Abstract
Salvage of nucleosides in the cytosol of human cells is carried out by deoxycytidine kinase (dCK) and thymidine kinase 1 (TK1). Whereas TK1 is only responsible for thymidine phosphorylation, dCK is capable of converting dC, dA, and dG into their monophosphate forms. Using structural data on dCK we predicted that select mutations at the active site would, in addition to making the enzyme faster, expand the catalytic repertoire of dCK to include thymidine. Specifically, we hypothesized that steric repulsion between the methyl group of the thymine base and Arg104 is the main factor preventing the phosphorylation of thymidine by wild type dCK. Here we present kinetic data on several dCK variants where Arg104 has been replaced by select residues, all performed in combination with the mutation of Asp133 to an alanine. We show that several hydrophobic residues at position 104 endow dCK with thymidine kinase activity. Depending on the exact nature of the mutations, the enzyme’s substrate preference is modified. The R104M-D133A double mutant is a pyrimidine-specific enzyme due to large Km values with purines. Crystal structure of the double mutant R104M-D133A in complex with the L-form of thymidine supplies a structural explanation for the ability of this variant to phosphorylate thymidine and thymidine analogs. The replacement of Arg104 by a smaller residue allows L-dT to bind deeper into the active site, making space for the C5-methyl group of the thymine base. The unique catalytic properties of several of the mutants make them good candidates for suicide gene/protein therapy applications.
As an initial enzyme in the cytoplasmic nucleoside salvage pathway, the physiological role of human deoxycytidine kinase (dCK) is to phosphorylate the nucleosides deoxycytidine (dC), deoxyadenosine (dA), and deoxyguanosine (dG) into their monophosphate forms. In humans, cytoplasmic salvage of the remaining base, thymidine (dT), is accomplished by thymidine kinase 1. The enzymes deoxyguanosine kinase and thymidine kinase 2 perform nucleoside salvage in the mitochondria (1). Interestingly, in the fruit fly a single enzyme that is homologous to dCK, referred to as deoxyribonucleoside kinase (dNK), is able to phosphorylate nucleosides containing any of the four bases (2). Moreover, dNK is considerably more active than human dCK (3). We were interested in understanding the reasons that make dNK a more efficient nucleoside kinase, and those that prevent dCK from being a thymidine kinase. Our long-term goal is to capitalize on such an understanding in order to engineer dCK to acquire improved activity and more diverse substrate specificity. Such engineered enzyme variants could play a role in therapeutic applications by efficiently activating unique nucleoside analog prodrugs.
Analysis of the sequences of human dCK and Drosophila melanogaster dNK reveals 31% identity (54% homology). Not surprisingly, the tertiary structures are very similar, with a root mean square deviation (RMSD) of 1.19 Å on 187 common Cα atoms. Nucleoside kinases such as dCK and dNK simultaneously bind the donor nucleotide and the acceptor nucleoside. We speculated that the kinetic differences between the two enzymes are due to variations in the nucleoside binding sites. Comparison of the nucleoside binding sites of dCK and dNK revealed three notable differences: in position 100, dNK has a valine whereas dCK an alanine; in position 104, a methionine versus an arginine; and in position 133, an alanine versus an aspartic acid. Residues at these positions are in proximity to the base moiety of the nucleoside. In previous work, we demonstrated that exchanging these three residues in dCK to the sequence as found in dNK results in a more active enzyme (4). However, this triple-mutant of dCK turned out to be less stable than the wild-type enzyme upon expression and purification from bacterial cells. We surmised that much of this instability is caused by the substitution at position 100 (A to V), but that this substitution was not critical for the improved kinetics of the enzyme. Indeed, the double mutant R104M-D133A maintains much of the improved kinetic properties of the triple mutant, and is more stable. Thus, this observation suggested a pivotal role for residues at position 104 and 133 in determining the catalytic prowess of the enzyme, and potentially its substrate specificity. Interestingly, a recent report from the Lutz group, using a genetic approach, independently identified these two residues as critical for endowing human dCK with thymidine kinase activity (5).
Here we report on the kinetic analysis of dCK variants that have been mutated to select residues at position 104 and an alanine at position 133. Our goal was to identify the optimal residue at position 104 in the background of an alanine at position 133. In addition, we present the crystal structure of the R104M-D133A double mutant in complex with the L-form of thymidine (L-dT). Human dCK, in addition to being able to phosphorylate both purines and pyrimidines, has the special ability to accept L-nucleosides as substrates (6). This structure provides an explanation for the ability of this variant to efficiently phosphorylate nucleosides with a thymine base.
Experimental Procedures
Materials
All general laboratory reagents were purchased from Fisher and Sigma. Pyruvate kinase and lactate dehydrogenase used for the kinetic assay were from Roche. Nucleosides and nucleotides were obtained from Sigma, except L-deoxythymidine (L-dT) that was purchased from ChemGenes Corporation. DNA samples were purified using the QIAprep purification kits (Qiagen) according to the manufacturer’s protocols. Oligonucleotides were purchased from Sigma.
Site-directed mutagenesis, protein expression, and purification
The various mutations of human dCK at position 104 and the alanine at position 133 were introduced using the QuikChange Site-Directed Mutagenesis Kit from Stratagene. The wild type dCK gene cloned into the pET14b vector providing a His6 tag at the N-terminus served as the initial template. Gene sequences were confirmed by sequencing. For expression, C41 (DE3) Escherichia coli was transformed with the dCK constructs in the pET14b vector, grown in 2YT media and induced with 0.1 mM IPTG over 4 h at 37 °C. Cells were harvested and the pellet lysed by sonication in a lysis buffer containing 50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol. Lysates were cleared by centrifugation at 30,000 × g for 1 hour at 4 °C and subjected to purification via Ni-NTA columns (GE Healthcare). After loading, the column was washed using a buffer containing 50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, and 50 mM imidazole. Subsequently, the protein was eluted with a buffer identical to the wash buffer but containing 250 mM imidazole. The eluted protein was aliquoted, frozen in liquid nitrogen, and stored at −80 °C until use.
Kinetic assay
The enzymatic activities of wild type dCK and mutants were determined with an NADH-dependent enzyme-coupled assay using a Carry UV spectrophotometer (7). Measurements were done at 37 °C in a buffer containing 100 mM Tris, pH 7.5,100 mM KCl, 10 mM MgCl2 and 1 mM ATP. The kobs values represent the rate of phosphorylation at 200 µM nucleoside. For kcat and Km determination, varying ranges of nucleoside concentrations were used (dC = 5 to 200 µM; AraC and gemcitabine = 20 to 200 µM; D-dT and L-dT = 20 to 500 µM; dA = 25 to 1000 µM; dG = 25 to 2000 µM). The concentration of dCK was 0.33 µM.
Crystallization, x-ray data collection, and refinement
Crystals of the L-dT/ADP complex were obtained by vapor diffusion from hanging drops (1 µl of protein/nucleoside/nucleotide mix plus 1 µl reservoir) using a reservoir solution that contained 0.90–1.5 M trisodium citrate dihydrate and 100 mM HEPES, pH 7.5. The crystals were grown at 22 °C. Diffraction data were collected from a single frozen crystal at the SER-CAT beam line BM-22 at the Advanced Photon Source, Argonne National Laboratory, and processed with XDS (8). The structure was solved by molecular replacement using the program MOLREP (9) and the dCK structure 1P5Z as search model. Refinement was carried out with REFMAC (10). Data collection and refinement statistics are presented in Table 5. The structure has been deposited in the PDB under accession number 3EXK.
Table 5.
Data collection and refinement statistics
| PDB ID | 3EXK |
| Beamline | SERCAT BM-22 |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Resolution Range (Å) | 30.0 – 2.3 |
| Reflections | |
| Observed | 93997 |
| Unique | 13710 |
| Completeness (%) | 99.3 (98.7)a |
| Rsym(%) | 8.9 (29.8) |
| I/σ(I) | 14.42 (6.93) |
| Space group | P43 21 2 |
| Unit cell (Å) | |
| a, b | 79.78 |
| c | 93.70 |
| Molecules per a.u. | 1 |
| Refinement statistics | |
| Rcryst (%) | 21.7 |
| Rfree (%) | 27.8 |
| Number of atoms | |
| Protein | 1838 |
| Nucleoside | 17 |
| ADP | 27 |
| Water | 63 |
| R.m.s. deviation | |
| Bond length (Å) | 0.013 |
| Bond angles (°) | 1.437 |
| Average B-factors (Å2) | |
| Protein | 45.0 |
| Main chain | 44.5 |
| Side chain | 45.4 |
| ADP | 35.5 |
| Nucleoside | 35.5 |
| Waters | 47.4 |
values for the highest resolution shell is in parenthesis
Results
Rationale for mutants generated
Our previous work, based on comparative analysis of human dCK to fruit fly dNK, identified positions 104 and 133 as important in determining the rate and substrate specificity of the enzyme (4). We speculated that for dCK the very high Km with D-dT of over 3 mM (5) is due to steric hindrance caused by the side chain of Arg104. The analogous position in dNK contains a methionine. Thus, exchanging the arginine with a shorter residue was predicted to make the necessary space for the methyl moiety present in the thymine base. However, we predicted that the single mutant R104M would be unstable due to the presence of Asp133. In the structure of wild-type dCK, a bidentate salt-bridge interaction is formed between Arg104 and Asp133. Solely exchanging the charged arginine residue with the hydrophobic methionine would leave the charge on Asp133 with no stabilizing partner. Consistent with this analysis, in dNK the methionine is paired with an alanine at the analogous position to dCK’s Asp 133. Therefore, we combined the R104M mutation with the D133A mutation. The terminology we use to designate the double mutants (DM) is in the form of DMXY, where X and Y are the single letter code for residues at positions 104 and 133, respectively. Since DMMA exhibited improved kinetic properties, we asked the question of what other substitutions at position 104 are tolerated. Small side chains were excluded from consideration on the premise that in exchanging the large arginine side chain, a small one would result in a cavity in the interior of the protein, which is likely to be destabilizing. We chose to focus on hydrophobic residues such as phenylalanine, leucine, isoleucine, and valine. Additionally, we tested a glutamine at this position, as it is uncharged and large enough to substitute the arginine, but small enough to make space for the thymine base. Lastly, we also tested the replacement of arginine by the more flexible lysine. Mutants were generated and purified in a single step by metal-affinity chromatography, yielding preparations of >95% purity. The N-terminal his-tag was not cleaved since previous experiments have shown that the tag does not influence the kinetic behavior of the protein (data not shown). To initially characterize the different mutants, we chose to use saturating concentration of ATP (1 mM) and a nucleoside concentration of 200 µM. The rationale for the latter is that we are seeking mutants that have relatively low nucleoside Km values.
Activity of mutants towards the physiological substrates of dCK – dC, dA, dG
In contrast to dCK, the enzyme dNK, in addition to being able to phosphorylate all four bases, is characterized by being generally much faster. For dNK, kcat varies between 14 to 20 sec−1 (3), whereas dCK is remarkably slow with the pyrimidine dC (kcat of 0.04 sec−1), and ~2.5 sec−1 with the purines dA and dG (see below). As many of the dCK mutants were designed to mimic the situation in dNK by having a hydrophobic residue at position 104, and an alanine at position 133, we anticipated a generally faster kinase. Table 1 shows that of the various hydrophobic permutations probed at position 104, the most active had either a methionine (as in dNK) or a leucine. At the sensitivity threshold of our assay, the mutant with a phenyl group at this position showed no activity, presumably due to its bulkiness poorly fitting in the space previously occupied by the more linear arginine. Mutants with other beta-branched side chains also showed little or no activity – the isoleucine variant being significantly lower than the leucine counterpart, and the valine variant below our activity cut-off.
Table 1.
Kinetic characterization of dCK variants with mutations at positions 104 and 133 with respect to the physiological nucleoside substrates
| Mutant | akobs D-dC | Rel kobs | kobs D-dG | Rel kobs | kobs D-dA | Rel kobs |
|---|---|---|---|---|---|---|
| WT | 0.03 ± 0.01b | 1 | 0.94 ± 0.18 | 1 | 1.19 ± 0.01 | 1 |
| DMMA | 1.84 ± 0.04 | 61 | 0.18 ± 0.01 | 0.19 | 0.37 ± 0.01 | 0.31 |
| DMLA | 0.27 ± 0.09 | 9 | 0.30 ± 0.01 | 0.32 | 3.45 ± 0.16 | 2.9 |
| DMIA | 0.06 ± 0.01 | 2 | 0.08 ± 0.01 | 0.08 | 0.21 ± 0.01 | 0.18 |
| DMQA | 0.08 ± 0.01 | 2.7 | 0.06 ± 0.01 | 0.06 | 0.18 ± 0.04 | 0.15 |
| DMVA | <0.005 | <0.005 | <0.005 | |||
| DMFA | <0.005 | <0.005 | <0.005 | |||
| DMKA | <0.005 | <0.005 | <0.005 |
kobs are in sec−1 determined for a nucleoside concentration of 200 µM
Standard deviation
Interestingly, whereas in the dNK enzyme kcat is relatively similar between the different nucleosides (14 to 20 sec−1 (3)), our most active mutants (DMMA and DMLA) showed selective rate enhancement relative to the rate of wild-type dCK. The DMMA variant is >60-fold faster (kobs determined at 200 µM nucleoside) with the pyrimidine dC in comparison to wild-type dCK. In contrast, kobs with the purines dA and dG are lower in the mutant. Combined with the fact that DMMA can also efficiently phosphorylate thymidine (see below), this shows that this variant greatly prefers pyrimidine substrates over purines. In contrast, the DMLA variant has preference for dA, with ~10-fold lower kobs values for dC and dG. The preferential behavior of this mutant towards dA can be rationalized by the unique binding mode we previously observed with this nucleoside (see Discussion). These results support our hypothesis that the nature of the residue at position 104 has significant ramifications as to the rate and substrate preference of the enzyme.
Activity of mutants towards the dC analogs AraC and gemcitabine
Human dCK plays an essential role in the activation of medicinally relevant nucleoside analogs. Examples include AraC and gemcitabine, which are extensively used for the treatment of hematological malignancies and some types of solid tumors. One of our goals is to generate dCK variants that have improved activity with these dC analogs. Wild-type dCK phosphorylates both of these dC analogs with ~10- to 13-fold higher kobs compared to dC (Table 2). As observed with the physiological nucleosides, the best double mutant variants for AraC and gemcitabine phosphorylation are DMMA and DMLA. DMIA and DMQA show approximately half the rate relative to wild-type dCK. The mutants that exhibited undetectable activity with the physiological substrates also have no activity with the nucleoside analogs.
Table 2.
Kinetic characterization of dCK variants with mutations at positions 104 and 133 with respect to the non-physiological substrates AraC and Gemcitabine
| Mutant | akobs AraC | Rel kobs | kobs Gem | Rel kobs |
|---|---|---|---|---|
| WT | 0.30 ± 0.04b | 1.00 | 0.39 ± 0.01 | 1.00 |
| DMMA | 0.84 ± 0.02 | 1.70 | 2.39 ± 0.07 | 5.80 |
| DMLA | 0.64 ± 0.06 | 0.35 | 2.04 ± 0.03 | 5.20 |
| DMIA | 0.07 ± 0.01 | 0.25 | 0.21 ± 0.08 | 0.53 |
| DMQA | 0.18 ± 0.01 | 0.60 | 0.17 ± 0.04 | 0.43 |
| DMVA | <0.005 | <0.005 | ||
| DMFA | <0.005 | <0.005 | ||
| DMKA | <0.005 | <0.005 |
kobs are in sec−1 determined for a nucleoside concentration of 200 µM
Standard deviation

In case of the significantly more active variants DMMA and DMLA, we note that the rate is noticeably faster with gemcitabine (5- to 6-fold) but only slightly better with AraC. In contrast, at the conditions of the measurements, wild-type dCK has very similar rates for both nucleoside analogs. Thus, these dCK variants have gained a preference for gemcitabine, despite the fact that the structural elements that differentiate between these agents are confined to the sugar moiety, and the site of the mutation at position 104 is distant from that location.
Activity of mutants towards thymidine
One of our goals was to increase the substrate repertoire of dCK to include thymidine. And indeed DMMA and DMLA are fast thymidine kinases, while DMIA and DMQA have low thymidine kinase activity (Table 3). Both mutants can phosphorylate the D- and L-form of thymidine. Similarly to the situation above, we observe that the exact nature of the residue at position 104 determines the specificity. With a methionine, the L-form is phosphorylated twice as fast as the D-form. Conversely, with a leucine at position 104, the D-form is faster by about 3-fold.
Table 3.
Kinetic characterization of dCK variants with mutations at positions 104 and 133 with respect to D- L-thymidine.
| Mutant | akobs D-dT | Rel kobs | kobs L-dT | Rel kobs |
|---|---|---|---|---|
| WT | 0.008 ± 0.001b | 1 | 0.017 ± 0.023 | 1 |
| DMMA | 1.15 ± 0.01 | 144 | 1.95 ± 0.09 | 115 |
| DMLA | 2.86 ± 0.09 | 358 | 1.07 ± 0.09 | 63 |
| DMIA | 0.40 ± 0.01 | 50 | 0.19 ± 0.02 | 10 |
| DMQA | 0.10 ± 0.06 | 12.5 | 0.15 ± 0.02 | 9 |
| DMVA | <0.005 | <0.005 | ||
| DMFA | <0.005 | <0.005 | ||
| DMKA | <0.005 | <0.005 |
kobs are in sec−1 determined for a nucleoside concentration of 200 µM
Standard deviation
Kinetic characterization of DMMA and DMLA
The experiments using a single nucleoside concentration of 200 µM identified the double mutants DMMA and DMLA as the most active ones. We proceeded to fully characterize these dCK variants kinetically with respect to the physiological substrates of dCK (dC, dA, dG), the nucleoside analogs AraC and gemcitabine, and both enantiomeric forms of thymidine (Table 4). In terms of efficiency, the highest value is attained for dC with DMMA. While the Km for dC is slightly increased in the mutant, this is offset by a dramatic 45-fold increase in kcat. Of note, the rate of dC phosphorylation by wild-type dCK is remarkably low. Recently, it was reported that dCK undergoes post-translational phosphorylation on Ser74 that increases its activity (11, 12). Using the dCK S74E mutant to mimic this phosphorylated state, we could show that this modification increases the rate of phosphorylation for dC by 11-fold, but not for dA or dG (13). The mechanism behind the selective rate enhancement of the S74E mutant is still not clear, as the serine is a part of the region called the insert, which is not observed in most crystal structures of dCK. Interestingly, the DMMA variant also selectively increases the rate of dC phosphorylation. The combination of the mutations present in DMMA and the S74E mutation results in a further increase in rate of dC phosphorylation to ~ 5 sec−1 (Hazra et al, unpublished results). This suggests that a different mechanism exists between that responsible for the rate enhancement by the S74E mutation, and that due to the R104M-D133A mutations.
Table 4.
Comparison of catalytic efficiencies of new mutants with wild type dCK
| Substrate | WT | R104M-D133A | R104L-D133A | ||||||
|---|---|---|---|---|---|---|---|---|---|
| kcat (sec−1) | Km (µM) |
kcat/Km (× 10−3) |
kcat (sec−1) | Km (µM) |
kcat/Km (× 10−3) |
kcat (sec−1) | Km (µM) |
kcat/Km (× 10−3) |
|
| D-dC | 0.04 ± 0.01a | <3 | >13.3 | 1.80 ± 0.04 | 5.70 ± 0.44 | 315.8 | 0.25 ± 0.14 | 8.0 ± 1.0 | 31.3 |
| D-dA | 2.13 ± 0.35 | 114.6 ± 3.5 | 18.6 | 4.51 ± 0.33 | 1040 ± 117 | 4.3 | 5.72 ± 0.21 | 162.3 ± 20.1 | 35.2 |
| D-dG | 2.60 ± 0.10 | 231.0 ± 20.0 | 11.3 | 1.73 ± 0.12 | 1865 ± 211 | 0.9 | 3.66 ± 0.01 | 2266 ± 823 | 1.6 |
| AraC | 0.34 ± 0.01 | 13.1 ± 1.1 | 26.0 | 1.43 ± 0.03 | 136.5 ± 10.0 | 10.5 | 0.79 ± 0.03 | 50.6 ± 6.1 | 15.6 |
| Gem | 0.39 ± 0.03 | 16.1 ± 3.5 | 24.2 | 2.68 ± 0.07 | 56.2 ± 16.8 | 47.7 | 2.01 ± 0.08 | 33.8 ± 5. 3 | 59.5 |
| D-dT | NA | NA | - | 1.74 ± 0.01 | 144.0 ± 10.1 | 12.1 | 3.20 ± 0.07 | 24.3 ± 3.6 | 131.7 |
| L-dT | NA | NA | - | 3.13 ± 0.10 | 138.0 ± 10.1 | 22.7 | 1.33 ± 0.01 | 17.8 ± 1.6 | 74.7 |
standard error
NA: not applicable, due to the very high Km (> 3 mM)
The specificity of DMMA to dC is due to the much-increased Km values for both dA and dG (8- to 10-fold over the Km with wild-type dCK). In contrast, in the case of DMLA, while the Km of dG is also dramatically higher, that for dA is comparable to wild-type dCK. Our recent structural analysis of dCK in complex with dA and dG (14) supplies a structural explanation for the different effect between dA and dG (see Discussion).
The enhancement in phosphorylation efficiency (kcat/Km) for gemcitabine by DMMA and DMLA, 2- and 2.5-fold, respectively, is due to a significant improvement in kcat, whereas Km counters this by a moderate increase. Interestingly, the efficiency of AraC phosphorylation by either mutant is below that of wild-type dCK. This demonstrates that whatever the mechanism for efficiency enhancement elicited by DMMA and DMLA, it is sensitive to the substituents at the 2’-position of the sugar ring. In contrast, the S74E mutant discussed previously does not significantly change the efficiency of AraC and gemcitabine phosphorylation (13). This is in agreement with our proposal that the mechanisms behind the modified activity for the S74E mutant and for the DMMA/DMLA variants are discrete.
Most interesting is the endowment of DMMA and DMLA with thymidine kinase activity. The Km for D-dT is reduced from over 3 mM with wild-type dCK (5) to less than 150 µM with DMMA and below 25 µM with DMLA. The DMLA variant also has a lower Km value for the L-form of thymidine than DMMA, and it is this fact that primarily makes this double mutant the most efficient dCK variant for thymidine kinase activity. As a methionine is one atom longer than a leucine, it is possible that this residue is still too long to optimally accommodate the C5- methyl group of the thymine base. However, due to methionine being a non-branched amino acid, thymine can still maintain a reasonable Km value (as measured for DMMA). The glutamine variant at position 104, DMQA, has much lower activity than DMLA. A glutamine, apart from being a polar residue, has the same length as a methionine but is branched at the delta-position. This branching is the likely cause for DMQA having only residual thymidine kinase activity. The structure of DMMA with L-dT (see below) supports this analysis, as the electron density for the tip of the methionine side chain is weak, consistent with high mobility/disorder caused by proximity of the C5-methyl group of L-dT.
Structural analysis of DMMA in complex with L-dT and ADP
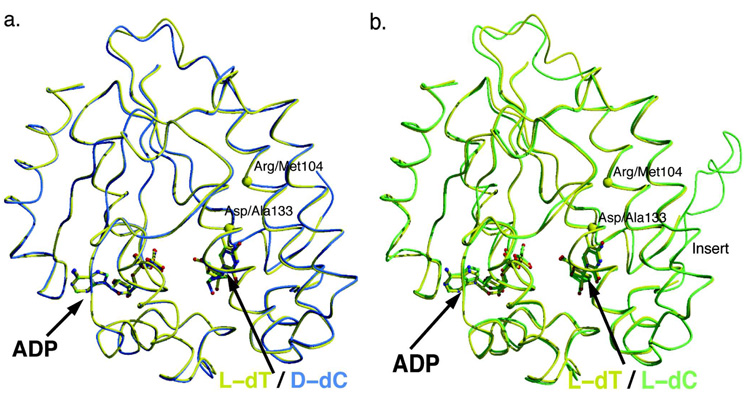
The above kinetic analyses of the dCK variants that concomitantly contain the D133A mutation and various substitutions of Arg104 revealed that DMMA and DMLA are generally endowed with higher activity and have gained the ability to phosphorylate nucleosides containing a thymine base. To understand how these mutations facilitate this we solved the 2.3 Å resolution crystal structure of DMMA in complex with ADP at the phosphoryl donor site and the L-form of thymidine at the nucleoside acceptor site (crystals with D-dT were also obtained but not pursued). Data collection and refinement statistics are presented in Table 5. As expected, the dual mutations did not change the overall structure of the enzyme. The RMSD between DMMA in complex with L-dT + ADP and wild-type dCK in complex with D-dC + ADP and is 0.40 Å on 226 atoms (Figure 1a), and with wild-type dCK in complex with L-dC + ADP is 0.64 Å on 226 atoms (Figure 1b). The local structure of regions proximal to positions 104 and 133 are also nearly identical with that in wild-type dCK. The few notable differences between the structures can be accounted by crystal contacts, and are not attributed to the mutations.
Figure 1.
The double mutant DMMA (R104M – D133A) adopts the same overall structure as wild type dCK. Superposition of the structure of DMMA in complex with L-dT and ADP (yellow) and in panel (a) wild-type dCK in complex with D-dC and ADP (cyan) or in panel (b) wild-type dCK in complex with L-dC and ADP (green) reveals that the double mutant maintains the structure of the wild-type protein. Yellow spheres mark the locations of the mutations.
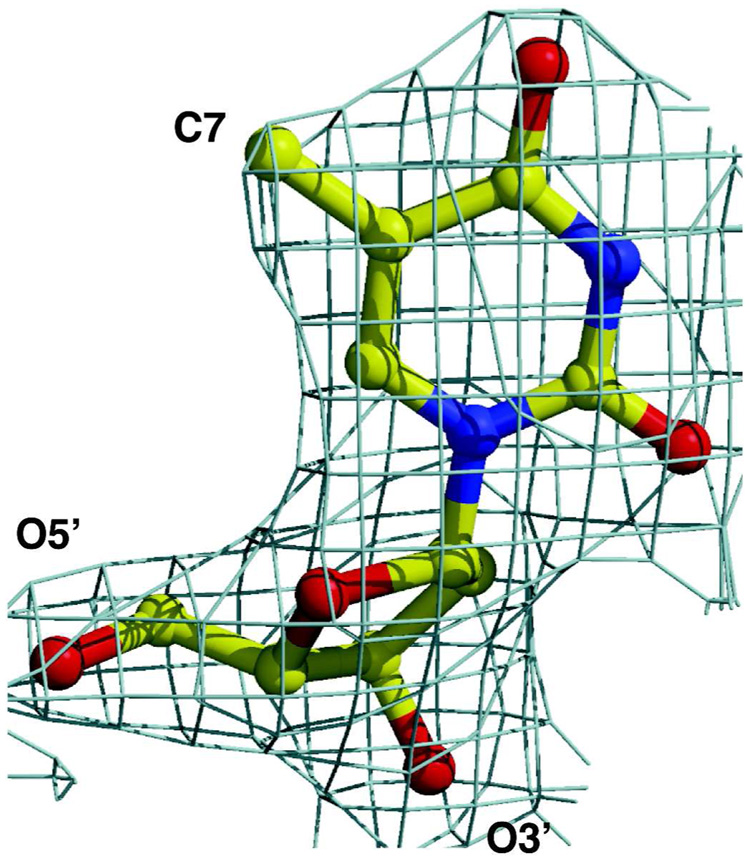
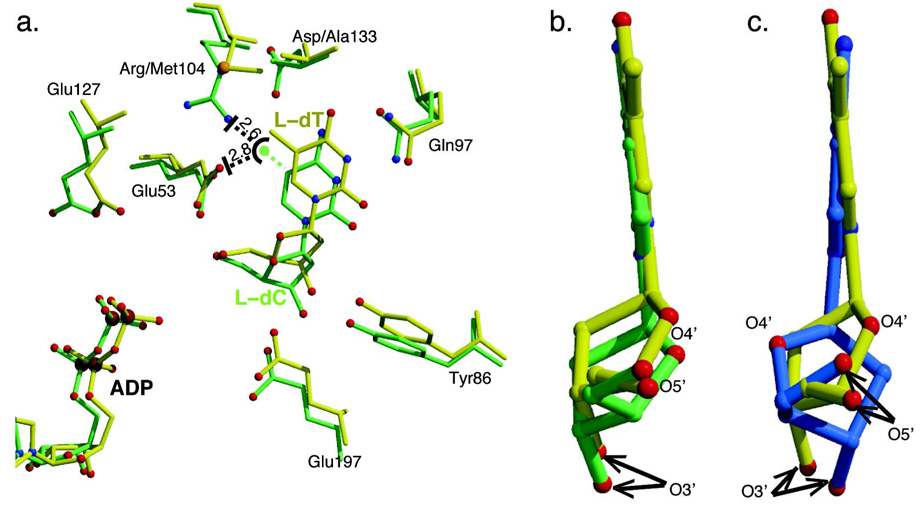
The electron density maps for the nucleotide ADP and the nucleoside L-dT are of good quality. The one for L-dT is shown in Figure 2. The thymine base is bound in the anti-conformation, thereby conserving the orientation seen with the dC (4). However, overlay of the DMMA complex with that of wild-type dCK with either the L-form or D-form of dC reveals a surprising result: L-dT binds deeper in the nucleoside binding site, by approximately 0.5 Å. Figure 3a shows the superposition of the DMMA complex with L-dT and the wild-type dCK complex with L-dC. Presumably, the space generated by substituting the arginine side chain by the shorter methionine allows L-dT to penetrate deeper into the pocket. The side chains of active site residues adjust their position to preserve the same interactions with L-dT as seen with dC. For example, Tyr86 and Glu197 track the shift of the nucleoside and maintain their interactions with the sugar’s 3’-hydroxyl group. Concomitantly, the ADP molecule shifts in the same direction as L-dT, and hence the change of position of Glu127.
Figure 2.
The 2Fo-Fc electron density map for L-dT, contoured at the 1.5 sigma level, is unambiguous.
Figure 3.
The mutations in DMMA allow L-dT to bind at the dCK active site. (a) Shown are active site residues of the DMMA/L-dT/ADP complex (yellow) overlaid with those from WT-dCK/L-dC/ADP (green). L-dT binds deeper in the active site in comparison to L-dC to avoid a steric clash between the 5-methyl group of the thymine base and the side chain of Glu53. Modeling of the thymine base 5-methyl group in the position taken up by L-dC, schematically shown as a dashed green ling and a sphere, shows that in wild-type dCK, this methyl group would be 2.8 Å away from Glu53, and 2.6 Å from Arg104. The mutation of Arg104 to a methionine permits L-dT to position itself deeper into the active site, avoiding the steric clash with Glu53. (b) A ~90 degrees rotated view relative to the orientation in (a) focusing on the nucleoside shows that the base of L-dT is parallel to that of L-dC. (c) In contrast, the analogous view of the overlay of L-dT structure on the D-dC structure (cyan) shows that the base in L-dT is tilted relative to the base orientation of D-dC. This base tilting is a consistent feature of dCK upon binding nucleosides of the L-chirality.
The side chain of Gln97 also adjusts somewhat to the deeper orientation of L-dT versus that seen with L-dC. However, the definitive orientation of the side chain of Gln97, that acts both as hydrogen-bond donor and acceptor, cannot be ascertained by the x-ray data at the resolution of our structure. In the complex with L-dC, the Gln97 orientation is unambiguous: based on distance considerations, the side chain carbonyl group interacts with the amino moiety of the base, and the amino group with N3 of the base. In cytosine, N3 is deprotonated, allowing it to accept a hydrogen-bond donor from the Gln97 amino group. In contrast, in the complex with L-dT, the situation is vague, as both orientations – interchanged by rotating the tip of the side chain by 180 degrees – make chemical sense (see Figure S1). In one orientation, identical to that adopted with L-dC, the carbonyl group of the side chain would interact with N3 (this is a productive interaction as the thymine N3 is protonated), and the amino group with the carbonyl group at position 2. The alternative orientation, the one depicted in Figure 3a, also has the carbonyl group of the Gln97 side chain interacting with N3, but now the amino group interacts with the carbonyl group at position 4 (Figure S1).
The deeper position of L-dT in the nucleoside-binding site of DMMA relative to that of L-dC in wild-type dCK clarifies why thymidine is a very poor substrate of the latter. If L-dT would bind in the same position as L-dC, its C5 methyl group (dashed green line and ball, Figure 3a) would clash with the side chains of Glu53 (2.8 Å) and Arg104 (2.6 Å). The replacement of Arg104 by a methionine creates the space into which, following the shift of the L-dT molecule deeper into the active site, the C5 methyl group can now fit. Importantly, the new position adopted by L-dT still allows the 5’-hydroxyl group to maintain its interaction to the side chain of the catalytic base Glu53. To achieve this, L-dT alters its sugar pucker to the 2’-endo-3’-endo conformation (whereas the 2’-endo-3’-exo conformation is observed in L-dC), and adjusts its exocyclic torsion angle (i.e. the position of the 5’-hydroxyl relative to the sugar ring), yet maintains a nearly identical glycosidic bond angle as observed with L-dC of ~ +160° (for definition of angles see Sabini et al (15)).
The capability of human dCK to phosphorylate L-nucleosides is directly related to the active site architecture that permits binding mode flexibility. Previous studies have shown that residues flanking the sugar moiety accommodate both enantiomers in the active site, albeit the base moiety must undergo a tilt (15, 16). The base moiety of L-nucleosides tilts by ~10 degrees relative to the position of their D-counterpart, yet the interactions with the base are preserved. Crucially, regardless of the enantiomeric form, the sugar 5’-hydroxyl group maintains close proximity to Glu53, the residue that functions as the catalytic base. In the DMMA complex with L-dT, while the nucleoside binds deeper into the binding site, L-dT (yellow in Figure 3b) retains the same tilt as seen for L-dC (green) when bound to wild type dCK. The base tilting of L-dT is apparent when compared to D-dC (blue) bound to wild-type dCK (Figure 3c). This supports our previous interpretation that base tilting of L-nucleosides is a consequence of active site constraints in the vicinity of the sugar moiety – lacking this tilt, the sugar would clash with Leu82. The new results show that even when the nucleoside enters deeper into the nucleoside-binding site, the same constraints on the sugar position necessitate the base to tilt relative to its position in D-nucleosides.
Discussion
The initial crystal structure of human dCK (4) allowed us to rationalize the lack of thymidine kinase activity by this enzyme, and to propose mutations that would add this ability. However, since it is difficult to predict the effect of mutations distant from the active site, the proposed mutations clustered near the nucleoside binding site. The work presented here reveals which residues at position 104 and an alanine at position 133 are compatible with thymidine kinase activity. But is the focus solely on the 104 and 133 positions justified? Recent work by Iyidogan and Lutz also addressed the issue of endowing dCK with thymidine kinase activity (5). This group used a thymidine kinase-deficient E. coli strain to select for dCK mutants with thymidine kinase activity. This screening process, which potentially included mutations throughout the dCK gene, identified positions 104 and 133 as the ones determining thymidine kinase activity. Thus, it seems that indeed the nature of the residues at these positions determines the substrate specificity of dCK.
One of the predictions we made based on the structural analysis is that it is imperative to mutate both Arg104 and Asp133 to gain thymidine kinase activity. Exchanging Arg104 is required to make space for the C5-methyl group of the thymine base. The exchange of Asp133 with an uncharged residue is needed to compensate for the removal of the charged Arg104 – in the crystal structure we observed that Arg104 and Asp133 form a salt bridge. The hypothesis that concomitant mutation of Asp133 is required is supported by data from Iyidogan and Lutz: a screen of all possible amino acids limited to position 104 using site-saturation libraries did not yield colonies in their genetic complementation experiments (5). On the other hand, in screens that included changes to both Arg104 and Asp133 colonies did appear, and the only mutations selected at position 104 were either methionine or glutamine. In contrast, our work shows that a leucine at position 104 (i.e. DMLA) yields the best thymidine kinase, with >10-fold higher efficiency than the analogous variant with a methionine (DMMA), followed by the isoleucine variant (DMIA) and the least active being the glutamine variant (DMQA). It is not clear why the leucine and isoleucine mutants did not show up in the genetic screen, but one possibility is that these variants are less stable.
How do the thymidine kinase-enabled dCK variants compare with the human thymidine kinases TK1 and TK2? Reported kinetic parameters for human TK1 agree that the Km for thymidine is ~0.5 µM, whereas values for kcat range from 0.28 sec−1 to 7 sec−1 (17–20). In terms of rate, the dCK mutants are only 2- to 4-times lower than TK1 compared to the highest reported TK1 rate. However, the dCK mutants are characterized by a measurably higher Km value - a factor of 50 for DMLA, and a factor of 300 for DMMA. Thus, the efficiency (kcat/Km) of the thymidine kinase reaction for the engineered dCK mutants is significantly lower than that of TK1. In contrast, in the comparison between the dCK mutants and TK2, the mutants do much better. The thymidine Km value is 15 µM for TK2, and its rate is only ~5% relative to TK1 (21). Thus, the efficiency of thymidine phosphorylation by TK2 is very similar to that of DMMA, and about 10-fold less than that of DMLA. In other words, the engineered dCK mutants are as good or better than TK2 when it comes to thymidine phosphorylation.
One interesting aspect of our study is the fact that the change in activity for a particular mutant is also dependent on the nature of the nucleoside. When considering efficiency (i.e. kcat/Km), DMMA is mainly a deoxycytidine kinase with some thymidine kinase activity (5% relative to dC activity). Both of the purine nucleosides are phosphorylated with very low efficiency, thereby turning this variant to a pyrimidine-specific kinase. Can our structural understanding of dCK shed light as to the reasons behind the low activity of DMMA with dA and dG? The lower activity with dG is due to a significant increase in the Km value (from 231 µM with wild type dCK to 1865 µM with DMMA, Table 4). The crystal structure of wild-type dCK in complex with dG reveals that Arg104 interacts directly with the guanine base (14). Thus, the high Km exhibited by DMMA (and DMLA for that matter) for dG can be attributed to the loss of this positive interaction. For dA, the effect of the mutations is mutant-specific: its Km values in DMMA increases ~10-fold compared to wild type dCK, but remains basically unchanged in DMLA. Since dA does not interact with Arg104, its mutation cannot explain this effect. Additionally, while Asp133 does interact with dA, the loss of this interaction due to the D133A mutation cannot explain the different behavior of the double mutants. Structures of DMMA and of DMLA in complex with dA are required to reveal this difference.
This study and that of Iyidogan and Lutz (5) show that two mutations suffice to confer thymidine kinase activity on dCK. This raises the question as to the cause behind the evolutionary pressure that selected for a human dCK enzyme lacking thymidine kinase activity, which is then complemented by the additional enzymes TK1 and TK2. One possible explanation has to do with the need to temporally and spatially control the activities of nucleoside salvage enzymes. In humans, both cytosolic dCK and mitochondrial TK2 are constitutively expressed, whereas cytosolic TK1 expression is limited to the S-phase of the cell cycle. This difference in expression is consistent with the continuous need for adenosine, guanosine, and cytosine triphosphates in the cytosol for diverse biological functions, whereas thymidine triphosphate is solely required for DNA replication. Thus, in a nonproliferating cell, there is no need for cytosolic thymidine nucleotides, and constitutive TK2 expression allows for mitochondrial DNA replication. Upon entry into the S-phase, the need for thymidine nucleotides for nuclear DNA replication is supplied by expressing TK1.
Finally, we would like to point out the potential use of the engineered dCK variants discussed here in suicide gene therapy strategies, or those that deliver the enzyme itself. Since TK1 does not phosphorylate L-nucleosides, whereas our dCK mutants (and cellular kinases further activating the monophosphates) do accept L-thymidine analogs, for example L-dT, the presence of the mutants in targeted cells would result in the transformation of such a thymidine analog into its cytotoxic triphosphate form. In this way, the targeted cells can be eliminated with minimal toxicity to cells not harboring the dCK mutants. While TK2 can phosphorylate L-nucleoside analogs, it does so relatively inefficiently, which explains the very low toxicity observed upon administration of L-dT, an analog used to treat chronic hepatitis B (22). The challenge is now to develop gene delivery, or protein delivery, technologies to allow for the selective introduction of the dCK mutants discussed here to cancer cells. Success in this endeavor would allow to efficiently block DNA replication using L-nucleoside analogs.
Supplementary Material
The possible conformations of the side chain of Gln97 are shown with distances to the cytosine base. This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgments
We thank the staff at the SER-CAT beamline for help in data collection.
This work was supported by an NIH grant (S.H., E.S., S.O., and A.L.) and the Max-Planck-Society (M.K.).
Abbreviations
- dCK
deoxycytidine kinase
- DMMA
dCK R104M-D133A double mutant
- DMLA
dCK R104L-D133A double mutant
- TK
thymidine kinase
- dNK
deoxyribonucleoside kinase from Drosophila melanogaster
Footnotes
The structure of human dCK in complex with L-dT and ADP has been deposited in the PDB under accession number 3EXK.
References
- 1.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 2.Munch-Petersen B, Piskur J, Sondergaard L. Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J Biol Chem. 1998;273:3926–3931. doi: 10.1074/jbc.273.7.3926. [DOI] [PubMed] [Google Scholar]
- 3.Knecht W, Sandrini MP, Johansson K, Eklund H, Munch-Petersen B, Piskur J. A few amino acid substitutions can convert deoxyribonucleoside kinase specificity from pyrimidines to purines. Embo J. 2002;21:1873–1880. doi: 10.1093/emboj/21.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat. Struct. Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 5.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;47:4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shewach DS, Liotta DC, Schinazi RF. Affinity of the antiviral enantiomers of oxathiolane cytosine nucleosides for human 2′-deoxycytidine kinase. Biochem Pharmacol. 1993;45:1540–1543. doi: 10.1016/0006-2952(93)90058-5. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal KC, Miech RP, Parks RE., Jr Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 1978;51:483–490. doi: 10.1016/s0076-6879(78)51066-5. [DOI] [PubMed] [Google Scholar]
- 8.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;24:795–800. [Google Scholar]
- 9.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 10.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 11.Smal C, Vertommen D, Bertrand L, Rider MH, van den Neste E, Bontemps F. Identification of phosphorylation sites on human deoxycytidine kinase after overexpression in eucaryotic cells. Nucleosides Nucleotides Nucleic Acids. 2006;25:1141–1146. doi: 10.1080/15257770600890194. [DOI] [PubMed] [Google Scholar]
- 12.Smal C, Van Den Neste E, Maerevoet M, Poire X, Theate I, Bontemps F. Positive regulation of deoxycytidine kinase activity by phosphorylation of Ser-74 in B-cell chronic lymphocytic leukaemia lymphocytes. Cancer Lett. 2007 doi: 10.1016/j.canlet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 13.McSorley T, Ort S, Hazra S, Lavie A, Konrad M. Mimicking phosphorylation of Ser-74 on human deoxycytidine kinase selectively increases catalytic activity for dC and dC analogues. FEBS Lett. 2008;582:720–724. doi: 10.1016/j.febslet.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabini E, Hazra S, Konrad M, Lavie A. Elucidation of different binding modes of purine nucleosides to human deoxycytidine kinase. J Med Chem. 2008;51:4219–4225. doi: 10.1021/jm800134t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabini E, Hazra S, Konrad M, Lavie A. The non-enantioselectivity property of human deoxycytidine kinase explained by structures of the enzyme in complex with L- and D-nucleosides. J. Med. Chem. 2007 doi: 10.1021/jm0700215. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabini E, Hazra S, Konrad M, Burley SK, Lavie A. Structural basis for activation of the therapeutic L-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase. Nucleic Acids Res. 2007;35:186–192. doi: 10.1093/nar/gkl1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berenstein D, Christensen JF, Kristensen T, Hofbauer R, Munch-Petersen B. Valine, not methionine, is amino acid 106 in human cytosolic thymidine kinase (TK1). Impact on oligomerization, stability, and kinetic properties. J Biol Chem. 2000;275:32187–32192. doi: 10.1074/jbc.M005325200. [DOI] [PubMed] [Google Scholar]
- 18.Birringer MS, Perozzo R, Kut E, Stillhart C, Surber W, Scapozza L, Folkers G. High-level expression and purification of human thymidine kinase 1: quaternary structure, stability, and kinetics. Protein Expr Purif. 2006;47:506–515. doi: 10.1016/j.pep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Li CL, Lu CY, Ke PY, Chang ZF. Perturbation of ATP-induced tetramerization of human cytosolic thymidine kinase by substitution of serine-13 with aspartic acid at the mitotic phosphorylation site. Biochem Biophys Res Commun. 2004;313:587–593. doi: 10.1016/j.bbrc.2003.11.147. [DOI] [PubMed] [Google Scholar]
- 20.Segura-Pena D, Lutz S, Monnerjahn C, Konrad M, Lavie A. Binding of ATP to TK1-like Enzymes Is Associated with a Conformational Change in the Quaternary Structure. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munch-Petersen B, Cloos L, Tyrsted G, Eriksson S. Diverging substrate specificity of pure human thymidine kinases 1 and 2 against antiviral dideoxynucleosides. J Biol Chem. 1991;266:9032–9038. [PubMed] [Google Scholar]
- 22.Ruiz-Sancho A, Sheldon J, Soriano V. Telbivudine: a new option for the treatment of chronic hepatitis B. Expert Opin Biol Ther. 2007;7:751–761. doi: 10.1517/14712598.7.5.751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The possible conformations of the side chain of Gln97 are shown with distances to the cytosine base. This material is available free of charge via the Internet at http://pubs.acs.org