.47 | 0.002996 | CCL4 | chemokine (c-c motif) ligand 4 | 213553_x_at | −1.83 | 0.004021 | APOC1 | apolipoprotein c-i |
| 205220_at | 2.72 | 0.003027 | GPR109B | g protein-coupled receptor 109b | 220491_at | −1.57 | 0.004027 | HAMP | hepcidin antimicrobial peptide |
| 216834_at | 2.24 | 0.003485 | RGS1 | regulator of g-protein signalling 1 | 204359_at | −1.83 | 0.004188 | FLRT2 | fibronectin leucine rich transmembrane protein 2 |
| 206745_at | 1.80 | 0.003645 | HOXC11 | homeobox c11 | 217807_s_at | −1.48 | 0.004252 | GLTSCR2 | glioma tumor suppressor candidate region gene 2 |
| 204254_s_at | 1.58 | 0.003671 | VDR | vitamin d (1,25- dihydroxyvitamin d3) receptor | 219952_s_at | −1.51 | 0.004269 | MCOLN1 | mucolipin 1 |
| 200054_at | 1.34 | 0.003724 | ZNF259 | zinc finger protein 259 | 214369_s_at | −1.36 | 0.004357 | RASGRP2 | ras guanyl releasing protein 2 (calcium and dag-regulated) |
| 202267_at | 2.48 | 0.003954 | LAMC2 | laminin, gamma 2 | 209616_s_at | −1.96 | 0.004451 | CES1 | carboxylesterase 1 (monocyte/macrophage serine esterase 1) |
| 207952_at | 4.12 | 0.003994 | IL5 | interleukin 5 (colony-stimulating factor, eosinophil) | 203167_at | −1.53 | 0.004531 | TIMP2 | timp metallopeptidase inhibitor 2 |
| 208450_at | 3.03 | 0.004025 | LGALS2 | lectin, galactoside-binding, soluble, 2 (galectin 2) | 218718_at | −1.74 | 0.004611 | PDGFC | spinal cord-derived growth factor; secretory growth factor-like protein fallotein |
| 218239_s_at | 1.38 | 0.004091 | GTPBP4 | gtp binding protein 4 | 205222_at | −2.53 | 0.004683 | EHHADH | enoyl-coenzyme a, hydratase/3-hydroxyacyl coenzyme a dehydrogenase |
| 210511_s_at | 5.01 | 0.004176 | INHBA | inhibin, beta a (activin a, activin ab alpha polypeptide) | 204394_at | −1.37 | 0.004684 | SLC43A1 | solute carrier family 43, member 1 |
| 210015_s_at | 2.32 | 0.004275 | MAP2 | microtubule-associated protein 2 | 40569_at | −1.30 | 0.004743 | ZNF42 | zinc finger protein 42 (myeloid-specific retinoic acid-responsive) |
| 201292_at | 2.09 | 0.004316 | TOP2A | topoisomerase (dna) ii alpha 170kda | 219267_at | −1.41 | 0.004851 | GLTP | glycolipid transfer protein |
| 202847_at | 1.30 | 0.004351 | PCK2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) | 206339_at | −1.71 | 0.004861 | CART | cocaine- and amphetamine-regulated transcript |
| 206632_s_at | 1.59 | 0.004372 | APOBEC3B | apolipoprotein b mrna editing enzyme, catalytic polypeptide-like 3b | 221364_at | −2.28 | 0.004933 | GRID2 | glutamate receptor, ionotropic, delta 2 |
| 208002_s_at | 1.43 | 0.004425 | ACOT7 | acyl-coa thioesterase 7 | 214132_at | −1.36 | 0.004975 | ATP5C1 | atp synthase, h+ transporting, mitochondrial f1 complex, gamma polypeptide 1 |
| 202252_at | 1.33 | 0.004492 | RAB13 | rab13, member ras oncogene family | 202391_at | −1.37 | 0.005083 | BASP1 | brain abundant, membrane attached signal protein 1 |
| 205242_at | 3.37 | 0.004539 | CXCL13 | chemokine (c-x-c motif) ligand 13 (b-cell chemoattractant) | 209990_s_at | −2.51 | 0.005271 | GABBR2 | gamma-aminobutyric acid (gaba) b receptor, 2 |
| 206513_at | 1.37 | 0.004563 | AIM2 | absent in melanoma 2 | 207345_at | −1.94 | 0.005697 | FST | follistatin |
| 204580_at | 2.69 | 0.004759 | MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 214743_at | −1.35 | 0.005728 | CUTL1 | cut-like 1, ccaat displacement protein (drosophila) |
| 220865_s_at | 1.37 | 0.004829 | PDSS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 | 213812_s_at | −1.38 | 0.006040 | CAMKK2 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| 219761_at | 2.14 | 0.004855 | CLEC1A | c-type lectin domain family 1, member a | 205486_at | −1.33 | 0.006059 | TESK2 | testis-specific kinase 2 |
| 206749_at | 2.91 | 0.004909 | CD1B | cd1b antigen | 203136_at | −1.60 | 0.006081 | RABAC1 | rab acceptor 1 (prenylated) |
| 201170_s_at | 1.38 | 0.004946 | BHLHB2 | basic helix-loop-helix domain containing, class b, 2 | 204131_s_at | −1.51 | 0.006086 | FOXO3A | forkhead box o3a |
| 218866_s_at | 1.42 | 0.004968 | POLR3K | polymerase (rna) iii (dna directed) polypeptide k, 12.3 kda | 209158_s_at | −1.35 | 0.006206 | PSCD2 | pleckstrin homology, sec7 and coiled-coil domains 2 (cytohesin–2) |
| 201798_s_at | 1.34 | 0.005042 | FER1L3 | fer-1-like 3, myoferlin (c. elegans) | 221246_x_at, 221748_s_at |
−1.42 | 0.006314 | TNS1 | tensin 1 |
| 200629_at | 1.50 | 0.005046 | WARS | interferon-induced protein 53 | 202108_at | −1.30 | 0.006356 | PEPD | peptidase d |
| 213415_at | 1.90 | 0.005165 | CLIC2 | chloride intracellular channel 2 | 204360_s_at | −1.44 | 0.006377 | NAGLU | n-acetylglucosaminidase, alpha- (sanfilippo disease iiib) |
| 203275_at | 1.32 | 0.005360 | IRF2 | interferon regulatory factor 2 | 217865_at | −1.32 | 0.006628 | RNF130 | ring finger protein 130 |
| 214512_s_at | 1.38 | 0.005550 | SUB1 | sub1 homolog (s. cerevisiae) | 219371_s_at | −1.53 | 0.006725 | KLF2 | kruppel-like factor 2 (lung) |
| 206461_x_at | 1.51 | 0.005684 | MT1H | metallothionein 1h | 202256_at | −1.46 | 0.007049 | CD2BP2 | cd2 antigen (cytoplasmic tail) binding protein 2 |
| 211333_s_at | 1.48 | 0.005789 | FASLG | fas ligand (tnf superfamily, member 6) | 211145_x_at | −2.22 | 0.007309 | IFNA21 | interferon, alpha 21 |
| 210538_s_at | 1.45 | 0.005790 | BIRC3 | baculoviral iap repeat-containing 3 | 221649_s_at | −1.35 | 0.007423 | PPAN | peter pan homolog (drosophila) |
| 204224_s_at | 1.49 | 0.006013 | GCH1 | gtp cyclohydrolase 1 (dopa-responsive dystonia) | 221378_at | −1.31 | 0.007478 | CER1 | cerberus 1, cysteine knot superfamily, homolog (xenopus laevis) |
| 203564_at | 1.52 | 0.006110 | FANCG | fanconi anemia, complementation group g | 206171_at | −1.51 | 0.007505 | ADORA3 | adenosine a3 receptor |
| 207586_at | 2.08 | 0.006245 | SHH | sonic hedgehog homolog (drosophila) | 205206_at | −1.72 | 0.007576 | KAL1 | kallmann syndrome 1 sequence |
| 221463_at | 2.63 | 0.006382 | CCL24 | chemokine (c-c motif) ligand 24 | 203414_at | −1.38 | 0.007979 | MMD | monocyte to macrophage differentiation-associated |
| 204748_at | 2.60 | 0.006552 | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin g/h synthase and cyclooxygenase) | 209541_at | −1.76 | 0.008232 | IGF1 | insulin-like growth factor 1 (somatomedin c) |
| 202743_at | 1.39 | 0.006584 | PIK3R3 | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 221065_s_at | −1.81 | 0.008237 | CHST8 | carbohydrate (n-acetylgalactosamine 4−0) sulfotransferase 8 |
| 209785_s_at | 1.35 | 0.006692 | PLA2G4C | phospholipase a2, group ivc (cytosolic, calcium-independent) | 208610_s_at | −1.41 | 0.008280 | SRRM2 | serine/arginine repetitive matrix 2 |
| 206975_at | 3.99 | 0.006853 | LTA | lymphotoxin alpha (tnf superfamily, member 1) | 213592_at | −1.91 | 0.008299 | AGTRL1 | angiotensin ii receptor-like 1 |
| 202499_s_at | 1.39 | 0.006958 | SLC2A3 | solute carrier family 2 (facilitated glucose transporter), member 3 | 201141_at | −1.44 | 0.008837 | GPNMB | glycoprotein (transmembrane) nmb |
| 201329_s_at | 1.49 | 0.006989 | ETS2 | v-ets erythroblastosis virus e26 oncogene homolog 2 (avian) | 220068_at | −2.11 | 0.009019 | VPREB3 | pre-b lymphocyte gene 3 |
| 204205_at | 1.35 | 0.007064 | APOBEC3G | apolipoprotein b mrna editing enzyme, catalytic polypeptide-like 3g | 201186_at | −1.39 | 0.009581 | LRPAP1 | low density lipoprotein receptor-related protein associated protein 1 |
| 218039_at | 1.35 | 0.007138 | NUSAP1 | nucleolar and spindle associated protein 1 | 204824_at | −1.56 | 0.009682 | ENDOG | endonuclease g |
| 214567_s_at | 1.51 | 0.007306 | XCL1 /// XCL2 | chemokine (c motif) ligand 2, chemokine (c motif) ligand 1 | 220762_s_at | −1.46 | 0.009853 | GNB1L | guanine nucleotide binding protein (g protein), beta polypeptide 1-like |
| 206278_at | 1.46 | 0.007320 | PTAFR | platelet-activating factor receptor | 209215_at | −1.33 | 0.009935 | TETRAN | tetracycline transporter-like protein |
| 204026_s_at | 1.35 | 0.007406 | ZWINT | zw10 interactor | 219963_at | −1.60 | 0.010000 | DUSP13 | dual specificity phosphatase 13 |
| 211138_s_at | 1.42 | 0.007457 | KMO | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 202838_at | −2.05 | 0.010039 | FUCA1 | fucosidase, alpha-l- 1, tissue |
| 201948_at | 1.48 | 0.007689 | GNL2 | guanine nucleotide binding protein-like 2 (nucleolar) | 202477_s_at | −1.36 | 0.010213 | TUBGCP2 | tubulin, gamma complex associated protein 2 |
| 202284_s_at | 1.51 | 0.007786 | CDKN1A | cyclin-dependent kinase inhibitor 1a (p21, cip1) | 206739_at | −2.24 | 0.010378 | HOXC5 | homeobox c5 |
| 202357_s_at | 1.82 | 0.007826 | CFB | complement factor b | 203113_s_at | −1.53 | 0.010380 | EEF1D | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| 214022_s_at | 1.35 | 0.007980 | IFITM1 | interferon induced transmembrane protein 1 (9–27) | 210123_s_at | −2.12 | 0.010514 | CHRNA7 /// CHRFAM7A /// LOC652740 | cholinergic receptor, nicotinic, alpha 7 |
| 203975_s_at | 1.54 | 0.008000 | CHAF1A | chromatin assembly factor 1, subunit a (p150) | 32837_at | −1.34 | 0.010565 | AGPAT2 | 1-acylglycerol-3-phosphate o-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| 206547_s_at | 2.70 | 0.008057 | PPEF1 | protein phosphatase, ef-hand calcium binding domain 1 | 217969_at | −1.34 | 0.010748 | C11orf2 | chromosome 11 open reading frame2 |
| 208075_s_at | 2.35 | 0.008074 | CCL7 | chemokine (c-c motif) ligand 7 | 204862_s_at | −1.38 | 0.010803 | NME3 | non-metastatic cells 3, protein expressed in |
| 206508_at | 2.28 | 0.008142 | TNFSF7 | tumor necrosis factor (ligand) superfamily, member 7 | 210205_at | −1.35 | 0.010859 | B3GALT4 | udp-gal:betaglcnac beta 1,3-galactosyltransferase, polypeptide 4 |
| 35150_at | 1.63 | 0.008147 | CD40 | cd40 antigen (tnf receptor superfamily member 5) | 203980_at | −1.83 | 0.011023 | FABP4 | fatty acid binding protein 4, adipocyte |
| 204070_at | 1.33 | 0.008335 | RARRES3 | retinoic acid receptor responder (tazarotene induced) 3 | 202201_at | −1.44 | 0.011165 | BLVRB | biliverdin reductase b (flavin reductase (nadph)) |
| 204440_at | 1.31 | 0.008599 | CD83 | cd83 antigen (activated b lymphocytes, immunoglobulin superfamily) | 217983_s_at | −1.53 | 0.011188 | RNASET2 | ribonuclease t2 |
| 209392_at | 1.57 | 0.008806 | ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 209236_at | −1.34 | 0.011251 | SLC23A2 | solute carrier family 23 (nucleobase transporters), member 2 |
| 214933_at | 1.36 | 0.008923 | CACNA1A | calcium channel, voltage-dependent, p/q type, alpha 1a subunit | 221116_at | −2.18 | 0.011341 | PPARL | pparl |
| 205266_at | 2.00 | 0.009015 | LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 207839_s_at | −1.33 | 0.012331 | C9orf127 | chromosome 9 open reading frame 127 |
| 201614_s_at | 1.35 | 0.009016 | RUVBL1 | ruvb-like 1 (e. coli) | 201820_at | −2.00 | 0.012345 | KRT5 | keratin 4 |
| 218662_s_at | 1.65 | 0.009357 | HCAP-G | chromosome condensation protein g | 221830_at | −1.38 | 0.012420 | RAP2A | rap2a, member of ras oncogene family |
| 220665_at | 2.32 | 0.009580 | LUZP4 | leucine zipper protein 4 | 204749_at | −2.44 | 0.012608 | NAP1L3 | nucleosome assembly protein 1-like 3 |
| 205570_at | 1.31 | 0.009613 | PIP5K2A | phosphatidylinositol-4-phosphate 5-kinase, type ii, alpha | 219264_s_at | −1.60 | 0.012642 | PPP2R3B | protein phosphatase 2 (formerly 2a), regulatory subunit b", beta |
| 217892_s_at | 1.31 | 0.009635 | LIMA1 | lim domain and actin binding 1 | 204595_s_at | −1.33 | 0.012951 | STC1 | stanniocalcin 1 |
| 204444_at | 1.62 | 0.009826 | KIF11 | kinesin family member 11 | 208013_s_at | −1.42 | 0.013342 | ACRV1 | acrosomal vesicle protein 1 |
| 201263_at | 1.35 | 0.009908 | TARS | threonyl-trna synthetase | 200665_s_at | −1.82 | 0.013513 | SPARC | secreted protein, acidic, cysteine-rich (osteonectin) |
| 202068_s_at | 1.33 | 0.010149 | LDLR | low density lipoprotein receptor (familial hypercholesterolemia) | 207643_s_at | −1.32 | 0.013568 | TNFRSF1A | tumor necrosis factor receptor superfamily, member 1a |
| 212378_at | 1.32 | 0.010160 | GART | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 203126_at | −1.34 | 0.013930 | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 |
| 204959_at | 1.31 | 0.010219 | MNDA | myeloid cell nuclear differentiation antigen | 221666_s_at | −1.31 | 0.013942 | PYCARD | pyd and card domain containing |
| 218350_s_at | 1.30 | 0.010640 | GMNN | geminin, dna replication inhibitor | 217996_at | −1.44 | 0.014560 | PHLDA1 | pleckstrin homology-like domain, family a, member 1 |
| 219255_x_at | 1.68 | 0.010644 | IL17RB | interleukin 17 receptor b | 219403_s_at | −1.44 | 0.014588 | HPSE | heparanase |
| 205476_at | 2.47 | 0.011419 | CCL20 | chemokine (c-c motif) ligand 20 | 61874_at | −1.52 | 0.014677 | C9orf7 | chromosome 9 open reading frame 7 |
| 208892_s_at | 1.38 | 0.011479 | DUSP6 | dual specificity phosphatase 6 | 208130_s_at | −1.78 | 0.014684 | TBXAS1 | thromboxane a synthase 1 (platelet, cytochrome p450, family 5, subfamily a) |
| 204023_at | 1.31 | 0.011734 | RFC4 | replication factor c (activator 1) 4, 37kda | 200785_s_at | −1.47 | 0.014746 | LRP1 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) |
| 220957_at | 1.33 | 0.012024 | CTAGE1 | cutaneous t-cell lymphoma-associated antigen 1 | 207117_at | −2.16 | 0.014850 | H-plk | krueppel-related zinc finger protein |
| 203105_s_at | 1.34 | 0.012173 | DNM1L | dynamin 1-like | 206028_s_at | −1.50 | 0.014914 | MERTK | c-mer proto-oncogene tyrosine kinase |
| 220658_s_at | 1.56 | 0.012531 | ARNTL2 | aryl hydrocarbon receptor nuclear translocator-like 2 | 209354_at | −1.32 | 0.014997 | TNFRSF14 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| 204998_s_at | 1.34 | 0.012554 | ATF5 | activating transcription factor 5 | 201064_s_at | −1.32 | 0.015387 | PABPC4 | poly(a) binding protein, cytoplasmic 4 (inducible form) |
| 204170_s_at | 1.44 | 0.013296 | CKS2 | cdc28 protein kinase regulatory subunit 2 | 204857_at | −1.39 | 0.015779 | MAD1L1 | mad1 mitotic arrest deficient-like 1 (yeast) |
| 209726_at | 1.56 | 0.013322 | CA11 | carbonic anhydrase xi | 221287_at | −1.84 | 0.015826 | RNASEL | ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent) |
| 202069_s_at | 1.38 | 0.013436 | IDH3A | isocitrate dehydrogenase 3 (nad+) alpha | 209185_s_at | −1.38 | 0.016378 | IRS2 | insulin receptor substrate 2 |
| 204531_s_at | 1.82 | 0.013582 | BRCA1 | breast cancer 1, early onset | 220727_at | −1.49 | 0.016475 | KCNK10 | potassium channel, subfamily k, member 10 |
| 221209_s_at | 1.90 | 0.013776 | OTOR | otoraplin | 201785_at | −1.97 | 0.016745 | RNASE1 | ribonuclease, rnase a family, 1 (pancreatic) |
| 201315_x_at | 1.34 | 0.014191 | IFITM2 | interferon induced transmembrane protein 2 (1–8d) | 209458_x_at, 214414_x_at | −1.52 | 0.017735 | HBA1 /// HBA2 | hemoglobin, alpha 1 |
| 208881_x_at | 1.33 | 0.014408 | IDI1 | isopentenyl-diphosphate delta isomerase 1 | 201876_at | −1.61 | 0.017903 | PON2 | paraoxonase 2 |
| 200886_s_at | 1.30 | 0.014546 | PGAM1 /// LOC642969 /// LOC643576 | phosphoglycerate mutase 1 (brain) | 216924_s_at | −1.97 | 0.018447 | DRD2 | dopamine receptor d2 |
| 203350_at | 1.42 | 0.014818 | AP1G1 | adaptor-related protein complex 1, gamma 1 subunit | 206196_s_at | −1.81 | 0.018469 | RPIP8 | rap2 interacting protein 8 |
| 204015_s_at | 2.04 | 0.014931 | DUSP4 | dual specificity phosphatase 4 | 207922_s_at | −1.31 | 0.018875 | MAEA | macrophage erythroblast attacher |
| 203805_s_at | 1.52 | 0.015213 | FANCA | fanconi anemia, complementation group a | 205911_at | −1.69 | 0.018983 | PTHR1 | parathyroid hormone receptor 1 |
| 203502_at | 1.38 | 0.015219 | BPGM | 2,3-bisphosphoglycerate mutase | 203910_at | −1.31 | 0.019419 | ARHGAP29 | rho gtpase activating protein 29 |
| 206254_at | 1.76 | 0.015446 | EGF | epidermal growth factor (beta-urogastrone) | 204592_at | −1.49 | 0.019475 | DLG4 | discs, large homolog 4 (drosophila) |
| 206096_at | 1.39 | 0.015587 | ZNF35 | zinc finger protein 35 (clone hf.10) | 205744_at | −2.07 | 0.019498 | DOC2A | double c2-like domains, alpha |
| 207904_s_at | 2.23 | 0.015741 | LNPEP | leucyl/cystinyl aminopeptidase | 206623_at | −2.16 | 0.019526 | PDE6A | phosphodiesterase 6a, cgmp-specific, rod, alpha |
| 208393_s_at | 1.31 | 0.015769 | RAD50 | rad50 homolog (s. cerevisiae) | 212191_x_at | −1.37 | 0.019681 | RPL13 | ribosomal protein l13 |
| 203276_at | 1.49 | 0.016097 | LMNB1 | lamin b1 | 204561_x_at | −1.44 | 0.019846 | APOC2 | apolipoprotein c-ii |
| 206561_s_at | 2.20 | 0.016231 | AKR1B10 | aldo-keto reductase family 1, member b11 (aldose reductase-like) | 201212_at | −1.58 | 0.019878 | LGMN | legumain |
| 210176_at | 1.38 | 0.016450 | TLR1 | toll-like receptor 1 | 202152_x_at | −1.38 | 0.020121 | USF2 | upstream transcription factor 2, c-fos interacting |
| 219148_at | 1.39 | 0.016529 | PBK | pdz binding kinase | 208982_at | −1.35 | 0.020229 | PECAM1 | platelet/endothelial cell adhesion molecule (cd31 antigen) |
| 210367_s_at | 1.40 | 0.016595 | PTGES | prostaglandin e synthase | 206816_s_at | −1.83 | 0.020456 | SPAG8 | sperm associated antigen 8 |
| 205500_at | 1.63 | 0.016907 | C5 | complement component 5 | 221123_x_at | −1.30 | 0.021205 | ZNF395 | hypothetical protein dkfzp434k1210 |
| 210007_s_at | 1.45 | 0.017219 | GPD2 | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) | 220066_at | −1.33 | 0.021232 | CARD15 | caspase recruitment domain family, member 15 |
| 208204_s_at | 1.83 | 0.017264 | CAV3 | caveolin 3 | 219752_at | −1.60 | 0.021342 | RASAL1 | ras protein activator like 1 (gap1 like) |
| 205403_at | 2.46 | 0.017286 | IL1R2 | interleukin 1 receptor, type ii | 216860_s_at | −1.77 | 0.021418 | GDF11 | growth differentiation factor 11 |
| 218854_at | 1.47 | 0.017922 | SART2 | squamous cell carcinoma antigen recognized by t cells 2 | 209030_s_at | −1.38 | 0.021664 | IGSF4 | immunoglobulin superfamily, member 4 |
| 210072_at | 1.82 | 0.017991 | CCL19 | chemokine (c-c motif) ligand 19 | 202187_s_at | −1.36 | 0.021770 | PPP2R5A | protein phosphatase 2, regulatory subunit b (b56), alpha isoform |
| 203454_s_at | 1.34 | 0.018216 | ATOX1 | atx1 antioxidant protein 1 homolog (yeast) | 207421_at | −2.30 | 0.022313 | CA5A | carbonic anhydrase va, mitochondrial |
| 219866_at | 2.10 | 0.018296 | CLIC5 | chloride intracellular channel 5 | 205131_x_at | −2.05 | 0.023117 | CLEC11A | c-type lectin domain family 11, member a |
| 214279_s_at | 2.24 | 0.018713 | NDRG2 | ndrg family member 2 | 205498_at | −2.27 | 0.023139 | GHR | growth hormone receptor |
| 206682_at | 1.46 | 0.018849 | CLEC10A | c-type lectin domain family 10, member a | 207741_x_at | −1.45 | 0.023415 | TPSAB1 /// TPSB2 /// LOC652751 | tryptase beta 2 |
| 209714_s_at | 1.55 | 0.019322 | CDKN3 | cyclin-dependent kinase inhibitor 3 (cdk2-associated dual specificity phosphatase) | 213213_at | −1.44 | 0.023500 | DIDO1 | death inducer-obliterator 1 |
| 202518_at | 1.30 | 0.019575 | BCL7B | b-cell cll/lymphoma 7b | 219799_s_at | −1.42 | 0.023653 | DHRS9 | dehydrogenase/reductase (sdr family) member 9 |
| 211367_s_at | 1.39 | 0.019698 | CASP1 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 220528_at | −2.33 | 0.023827 | VNN3 | vanin 3 |
| 212671_s_at | 1.40 | 0.020228 | HLA-DQA1 /// HLA-DQA2 /// LOC650946 | major histocompatibility complex, class ii, dq alpha 1 | 219607_s_at | −1.86 | 0.024188 | MS4A4A | membrane-spanning 4-domains, subfamily a, member 4 |
| 203409_at | 1.56 | 0.020373 | DDB2 | lim homeobox 3, damage-specific dna binding protein 2, 48kda | 208472_at | −1.42 | 0.024357 | ZNFN1A4 | zinc finger protein, subfamily 1a, 4 (eos) |
| 200986_at | 1.36 | 0.021457 | SERPING1 | serpin peptidase inhibitor, clade g (c1 inhibitor), member 1, (angioedema, hereditary) | 214636_at | −1.55 | 0.024501 | CALCB | calcitonin-related polypeptide, beta |
| 220042_x_at | 2.32 | 0.021477 | HIVEP3 | human immunodeficiency virus type i enhancer binding protein 3 | 203973_s_at | −1.40 | 0.024666 | CEBPD | ccaat/enhancer binding protein (c/ebp), delta |
| 204408_at | 1.41 | 0.021502 | APEX2 | apex nuclease (apurinic/apyrimidinic endonuclease) 2 | 202812_at | −1.35 | 0.024799 | GAA | glucosidase, alpha; acid (pompe disease, glycogen storage disease type ii) |
| 218036_x_at | 1.30 | 0.021537 | NMD3 | nmd3 homolog (s. cerevisiae) | 204042_at | −1.32 | 0.025662 | WASF3 | was protein family, member 3 |
| 221034_s_at | 1.44 | 0.021674 | TEX13B | testis expressed sequence 13b | 202450_s_at | −1.34 | 0.025802 | CTSK | cathepsin k (pycnodysostosis) |
| 205598_at | 1.90 | 0.021893 | TRAIP | traf interacting protein | 201050_at | −1.43 | 0.025854 | PLD3 | phospholipase d family, member 3 |
| 205347_s_at | 1.39 | 0.021921 | TMSL8 | thymosin-like 8 | 209978_s_at | −2.28 | 0.025937 | LPA /// PLG | plasminogen, lipoprotein, lp(a) |
| 220386_s_at | 1.43 | 0.022175 | EML4 | echinoderm microtubule associated protein like 4 | 201753_s_at | −1.35 | 0.026385 | ADD3 | adducin 3 (gamma) |
| 217738_at | 1.35 | 0.022376 | PBEF1 /// LOC646309 /// RP11-92J19.4 | pre-b-cell colony enhancing factor 1 | 219892_at | −1.43 | 0.026459 | TM6SF1 | transmembrane 6 superfamily member 1 |
| 207662_at | 1.64 | 0.023056 | TBX1 | t-box 1 | 205844_at | −1.74 | 0.026498 | VNN1 | vanin 1 |
| 203362_s_at | 1.41 | 0.023308 | MAD2L1 | mad2 mitotic arrest deficient-like 1 (yeast) | 207582_at | −2.12 | 0.027216 | PIN1L | protein (peptidylprolyl cis/trans isomerase) nima-interacting 1-like |
| 206325_at | 1.83 | 0.023449 | SERPINA6 | serpin peptidase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 6 | 203729_at | −1.41 | 0.027633 | EMP3 | epithelial membrane protein 3 |
| 220938_s_at | 1.59 | 0.023571 | GMEB1 | glucocorticoid modulatory element binding protein 1 | 204079_at | −1.30 | 0.027635 | TPST2 | tyrosylprotein sulfotransferase 2 |
| 201202_at | 1.40 | 0.023620 | PCNA | proliferating cell nuclear antigen | 208511_at | −2.68 | 0.027688 | PTTG3 | pituitary tumor-transforming 3 |
| 201507_at | 1.38 | 0.023999 | PFDN1 | prefoldin subunit 1 | 209661_at | −1.90 | 0.027937 | KIFC3 | kinesin family member c3 |
| 37950_at | 1.36 | 0.024052 | PREP | prolyl endopeptidase | 206106_at | −1.63 | 0.028087 | MAPK12 | mitogen-activated protein kinase 12 |
| 204835_at | 1.66 | 0.024112 | POLA | polymerase (dna directed), alpha | 203028_s_at | −1.48 | 0.028274 | CYBA | cytochrome b-245, alpha polypeptide |
| 202780_at | 1.40 | 0.024117 | OXCT1 | 3-oxoacid coa transferase 1 | 219440_at | −2.54 | 0.028291 | RAI2 | retinoic acid induced 2 |
| 219424_at | 1.97 | 0.024648 | EBI3 | epstein-barr virus induced gene 3 | 203996_s_at | −1.71 | 0.028734 | C21orf2 | chromosome 21 open reading frame 2 |
| 204769_s_at | 1.35 | 0.025000 | TAP2 | transporter 2, atp-binding cassette, sub-family b (mdr/tap) | 221061_at | −2.63 | 0.029166 | PKD2L1 | polycystic kidney disease 2-like 1 |
| 210119_at | 2.62 | 0.026057 | KCNJ15 | potassium inwardly-rectifying channel, subfamily j, member 15 | 204367_at | −1.30 | 0.029203 | SP2 | sp2 transcription factor |
| 207143_at | 1.93 | 0.026156 | CDK6 | cyclin-dependent kinase 6 | 213436_at | −1.73 | 0.029349 | CNR1 | cannabinoid receptor 1 (brain) |
| 218219_s_at | 1.38 | 0.026250 | LANCL2 | lanc lantibiotic synthetase component c-like 2 (bacterial) | 38157_at | −1.64 | 0.030123 | DOM3Z | dom-3 homolog z (c. elegans) |
| 219645_at | 1.38 | 0.026851 | CASQ1 | calsequestrin 1 (fast-twitch, skeletal muscle) | 210423_s_at | −1.56 | 0.030184 | SLC11A1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| 201710_at | 1.85 | 0.027145 | MYBL2 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | 201911_s_at | −2.04 | 0.031248 | FARP1 | ferm, rhogef (arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| 203859_s_at | 1.49 | 0.027287 | PALM | paralemmin | 220782_x_at | −1.82 | 0.031968 | KLK12 | kallikrein 12 |
| 204297_at | 1.32 | 0.027612 | PIK3C3 | phosphoinositide-3-kinase, class 3 | 207482_at | −1.62 | 0.032061 | C20orf10 | chromosome 20 open reading frame 10 |
| 219971_at | 1.37 | 0.028335 | IL21R | interleukin 21 receptor | 204697_s_at | −2.06 | 0.032591 | CHGA | chromogranin a (parathyroid secretory protein 1) |
| 211297_s_at | 1.37 | 0.028415 | CDK7 | cyclin-dependent kinase 7 (mo15 homolog, xenopus laevis, cdk-activating kinase) | 206873_at | −1.68 | 0.032847 | CA6 | carbonic anhydrase vi |
| 204126_s_at | 2.09 | 0.028496 | CDC45L | cdc45 cell division cycle 45-like (s. cerevisiae) | 207914_x_at | −1.54 | 0.033022 | EVX1 | eve, even-skipped homeobox homolog 1 (drosophila) |
| 207455_at | 1.46 | 0.028568 | P2RY1 | purinergic receptor p2y, g-protein coupled, 1 | 218206_x_at | −1.37 | 0.033390 | SCAND1 | scan domain containing 1 |
| 206765_at | 1.33 | 0.028640 | KCNJ2 | potassium inwardly-rectifying channel, subfamily j, member 2 | 217975_at | −1.50 | 0.034679 | WBP5 | ww domain binding protein 5 |
| 206175_x_at | 2.21 | 0.028671 | ZNF222 | zinc finger protein 222 | 219327_s_at | −1.60 | 0.035562 | GPRC5C | g protein-coupled receptor, family c, group 5, member c |
| 206173_x_at | 2.11 | 0.028761 | GABPB2 | ga binding protein transcription factor, beta subunit 1, 53kda | 206206_at | −1.39 | 0.035730 | CD180 | cd180 antigen |
| 205463_s_at | 1.84 | 0.029164 | PDGFA | platelet-derived growth factor alpha polypeptide | 222239_s_at | −1.31 | 0.035916 | INTS6 | dkfzp434b105 protein |
| 206407_s_at | 2.20 | 0.029186 | CCL13 | chemokine (c-c motif) ligand 13 | 220189_s_at | −1.44 | 0.036040 | MGAT4B | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-n-acetylglucosaminyltransferase, isozyme b |
| 210772_at | 1.37 | 0.029514 | FPRL1 | formyl peptide receptor-like 1 | 203476_at | −1.59 | 0.036505 | TPBG | trophoblast glycoprotein |
| 211200_s_at | 2.47 | 0.029575 | EFCAB2 | ef-hand calcium binding domain 2 | 221005_s_at | −1.55 | 0.037185 | PTDSS2 | phosphatidylserine synthase 2 |
| 203592_s_at | 1.54 | 0.029679 | FSTL3 | follistatin-like 3 (secreted glycoprotein) | 209822_s_at | −1.37 | 0.037473 | VLDLR | very low density lipoprotein receptor |
| 203052_at | 1.39 | 0.029744 | C2 | complement component 2 | 205638_at | −1.71 | 0.037898 | BAI3 | brain-specific angiogenesis inhibitor 3 |
| 211499_s_at | 1.67 | 0.029863 | MAPK11 | mitogen-activated protein kinase 11 | 220777_at | −1.74 | 0.038565 | KIF13A | kinesin family member 13a |
| 204162_at | 1.99 | 0.030006 | KNTC2 | kinetochore associated 2 | 210139_s_at | −1.33 | 0.038607 | PMP22 | peripheral myelin protein 22 |
| 201737_s_at | 1.35 | 0.030278 | MARCH6 | membrane-associated ring finger (c3hc4) 6 | 221324_at | −1.32 | 0.038629 | TAS2R1 | taste receptor, type 2, member 1 |
| 210549_s_at | 2.57 | 0.030537 | CCL23 | chemokine (c-c motif) ligand 23 | 210884_s_at | −1.62 | 0.039295 | LOC653423 | sperm associated antigen 11 |
| 218931_at | 1.90 | 0.030914 | RAB17 | rab17, member ras oncogene family | 207739_s_at | −2.22 | 0.039456 | GAGE1 /// GAGE2 /// GAGE3 /// GAGE4 /// GAGE5 /// GAGE6 /// GAGE7 /// GAGE7B /// GAGE8 /// LOC645009 /// LOC645037 /// LOC645073 /// LOC645093 | g antigen 1, g antigen 3, g antigen 4, g antigen 2, g antigen 5, g antigen 8, g antigen 6 |
| 204304_s_at | 1.48 | 0.030950 | PROM1 | prominin 1 | 209737_at | −1.42 | 0.039512 | MAGI2 | membrane associated guanylate kinase, ww and pdz domain containing 2 |
| 205450_at | 2.53 | 0.030967 | PHKA1 | phosphorylase kinase, alpha 1 (muscle) | 32094_at | −1.72 | 0.039607 | CHST3 | carbohydrate (chondroitin 6) sulfotransferase 3 |
| 204504_s_at | 1.55 | 0.031187 | HIRIP3 | hira interacting protein 3 | 41660_at | −1.79 | 0.039999 | CELSR1 | cadherin, egf lag seven-pass g-type receptor 1 (flamingo homolog, drosophila) |
| 209890_at | 1.38 | 0.031529 | TSPAN5 | tetraspanin 5 | 206610_s_at | −1.92 | 0.040448 | F11 | coagulation factor xi (plasma thromboplastin antecedent) |
| 34187_at | 1.89 | 0.031861 | RBMS2 | rna binding motif, single stranded interacting protein 2 | 209988_s_at | −1.57 | 0.040611 | ASCL1 | achaete-scute complex-like 1 (drosophila) |
| 218943_s_at | 1.58 | 0.032067 | DDX58 | dead (asp-glu-ala-asp) box polypeptide 58 | 219922_s_at | −1.47 | 0.040966 | LTBP3 | latent transforming growth factor beta binding protein 3 |
| 202240_at | 1.53 | 0.032437 | PLK1 | polo-like kinase 1 (drosophila) | 205054_at | −1.78 | 0.041014 | NEB | nebulin |
| 205716_at | 1.39 | 0.033296 | MCFP | mitochondrial carrier family protein | 206404_at | −1.45 | 0.041028 | FGF9 | fibroblast growth factor 9 (glia-activating factor) |
| 208005_at | 1.69 | 0.033662 | NTN1 | netrin 1 | 203305_at | −1.59 | 0.041286 | F13A1 | coagulation factor xiii, a1 polypeptide |
| 220059_at | 1.53 | 0.033861 | BRDG1 | bcr downstream signaling 1 | 201656_at | −1.38 | 0.041995 | ITGA6 | integrin, alpha 6 |
| 204767_s_at | 1.39 | 0.034527 | FEN1 | flap structure-specific endonuclease 1 | 205384_at | −1.96 | 0.042828 | FXYD1 | fxyd domain containing ion transport regulator 1 (phospholemman) |
| 210280_at | 2.18 | 0.034875 | MPZ | myelin protein zero (charcot-marie-tooth neuropathy 1b) | 203618_at | −1.34 | 0.043195 | FAIM2 | fas apoptotic inhibitory molecule 2 |
| 213492_at | 1.64 | 0.034876 | COL2A1 | collagen, type ii, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 210117_at | −2.03 | 0.044321 | SPAG1 | sperm associated antigen 1 |
| 212020_s_at | 1.51 | 0.035187 | MKI67 | antigen identified by monoclonal antibody ki-67 | 206498_at | −1.85 | 0.044628 | OCA2 | oculocutaneous albinism ii (pink-eye dilution homolog, mouse) |
| 206247_at | 1.32 | 0.035337 | MICB | mhc class i polypeptide-related sequence b | 202621_at | −1.41 | 0.045125 | IRF3 | interferon regulatory factor 3 |
| 212657_s_at | 1.61 | 0.035551 | IL1RN | interleukin 1 receptor antagonist | 205881_at | −2.10 | 0.045147 | ZNF74 | zinc finger protein 74 (cos52) |
| 215171_s_at | 1.31 | 0.035701 | TIMM17A | translocase of inner mitochondrial membrane 17 homolog a (yeast) | 204449_at | −1.40 | 0.045296 | PDCL | phosducin-like |
| 210048_at | 1.31 | 0.036177 | NAPG | n-ethylmaleimide-sensitive factor attachment protein, gamma | 209657_s_at | −1.38 | 0.045366 | HSF2 | heat shock transcription factor 2 |
| 221507_at | 1.35 | 0.036194 | TNPO2 | transportin 2 (importin 3, karyopherin beta 2b) | 208000_at | −1.41 | 0.045916 | GML | gpi anchored molecule like protein |
| 208118_x_at | 1.31 | 0.036247 | LAT1-3TM /// IMAA /// LOC440345 /// LOC440354 /// LOC595101 /// LOC641298 /// LOC646866 | slc7a5 pseudogene, hypothetical gene loc283846, kiaa0220-like protein | 206251_s_at | −2.18 | 0.046131 | AVPR1A | arginine vasopressin receptor 1a |
| 219125_s_at | 1.35 | 0.036524 | RAG1AP1 | recombination activating gene 1 activating protein 1 | 207503_at | −2.22 | 0.046146 | TCP10 | t-complex 10 (mouse) |
| 204092_s_at | 1.49 | 0.037044 | AURKA | aurora kinase a | 202191_s_at | −1.33 | 0.046843 | GAS7 | growth arrest-specific 7 |
| 202627_s_at | 1.79 | 0.037267 | SERPINE1 | serpin peptidase inhibitor, clade e (nexin, plasminogen activator inhibitor type 1), member 1 | 205200_at | −1.72 | 0.047002 | CLEC3B | c-type lectin domain family 3, member b |
| 208478_s_at | 1.51 | 0.037303 | BAX | bcl2-associated x protein | 202178_at | −1.88 | 0.047241 | PRKCZ | protein kinase c, zeta |
| 207473_at | 1.40 | 0.037435 | MLN | motilin | 217871_s_at | −1.31 | 0.047313 | MIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| 207329_at | 2.09 | 0.037908 | MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) | 221309_at | −1.90 | 0.047786 | RBM17 | rna binding motif protein 17 |
| 219199_at | 1.64 | 0.037979 | AFF4 | af4/fmr2 family, member 4 | 205050_s_at | −1.54 | 0.047840 | MAPK8IP2 | mitogen-activated protein kinase 8 interacting protein 2 |
| 38037_at | 1.51 | 0.038257 | HBEGF | heparin-binding egf-like growth factor | 203643_at | −1.55 | 0.048239 | ERF | ets2 repressor factor |
| 33767_at | 1.39 | 0.038470 | NEFH | neurofilament, heavy polypeptide 200kda | 203422_at | −1.33 | 0.049132 | POLD1 | polymerase (dna directed), delta 1, catalytic subunit 125kda |
| 207882_at | 1.38 | 0.038622 | HSAJ2425 | p65 protein | 204811_s_at | −1.35 | 0.049138 | CACNA2D2 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| 204864_s_at | 1.55 | 0.038725 | IL6ST | interleukin 6 signal transducer (gp130, oncostatin m receptor) | 209264_s_at | −1.54 | 0.049162 | TSPAN4 | tetraspanin 4 |
| 204668_at | 1.39 | 0.039002 | RNF24 | ring finger protein 24 | 207929_at | −1.44 | 0.049398 | GRPR | gastrin-releasing peptide receptor |
| 207016_s_at | 1.50 | 0.039529 | ALDH1A2 | aldehyde dehydrogenase 1 family, member a2 | 219714_s_at | −1.90 | 0.049494 | CACNA2D3 | calcium channel, voltage-dependent, alpha 2/delta 3 subunit |
| 207346_at | 2.21 | 0.039649 | STX2 | epimorphin | |||||
| 32128_at | 1.50 | 0.039852 | CCL18 | chemokine (c-c motif) ligand 18 (pulmonary and activation-regulated) | |||||
| 206932_at | 1.67 | 0.040074 | CH25H | cholesterol 25-hydroxylase | |||||
| 205225_at | 1.33 | 0.040378 | ESR1 | estrogen receptor 1 | |||||
| 202416_at | 1.32 | 0.040835 | DNAJC7 | dnaj (hsp40) homolog, subfamily c, member 7 | |||||
| 220315_at | 1.36 | 0.041177 | PARP11 | poly (adp-ribose) polymerase family, member 11 | |||||
| 206999_at | 2.28 | 0.041871 | IL12RB2 | interleukin 12 receptor, beta 2 | |||||
| 220993_s_at | 2.19 | 0.042266 | GPR63 | g protein-coupled receptor 63 | |||||
| 221170_at | 1.60 | 0.042356 | HRH4 | histamine receptor h4 | |||||
| 207845_s_at | 1.39 | 0.042369 | ANAPC10 | anaphase promoting complex subunit 10 | |||||
| 204401_at | 1.40 | 0.042558 | KCNN4 | potassium intermediate/small conductance calcium-activated channel, subfamily n, member 4 | |||||
| 207124_s_at | 1.53 | 0.042696 | GNB5 | guanine nucleotide binding protein (g protein), beta 5 | |||||
| 219187_at | 1.85 | 0.042908 | FKBPL | fk506 binding protein like | |||||
| 203952_at | 1.76 | 0.042917 | ATF6 | activating transcription factor 6 | |||||
| 36742_at | 1.57 | 0.043363 | TRIM15 | tripartite motif-containing 15 | |||||
| 218117_at | 1.35 | 0.043508 | RBX1 | ring-box 1 | |||||
| 208592_s_at | 1.49 | 0.043731 | CD1E | cd1e antigen, e polypeptide | |||||
| 221106_at | 1.98 | 0.044473 | SLC22A17 | solute carrier family 22 (organic cation transporter), member 17 | |||||
| 207275_s_at | 1.34 | 0.044487 | ACSL1 | fatty-acid-coenzyme a ligase, long-chain 1 | |||||
| 202403_s_at | 2.21 | 0.044527 | COL1A2 | collagen, type i, alpha 2 | |||||
| 220633_s_at | 1.30 | 0.044556 | HP1BP3 | heterochromatin protein 1, binding protein 3 | |||||
| 208916_at | 1.36 | 0.044687 | SLC1A5 | solute carrier family 1 (neutral amino acid transporter), member 5 | |||||
| 203881_s_at | 1.76 | 0.045235 | DMD | dystrophin (muscular dystrophy, duchenne and becker types) | |||||
| 203948_s_at | 1.89 | 0.045239 | MPO | myeloperoxidase | |||||
| 203968_s_at | 1.48 | 0.045435 | CDC6 | cdc6 cell division cycle 6 homolog (s. cerevisiae) | |||||
| 209546_s_at | 1.46 | 0.045614 | APOL1 | apolipoprotein l, 1 | |||||
| 217847_s_at | 2.47 | 0.045686 | THRAP3 | thyroid hormone receptor associated protein 3 | |||||
| 206976_s_at | 1.31 | 0.046433 | HSPH1 | heat shock 105kda/110kda protein 1 | |||||
| 210133_at | 1.87 | 0.046451 | CCL11 | chemokine (c-c motif) ligand 11 | |||||
| 219540_at | 1.31 | 0.046489 | ZNF267 | zinc finger protein 267 | |||||
| 203287_at | 1.33 | 0.046497 | LAD1 | ladinin 1 | |||||
| 220091_at | 1.49 | 0.047676 | SLC2A6 | solute carrier family 2 (facilitated glucose transporter), member 6 | |||||
| 213800_at | 2.30 | 0.047921 | CFH | complement factor h | |||||
| 200793_s_at | 1.37 | 0.048667 | ACO2 | aconitase 2, mitochondrial | |||||
| 202732_at | 1.34 | 0.048888 | PKIG | protein kinase (camp-dependent, catalytic) inhibitor gamma | |||||
| 39402_at | 1.56 | 0.049565 | IL1B | interleukin 1, beta | |||||
DE= differentially expressed
F.C.= fold change post versus pre vaccination
Correlation of gene expression with neutralizing antibody titers after vaccination
Durability of protection after vaccination has not been established, but it is possible that individuals developing higher neutralizing titers upon immunization will have longer lasting protection against HPV infection. If so, understanding the immunological profile of individuals who develop higher neutralizing titers upon immunization might help elucidate the molecular mechanisms of long-term protection. With this in mind, we compared probeset expression levels induced by vaccination against neutralizing titers at one month after second vaccination (Month 2) and one or six months after the third vaccination (Month 7 or 12, respectively). Only individuals negative for neutralizing antibodies before vaccination were included. For this analysis, we considered the subset of 624 probesets that were differentially expressed at a p <0.05.
Correlations were typically lower when vaccination-induced changes in probeset expression levels (measured at Month 2) were compared against neutralizing antibody levels at Month 2 (98% of correlations were between −0.50 and +0.50 at Month 2, compared to 87% when Month 7 and 84% when Month 12 neutralizing antibody levels were used). Table IV summarizes results for the subset of 22 probesets observed to have a correlation with neutralizing antibodies of >0.60 or <−0.60 at both Month 7 and Month 12 after vaccination. A complete list of results is provided in Supplementary Table III. The highest positive correlations at both month 7 and 12 after vaccination correlation (>0.70 at both time points) was observed for the following probesets: CCND2 (month 7 r=0.88; month 12 r=0.82), LGALS2 (month 7 r=0.86; month 12 r=0.74), and IL1RN (month 7=0.75; month 12= 0.73).
Confirmation of a subset of differentially expressed genes by quantitative RT-PCR
To confirm the microarray results, a subset of 22 differentially expressed genes (defined in Methods) was selected for confirmation by PCR, using samples from an independent group of vaccine recipients (n=9). As shown in Table V, the expression pattern of the selected genes concurred with the microarray data for 15 out of 22 of the analyzed probesets (GZMB, IFNG, HSD11B1, INDO, IL6, IL3, CSF2, CYP27B1, IL2, IL5, INHBA, MMP12, CD1B, CCL7 and LIF). For a smaller group of 6 probesets, the frequency of up-regulated probesets was lower using PCR than observed with microarray (GGTLA1, KIF20A, AICDA, CXCL13, LTA and CCL13). Finally, no up-regulation (mean fold change = 1.4) was detected by PCR for one of the selected probesets (GOLPH2), while 61% of samples showed an up-regulation by microarray analysis.
Table V.
Confirmation of Microarray Results with Real-Time PCR in an independent sample (n=9)
| Gene | Gene Name | Frequency (>2-fold) | Mean Fold Change | Mean Fold Change-Only Responders | p-value-Microarray | |||
|---|---|---|---|---|---|---|---|---|
| Microarray | RT-PCR | Microarray | RT-PCR | Microarray | RT-PCR | |||
| GZMB | granzyme b | 12 of 18 (67%) | 7 of 9 (78%) | 2.3 | 3.0 | 3.0 | 3.6 | 0.00002 |
| IFNG | interferon, gamma | 12 of 18 (67%) | 5 of 8 (62%) | 2.6 | 5.0 | 4.0 | 7.0 | 0.00002 |
| HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 | 15 of 18 (83%) | 7 of 8 (88%) | 3.5 | 4.0 | 4.8 | 4.4 | 0.00005 |
| INDO | indoleamine-pyrrole 2,3 dioxygenase | 11 of 18 (61%) | 5 of 9 (56%) | 2.5 | 2.4 | 3.5 | 3.5 | 0.00006 |
| IL6 | interleukin 6 | 9 of 18 (50%) | 6 of 8 (75%) | 2.2 | 4.5 | 3.5 | 5.7 | 0.00030 |
| IL3 | interleukin 3 (colony-stimulating factor, multiple) | 11 of 18 (61%) | 5 of 9 (56%) | 4.0 | 2.9 | 9.3 | 4.2 | 0.00030 |
| CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | 12 of 18 (67%) | 5 of 8 (62%) | 3.3 | 3.5 | 6.6 | 4.7 | 0.00125 |
| CYP27B1 | cytochrome p450, family 27, subfamily b, polypeptide 1 | 13 of 18 (72%) | 7 of 9 (78%) | 2.4 | 3.2 | 3.8 | 3.8 | 0.00133 |
| IL2 | interleukin 2 | 11 of 18 (61%) | 4 of 8 (50%) | 3.9 | 3.7 | 9.0 | 6.4 | 0.00187 |
| IL5 | interleukin 5 (colony-stimulating factor, eosinophil) | 11 of 18 (61%) | 7 of 8 (88%) | 4.1 | 67.5 | 10.8 | 77.0 | 0.00399 |
| INHBA | inhibin, beta a (activin a, activin ab alpha polypeptide) | 15 of 18 (83%) | 7 of 9 (78%) | 5.0 | 9.2 | 9.0 | 11.5 | 0.00418 |
| MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 12 of 18 (67%) | 5 of 8 (62%) | 2.7 | 39.7 | 6.5 | 62.8 | 0.00476 |
| CD1B | cd1b antigen | 11 of 18 (61%) | 6 of 8 (75%) | 2.9 | 5.7 | 5.7 | 7.3 | 0.00491 |
| CCL7 | chemokine (c-c motif) ligand 7 | 10 of 18 (56%) | 7 of 9 (78%) | 2.4 | 49.2 | 5.9 | 63.1 | 0.00807 |
| LIF | leukemia inhibitory factor | 10 of 18 (56%) | 5 of 9 (56%) | 2.0 | 2.6 | 4.0 | 3.4 | 0.00901 |
| GGTLA1 | gamma-glutamyltransferase-like activity 1 | 13 of 18 (72%) | 3 of 8 (38%) | 3.9 | 5.0 | 6.3 | 12.0 | 0.00006 |
| KIF20A | kinesin family member 20a | 13 of 18 (72%) | 3 of 9 (33%) | 4.6 | 1.7 | 7.8 | 2.9 | 0.00015 |
| AICDA | activation-induced cytidine deaminase | 10 of 18 (56%) | 4 of 9 (44%) | 2.9 | 2.2 | 5.3 | 3.6 | 0.00033 |
| CXCL13 | chemokine (c-x-c motif) ligand 13 (b-cell chemoattractant) | 10 of 18 (56%) | 3 of 9 (33%) | 3.4 | 2.3 | 9.4 | 3.9 | 0.00454 |
| LTA | lymphotoxin alpha (tnf superfamily, member 1) | 12 of 18 (67%) | 4 of 9 (44%) | 4.0 | 2.4 | 11.7 | 3.4 | 0.00685 |
| CCL13 | chemokine (c-c motif) ligand 13 | 13 of 18 (72%) | 3 of 8 (38%) | 2.2 | 10.0 | 4.1 | 24.8 | 0.02919 |
| GOLPH2 | golgi phosphoprotein 2 | 11 of 18 (61%) | 0 of 8 (0%) | 3.1 | 1.4 | 5.5 | N/A | 0.00020 |
Analysis of pre-vaccination response to HPV-16 L1 VLP
Finally, we evaluated the direct effect of HPV-16 L1VLP on gene expression of PBMC obtained prior to vaccination by comparing levels of gene expression in PBMC incubated with HPV-16 L1 VLP against media-treated cells. This comparison allowed us to analyze the primary, vaccine-independent response to HPV-16 L1 VLP.
One-hundred thirty nine probesets were differentially expressed at a significance level of p<0.001 (106 up-regulated, 33 down-regulated) in pre-vaccination PBMC incubated with HPV-16 L1 VLP (Supplementary Table IV). An additional 258 probesets were differentially expressed at the 0.001<p<0.05 level (154 up-regulated, 104 down-regulated). Pathway enrichment evaluation was suggestive of enrichment for probesets mapping to genes from inflammatory, immune response, chemotaxis, signal transduction and cell proliferation pathways, as summarized in Figure 2. Interestingly, at least 26 out of the 260 up-regulated probesets are interferon-induced genes, or play a role in interferon signaling. There was an overlap between the probesets directly induced by HPV-16 L1 VLP prior to vaccination and those differentially expressed in post-vaccination samples (n = 91 or 9 up-regulated probesets, and 45 or 7 down-regulated probesets when using p<0.05 or p<0.001, respectively; Figure 3). The overlapped probesets belong to the most enriched pathways observed in HPV-16 L1 VLP-treated PBMC (for example, IFN-γ, IL-6, Granzyme B, INDO and several chemokines). The up-regulated, overlapping probesets significant at p<0.001 were INDO, LAMP3, TYMS, ALAS1, RRM2, CD38, IL6, IFNG and AKAP2. Down-regulated overlapping probesets significant at p<0.001 included CD9, HS3ST2, SORL1, ADFP, DFNA5, EPAS1 and ALDH3A2. A complete list of overlapping differentially expressed probesets is shown in supplementary Table IV.
Figure 2. Expression profiles of probesets significantly modulated following incubation of pre-vaccination PBMCs with HPV-16 L1 VLP.
PBMC from pre-vaccination samples were incubated with HPV-16 L1 VLP as indicated in Materials and Methods. Functional annotation of probesets differentially expressed was done using DAVID. Upregulated genes are shown in red and downregulated genes in green.
Figure 3. Number of probesets with increased or decreased expression in HPV-16 L1 VLP-treated PBMC from vaccine recipients induced by vaccination (vaccination-specific effect) or independent of vaccination (direct effect).
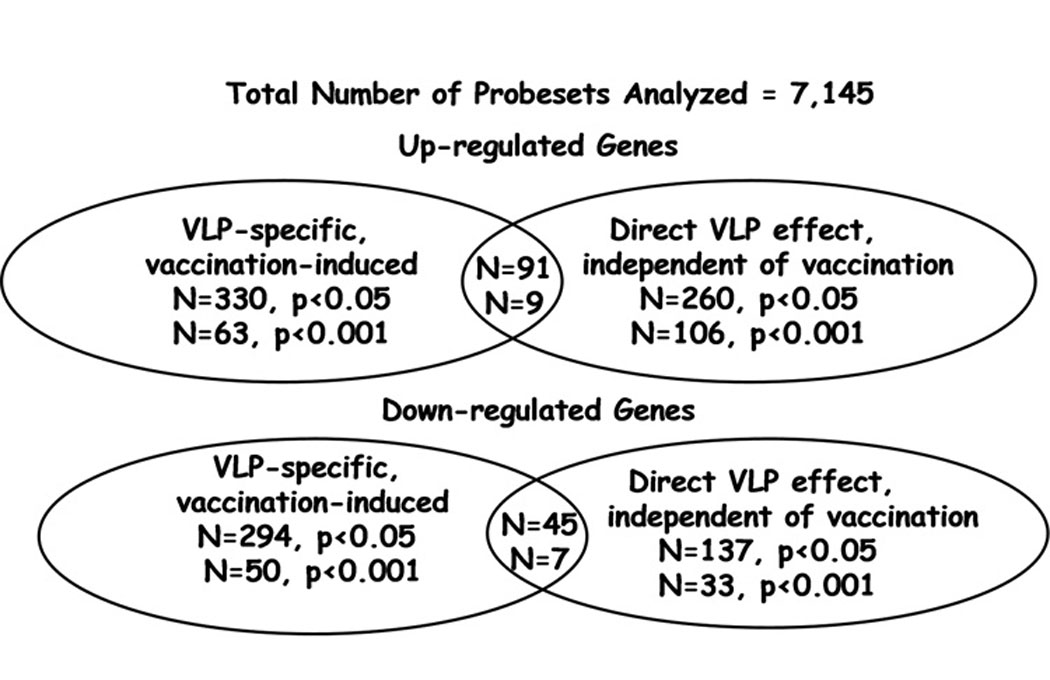
Number of probesets overlapping the lists of vaccination-specific probesets and the ones modulated by a direct HPV-16 L1VLP effect are also shown.
Discussion
In this study, we characterized the gene expression pattern of recall immune responses to the HPV-16 L1 VLP by comparing pre- and post-vaccination gene expression patterns of PBMCs incubated with HPV-16 L1 VLP antigen to better characterize the cellular and innate immune responses in vaccination against HPV. Although the effector mechanism of protection for this vaccine is believed to be neutralizing antibodies, our results show that multiple pathways within the cellular and innate arms of the immune system are targeted upon vaccination. The exact role of these pathways in protection or duration of protection is still unknown but based on the results presented here deserves further investigation. The analysis of correlation between differentially expressed genes and neutralizing antibody levels allowed us to identify genes (in particular CCND2, LGALS2 and IL-1RN) that may be predictors of prolonged antibody responses. New studies are necessary to better determine which pre- and post-vaccination clinical, immunological and expression profiles are most associated with long-term protection against HPV.
We took two different approaches to data analysis: a conservative approach that takes into account the potential high false discovery rate that may accompany the high number of statistical comparisons performed, and an exploratory approach to define additional biological pathways and probesets altered in a recall immune response to the vaccine. Using the conservative approach, we pre-selected pathways based on overall knowledge of microbial immune responses. This reduced the number of comparisons made and focused our analysis on the strongest a priori pathways expected to be involved in immune responses to vaccination. Our results suggest that the response to HPV-16 L1 VLP in vaccine recipients involves modulation of genes within the cell cycle, cytokine, defense, inflammation and interferon pathways. This finding is consistent with the induction of various arms of the immune response (42, 43) and with our previous data that a recall response to HPV-16 L1 VLP involves a broad and complex pattern of cytokines and chemokines (16, 17).
No significant enrichment was observed for the signal transduction pathway in our conservative analysis despite differential expression in genes that belonged to this pathway. A possible explanation for this observation might be that the definition of this pathway is broad (1577 probesets), and that the differentially expressed probesets within the pathway represent a small subset of the total number. In our exploratory analysis, the signal transduction pathway was found to be over-represented. This information may be used for hypothesis generation in future studies.
Within the pathways evaluated, we identified up-regulation of genes involved in cytotoxic (CTSC, GZMA, GZMB, TNFSF10, FASLG) and important immuno-regulatory functions (INDO, HSD11B1, CTLA4, SOCS1). These results suggest that the immune response to HPV-16 L1 VLP induces a complex modulation of various components of the cell-mediated immune response, including cytokine secretion (T-helper cells), cytotoxic response, and activation of feedback mechanisms involving immunoregulatory genes. These results raise the need for future studies determining the contributing roles of these pathways in immunogenicity induced by VLPs. A better understanding of these pathways may be also useful in the design of improved vaccine strategies. In addition, a substantial fraction of the differentially expressed probesets was down-regulated after vaccination including genes that fell into cell death, immune response, signalling and metabolism pathways. Because we used PBMCs, the transcriptional change is representative of the changes from various cell types comprising the PBMC population and the cross-talk of extracellular signals, such as cytokines induced directly or indirectly by the VLPs. Interestingly, expression of a number of these down-regulated genes and corresponding proteins, such as CD9, CD14, ly86, LEF-1, CD26 have been shown to be down-modulated in the context of cell activation and/or inflammatory signals (44, 45, 46, 47). Such changes may relate to the shutting down specific elements in order to better mount a protective immune response to HPV vaccine and avoid excessive inflammatory signals or limit feedback signaling. However, the underlying mechanisms and impact of these transcriptional changes in the development of an immune response to vaccination are not known.
When we examined the correlation of probeset expression levels with neutralizing antibody titers, more probesets correlated to antibody levels at months 7 and 12 post vaccination than at month 2. The highest correlations at month 7 and 12 were obtained for cyclin d2 (CCND2), which plays an essential role controlling the cell cycle at the G1/S transition (48); galectin 2 (LGALS2), a galactoside-binding lectin that has been shown to promote apoptosis in activated T-cells and to shift immune responses to a Th2 profile (49); and IL-1 receptor antagonist (IL_1RN) a regulator of IL-1 induced inflammatory response. In fact, cyclin D2 and AICDA are components of signaling cascades needed for B cell proliferation and antibody production (50, 51). However, the potential role of galectin-2 and IL-1 receptor antagonist in B cell responses is unclear and could be of indirect nature since these are immune regulatory molecules. Confirmation of the role of the products of these genes in B cell memory responses to the vaccine would be informative in the future.
To confirm the microarray results, we selected PBMCs from an independent group of vaccine recipients and measured RNA expression for a subset of 22 probesets representing various biological pathways targeted by vaccination. We observed a comparable frequency of responders for 68% of analyzed genes (15 of 22). For an additional 27% (6 of 22) of analyzed genes, differential expression was similar to those observed by microarray testing, but at a lower frequency. We were unable to observe evidence of differential expression as noted by microarray for 1 of the 22 selected genes. These results validate our microarray findings, and suggest that the HPV-16 L1 VLP-specific response after vaccination involves activation of a complex cell-mediated immune response.
Our results demonstrate that the immune response to vaccination is heterogeneous among vaccine recipients at the gene expression level. For all the probesets analyzed, only a fraction of the participants were considered responders when looking at one gene at a time. The highest frequency was observed for HSD11B1 (15 of 18 donors in microarray, and 7 of 8 with RT-PCR). However when “responders” are defined by the global expression profile of multiple genes at one time the response rate is high (data not shown). Whether this heterogeneity of response will have an impact on long-term memory and quality of the immune responses over time needs to be addressed in further studies. It is possible that heterogeneity of response may be associated with previous exposures to HPV. Studies of comparison of expression profiles induced by vaccination in naturally infected versus naïve individuals are warranted. In addition, identification of gene signatures associated with infection or vaccination will also be important.
In addition to the evaluation of vaccine-specific immune responses, we characterized the vaccination-independent responses to VLP by comparing HPV-16 L1 VLP-incubated cells with media controls in pre-vaccination samples. Our analysis revealed that incubation with HPV-16 L1 VLP prior to vaccination induces up-regulation of the defense response, cytokine/inflammation, signal transduction, and cell proliferation pathways. If the responses elicited by HPV-16 L1 VLP in the post and pre-vaccination responses are compared, about 30% of the probesets modulated by VLP (136 out of 397; p<0.05) prior to vaccination overlap with the HPV-16 L1 VLP specific responses induced by vaccination. It is interesting to note that for most of the overlapping probesets, responses observed after incubation with HPV-16 L1 VLP are stronger than before vaccination. This probably reflects an increase in the number of responding cells after vaccination. In contrast to the vaccination-specific responses, a strong induction of an interferon response was observed in the vaccination-independent responses. These findings are important considering the current potential applications of VLP as vectors for antigen delivery and as vaccines for various pathogens and tumors (52).
Peripheral blood leukocytes (PBMCs) are a valuable tool to study immune responses to various diseases (29, 30, 53). Use of PBMCs, however, has some limitations that need to be taken into account. PBMCs are heterogeneous in their cell composition. For this reason, an enhanced or reduced gene expression might be restricted to particular cell populations. In addition, the response to stimulation with HPV-16 L1 VLP in an ex vivo system might differ from responses that occur in disease-relevant tissues. Also, since the number of memory cells in PBMCs is low, a recall response could be diluted by the response from other cell populations. Thus, it is conceivable that large alterations in the expression of a unique gene from an underrepresented cell type may be overlooked upon examination of the PBMC population. Despite these considerations, we confirmed a group of genes using RT-PCR analysis, and a good correlation for a number of genes with neutralizing antibody levels was observed. Moreover, we found no correlation between levels of gene expression and the percentage of leukocyte subpopulations, suggesting that differences seen were not likely to be due to changes in cell populations. In fact, flow cytometric analysis of main lymphocyte subsets revealed no major cell subset differences between baseline (month 0) and month 2 samples.
An additional potential limitation of our study is the fact that results observed may not be directly extrapolated to patterns that will be obtained after vaccination with the commercially available HPV L1 VLP vaccines, because participants in our study were immunized with HPV-16 L1VLP without an adjuvant. In addition, the HPV-16 L1 VLP vaccine used in our study is monovalent and produced in a recombinant baculovirus system. However, our results provide interesting and unique information on HPV-16 L1 VLP-specific changes, independent of adjuvant effects.
In conclusion, we used gene expression profiling to identify important adaptive and innate immune responses induced by vaccination with HPV-16 L1 VLP vaccine. We also identified differentially expressed genes predictive of neutralizing antibody titers. Interestingly, the host transcriptional response to HPV-16 L1 VLPs following vaccination varies considerably among donors, raising the need for further studies to better understand the potential impact of those differences in APC function, T cell and B cell responses and how these gene signatures predict immunogenicity and long term outcome.
Supplementary Material
Acknowledgments
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Grant Support:
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health (N01-CO-12400).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 3.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy DR, Schiller JT. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FUTURE II study group. Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N Engl J Med. 2007;356:1915–1927. doi: 10.1056/NEJMoa061741. [DOI] [PubMed] [Google Scholar]
- 6.Garland SM, Hernandez-Avila M, Wheeler CM, Perez G, Harper DM, Leodolter S, Tang GW, Ferris DG, Steben M, Bryan J, Taddeo FJ, Railkar R, Esser MT, Sings HL, Nelson M, Boslego J, Sattler C, Barr E, Koutsky LA. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N Engl J Med. 2007;356:1928–1943. doi: 10.1056/NEJMoa061760. [DOI] [PubMed] [Google Scholar]
- 7.Paavonen J, Jenkins D, Bosch FX, Naud P, Salmeron J, Wheeler CM, Chow SN, Apter DL, Kitchener HC, Castellsague X, de Carvalho NS, Skinner SR, Harper DM, Hedrick JA, Jaisamrarn U, Limson GA, Dionne M, Quint W, Spiessens B, Peeters P, Struyf F, Wieting SL, Lehtinen MO, Dubin G. Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: an interim analysis of a phase III double-blind, randomised controlled trial. Lancet. 2007;369:2161–2170. doi: 10.1016/S0140-6736(07)60946-5. [DOI] [PubMed] [Google Scholar]
- 8.Brown DR, Fife KH, Wheeler CM, Koutsky LA, Lupinacci LM, Railkar R, Suhr G, Barr E, Dicello A, Li W, Smith JF, Tadesse A, Jansen KU. Early assessment of the efficacy of a human papillomavirus type 16 L1 virus-like particle vaccine. Vaccine. 2004;22:2936–2942. doi: 10.1016/j.vaccine.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 9.Fife KH, Wheeler CM, Koutsky LA, Barr E, Brown DR, Schiff MA, Kiviat NB, Jansen KU, Barber H, Smith JF, Tadesse A, Giacoletti K, Smith PR, Suhr G, Johnson DA. Dose-ranging studies of the safety and immunogenicity of human papillomavirus Type 11 and Type 16 virus-like particle candidate vaccines in young healthy women. Vaccine. 2004;22:2943–2952. doi: 10.1016/j.vaccine.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 10.Poland GA, Jacobson RM, Koutsky LA, Tamms GM, Railkar R, Smith JF, Bryan JT, Cavanaugh PF, Jr, Jansen KU, Barr E. Immunogenicity and reactogenicity of a novel vaccine for human papillomavirus 16: a 2-year randomized controlled clinical trial. Mayo Clin Proc. 2005;80:601–610. doi: 10.4065/80.5.601. [DOI] [PubMed] [Google Scholar]
- 11.Harper DM, Franco EL, Wheeler CM, Moscicki AB, Romanowski B, Roteli-Martins CM, Jenkins D, Schuind A, Costa Clemens SA, Dubin G. Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: follow-up from a randomised control trial. Lancet. 2006;367:1247–1255. doi: 10.1016/S0140-6736(06)68439-0. [DOI] [PubMed] [Google Scholar]
- 12.Mao C, Koutsky LA, Ault KA, Wheeler CM, Brown DR, Wiley DJ, Alvarez FB, Bautista OM, Jansen KU, Barr E. Efficacy of human papillomavirus-16 vaccine to prevent cervical intraepithelial neoplasia: a randomized controlled trial. Obstet Gynecol. 2006;107:18–27. doi: 10.1097/01.AOG.0000192397.41191.fb. [DOI] [PubMed] [Google Scholar]
- 13.Villa LL, Costa RL, Petta CA, Andrade RP, Paavonen J, Iversen OE, Olsson SE, Hoye J, Steinwall M, Riis-Johannessen G, Andersson-Ellstrom A, Elfgren K, Krogh G, Lehtinen M, Malm C, Tamms GM, Giacoletti K, Lupinacci L, Railkar R, Taddeo FJ, Bryan J, Esser MT, Sings HL, Saah AJ, Barr E. High sustained efficacy of a prophylactic quadrivalent human papillomavirus types 6/11/16/18 L1 virus-like particle vaccine through 5 years of follow-up. Br J Cancer. 2006;95:1459–1466. doi: 10.1038/sj.bjc.6603469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emeny RT, Wheeler CM, Jansen KU, Hunt WC, Fu TM, Smith JF, MacMullen S, Esser MT, Paliard X. Priming of human papillomavirus type 11-specific humoral and cellular immune responses in college-aged women with a virus-like particle vaccine. J Virol. 2002;76:7832–7842. doi: 10.1128/JVI.76.15.7832-7842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans TG, Bonnez W, Rose RC, Koenig S, Demeter L, Suzich JA, O'Brien D, Campbell M, White WI, Balsley J, Reichman RC. A Phase 1 study of a recombinant viruslike particle vaccine against human papillomavirus type 11 in healthy adult volunteers. J Infect Dis. 2001;183:1485–1493. doi: 10.1086/320190. [DOI] [PubMed] [Google Scholar]
- 16.Pinto LA, Castle PE, Roden RB, Harro CD, Lowy DR, Schiller JT, Wallace D, Williams M, Kopp W, Frazer IH, Berzofsky JA, Hildesheim A. HPV-16 L1 VLP vaccine elicits a broad-spectrum of cytokine responses in whole blood. Vaccine. 2005;23:3555–3564. doi: 10.1016/j.vaccine.2005.01.146. [DOI] [PubMed] [Google Scholar]
- 17.Pinto LA, Edwards J, Castle PE, Harro CD, Lowy DR, Schiller JT, Wallace D, Kopp W, Adelsberger JW, Baseler MW, Berzofsky JA, Hildesheim A. Cellular immune responses to human papillomavirus (HPV)-16 L1 in healthy volunteers immunized with recombinant HPV-16 L1 virus-like particles. J Infect Dis. 2003;188:327–338. doi: 10.1086/376505. [DOI] [PubMed] [Google Scholar]
- 18.Yao Q, Zhang R, Guo L, Li M, Chen C. Th cell-independent immune responses to chimeric hemagglutinin/simian human immunodeficiency virus-like particles vaccine. J Immunol. 2004;173:1951–1958. doi: 10.4049/jimmunol.173.3.1951. [DOI] [PubMed] [Google Scholar]
- 19.Zinkernagel RM, Bachmann MF, Kundig TM, Oehen S, Pirchet H, Hengartner H. On immunological memory. Annu Rev Immunol. 1996;14:333–367. doi: 10.1146/annurev.immunol.14.1.333. [DOI] [PubMed] [Google Scholar]
- 20.de Jong D, Rosenwald A, Chhanabhai M, Gaulard P, Klapper W, Lee A, Sander B, Thorns C, Campo E, Molina T, Norton A, Hagenbeek A, Horning S, Lister A, Raemaekers J, Gascoyne RD, Salles G, Weller E. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications--a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25:805–812. doi: 10.1200/JCO.2006.09.4490. [DOI] [PubMed] [Google Scholar]
- 21.Dodd LE, Sengupta S, Chen IH, den Boon JA, Cheng YJ, Westra W, Newton MA, Mittl BF, McShane L, Chen CJ, Ahlquist P, Hildesheim A. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:2216–2225. doi: 10.1158/1055-9965.EPI-06-0455. [DOI] [PubMed] [Google Scholar]
- 22.Kendrick JE, Conner MG, Huh WK. Gene expression profiling of women with varying degrees of cervical intraepithelial neoplasia. J Low Genit Tract Dis. 2007;11:25–28. doi: 10.1097/01.lgt.0000230124.68996.38. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, Cheng YJ, Westra WH, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. Genome-Wide Expression Profiling Reveals EBV-Associated Inhibition of MHC Class I Expression in Nasopharyngeal Carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 24.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 25.Efron B, Tibshirani R. On Testing the Significance of Sets of Genes. The Annals of Applied Statistics. 2007;1:107–129. [Google Scholar]
- 26.Cliff JM, Andrade IN, Mistry R, Clayton CL, Lennon MG, Lewis AP, Duncan K, Lukey PT, Dockrell HM. Differential gene expression identifies novel markers of CD4+ and CD8+ T cell activation following stimulation by Mycobacterium tuberculosis. J Immunol. 2004;173:485–493. doi: 10.4049/jimmunol.173.1.485. [DOI] [PubMed] [Google Scholar]
- 27.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 28.Kawada J, Kimura H, Kamachi Y, Nishikawa K, Taniguchi M, Nagaoka K, Kurahashi H, Kojima S, Morishima T. Analysis of gene-expression profiles by oligonucleotide microarray in children with influenza. J Gen Virol. 2006;87:1677–1683. doi: 10.1099/vir.0.81670-0. [DOI] [PubMed] [Google Scholar]
- 29.Ockenhouse CF, Bernstein WB, Wang Z, Vahey MT. Functional genomic relationships in HIV-1 disease revealed by gene-expression profiling of primary human peripheral blood mononuclear cells. J Infect Dis. 2005;191:2064–2074. doi: 10.1086/430321. [DOI] [PubMed] [Google Scholar]
- 30.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blumerman SL, Herzig CT, Wang F, Coussens PM, Baldwin CL. Comparison of gene expression by co-cultured WC1+ gammadelta and CD4+ alphabeta T cells exhibiting a recall response to bacterial antigen. Mol Immunol. 2007;44:2023–2035. doi: 10.1016/j.molimm.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Mueller A, O'Rourke J, Chu P, Kim CC, Sutton P, Lee A, Falkow S. Protective immunity against Helicobacter is characterized by a unique transcriptional signature. Proc Natl Acad Sci U S A. 2003;100:12289–12294. doi: 10.1073/pnas.1635231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Mitoma F, Alvarez-Maya I, Dabrowski A, Jaffey J, Frost R, Aucoin S, Kryworuchko M, Lapner M, Tadesse H, Giulivi A. Transcriptional analysis of human peripheral blood mononuclear cells after influenza immunization. J Clin Virol. 2004;31:100–112. doi: 10.1016/j.jcv.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Fuller CL, Brittingham KC, Porter MW, Hepburn MJ, Petitt PL, Pittman PR, Bavari S. Transcriptome analysis of human immune responses following live vaccine strain (LVS) Francisella tularensis vaccination. Mol Immunol. 2007;44:3173–3184. doi: 10.1016/j.molimm.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giacalone MJ, Sabbadini RA, Chambers AL, Pillai S, Berkley NL, Surber MW, McGuire KL. Immune responses elicited by bacterial minicells capable of simultaneous DNA and protein antigen delivery. Vaccine. 2006;24:6009–6017. doi: 10.1016/j.vaccine.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harro CD, Pang YY, Roden RB, Hildesheim A, Wang Z, Reynolds MJ, Mast TC, Robinson R, Murphy BR, Karron RA, Dillner J, Schiller JT, Lowy DR. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J Natl Cancer Inst. 2001;93:284–292. doi: 10.1093/jnci/93.4.284. [DOI] [PubMed] [Google Scholar]
- 37.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto LA, Trivett MT, Wallace D, Higgins J, Baseler M, Terabe M, Belyakov IM, Berzofsky JA, Hildesheim A. Fixation and cryopreservation of whole blood and isolated mononuclear cells: Influence of different procedures on lymphocyte subset analysis by flow cytometry. Cytometry B Clin Cytom. 2005;63:47–55. doi: 10.1002/cyto.b.20038. [DOI] [PubMed] [Google Scholar]
- 39.Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger Kjaer S, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology. 2004;321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolf MP, Fausch SC, Da Silva DM, Kast WM. Human dendritic cells are activated by chimeric human papillomavirus type-16 virus-like particles and induce epitope-specific human T cell responses in vitro. J Immunol. 2001;166:5917–5924. doi: 10.4049/jimmunol.166.10.5917. [DOI] [PubMed] [Google Scholar]
- 43.Yang R, Murillo FM, Cui H, Blosser R, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. Papillomavirus-like particles stimulate murine bone marrow-derived dendritic cells to produce alpha interferon and Th1 immune responses via MyD88. J Virol. 2004;78:11152–11160. doi: 10.1128/JVI.78.20.11152-11160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang XQ, Evans GF, Alfaro ML, Zuckerman SH. Down-regulation of macrophage CD9 expression by interferon-gamma. 1: Biochem Biophys Res Commun. 2002;290(3):891–897. doi: 10.1006/bbrc.2001.6293. [DOI] [PubMed] [Google Scholar]
- 45.Arndt M, Lendeckel U, Spiess A, Faust J, Neubert K, Reinhold D, Ansorge S. Dipeptidyl peptidase IV (DP IV/CD26) mRNA expression in PWM-stimulated T-cells is suppressed by specific DP IV inhibition, an effect mediated by TGF-beta(1) Biochem Biophys Res Commun. 2000;274(2):410–414. doi: 10.1006/bbrc.2000.3144. [DOI] [PubMed] [Google Scholar]
- 46.Begum NA, Tsuji S, Nomura M, Shida K, Azuma I, Hayashi A, Matsumoto M, Seya T, Toyoshima K. Human MD-1 homologue is a BCG-regulated gene product in monocytes: its identification by differential display. Biochem Biophys Res Commun. 1999;256(2):325–329. doi: 10.1006/bbrc.1999.0329. [DOI] [PubMed] [Google Scholar]
- 47.Hebenstreit D, Giaisi M, Treiber MK, Zhang XB, Mi HF, Horejs-Hoeck J, Andersen KG, Krammer PH, Duschl A, Li-Weber M. LEF-1 negatively controls interleukin-4 expression through a proximal promoter regulatory element. J Biol Chem. 2008;283(33):22490–22907. doi: 10.1074/jbc.M804096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 49.Sturm A, Lensch M, Andre S, Kaltner H, Wiedenmann B, Rosewicz S, Dignass AU, Gabius HJ. Human galectin-2: novel inducer of T cell apoptosis with distinct profile of caspase activation. J Immunol. 2004;173:3825–3837. doi: 10.4049/jimmunol.173.6.3825. [DOI] [PubMed] [Google Scholar]
- 50.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catalan N, Selz F, Imai K, Revy P, Fischer A, Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 52.Ramqvist T, Andreasson K, Dalianis T. Vaccination, immune and gene therapy based on virus-like particles against viral infections and cancer. Expert Opin Biol Ther. 2007;7:997–1007. doi: 10.1517/14712598.7.7.997. [DOI] [PubMed] [Google Scholar]
- 53.Lempicki RA, Polis MA, Yang J, McLaughlin M, Koratich C, Huang DW, Fullmer B, Wu L, Rehm CA, Masur H, Lane HC, Sherman KE, Fauci AS, Kottilil S. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193:1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.