Abstract
Spontaneous mouse models of cancer show promise to more accurately recapitulate human disease and predict clinical efficacy. Transgenic mice or viral vectors have been required to generate spontaneous models of glioma, a lethal brain tumor, because nonviral gene transfer is typically transient. To overcome this constraint we used the Sleeping Beauty transposable element to achieve chromosomal integration of human oncogenes into endogenous brain cells of immunocompetent mice. Genetically engineered, spontaneous brain tumors were induced with plasmid DNA in a matter of weeks in three separate mouse strains. The phenotype of tumors was influenced by the combination of oncogenes delivered, resembling human astrocytoma or glioblastoma in the majority of cases. At least five different genes can be co-transfected simultaneously including reporters, allowing measurement of tumor viability by in vivo imaging. This model can accelerate brain tumor research in a variety of ways such as generation of “humanized” models for high throughput drug screening and candidate gene validation with exceptional speed and flexibility.
Keywords: glioblastoma, astrocytoma, Sleeping Beauty, spontaneous mouse model, EGFRvIII, bioluminescence
Introduction
Malignant glioma (MG) is a devastating primary brain tumor. Gliomas are classified according the World Health Organization (WHO) criteria ranging from grade I, a typically treatable tumor, to grade IV, a glioblastoma multiforme (GBM) (1). GBM is a lethal brain tumor claiming over 12,000 lives each year in the United States (2). Prognosis for patients with high-grade MG is poor and has remained relatively unchanged for decades. MG often displays marked genetic and phenotypic heterogeneity (3). For example, as many as three different alleles of the Trp53 tumor suppressor gene are detectable in a single GBM; such tumors contain mixed regions appearing as high or low grade histologically and exhibit heterogeneous expression of the epidermal growth factor receptor (EGFR) immunohistochemically (4, 5). The heterogeneity of MG may account for the failure of therapies with a single mechanism of action. Animal models that recapitulate the complexity of human MG would be useful to better understand glioma biology and predict therapeutic response.
The most widely used rodent model of MG involves intracerebral transplantation of glioma cells that have been cultured. extensively in serum. Recent studies have brought this traditional glioma model into question because glioma cells cultured in serum do not recapitulate the genotype, phenotype, or global gene expression of the primary tumor (6). In contrast, glioma cells cultured in serum free media supplemented cytokines more faithfully mimic the genotype and phenotype of the primary tumor (6). Nonetheless, cell transplantation xenograft models lack a functional immune system, which plays a central role in the efficacy of radiation and chemotherapy (7). There has been great interest in spontaneous models of MG in which the tumor evolves with the host immune system with the expectation that they will more faithfully mimic human disease progression and better predict clinical efficacy.
A number of genetically engineered mice (GEM) are available harboring constitutive or conditional alleles of genes associated with MG development (8). Some models combine various GEM to test cooperativity of particular mutations in tumor development (9–11). Spontaneous MG can also be induced by intracerebral injection of a retroviral vector encoding platelet derived growth factor (12, 13). Hybrid models have also been created by utilizing GEM that express a receptor for a replication-competent ALV splice acceptor (RCAS) retroviral vector in a tissue specific pattern, leading to MG after avian retroviral oncogene transfer (14, 15). This GEM has been bred to a second GEM expressing firefly luciferase (FLuc) in mitotic cells to allow tumor growth to be non-invasively monitored using bioluminescent imaging (16). Bioluminescent imaging provides unparalleled convenience and speed for determining the efficacy of therapeutic agents in living mice (17). To date all spontaneous mouse models of MG have required the use of GEM or viral vectors. Production of new GEM or viral vectors can take months to years to develop and characterize. In some models, mice develop tumors with incomplete penetrance and exhibit relatively long survival times making them inconvenient for preclinical trials. We sought to develop a more flexible, rapid, spontaneous MG model that was independent of strain background while retaining the ability to monitor tumor viability with bioluminescence.
Several investigators have previously achieved non-viral transfection of the murine brain with polyethylenimine/plasmid DNA (PEI/DNA) complexes (20–23). Unfortunately, gene expression following plasmid DNA transfection is typically transient. To overcome this constraint we utilized the Sleeping Beauty (SB) transposable element delivered as plasmid DNA to achieve chromosomal integration and long-term expression (21). SB is a synthetic transposable element that was created by genetically engineering inactive transposase gene sequences isolated from salmonid fish (24). SB is a two-part system comprised of a transposon DNA substrate and a transposase enzyme. SB transposase mediates “cut and paste” excision and insertion of transposon DNA into a TA dinucleotide of the host genomic DNA (25). The gene encoding the SB transposase enzyme can be provided on the plasmid DNA backbone or on a separate plasmid relative to transposon DNA.
Here we show that injection of PEI/DNA complexes into the lateral cerebral ventricle of neonatal mice leads to SB-dependant long-term gene expression. Using this method we describe the development of a series of spontaneous MG models that express several combinations of reporter genes, human oncogenes, and inhibitors of tumor suppressor function. This model is readily adaptable for rapid high throughput drug screening, candidate gene validation, and basic biology studies that could accelerate brain tumor research in a variety of ways.
Methods
Animal Care
Mice were purchased from the Jackson Laboratories or Charles River Corporation for all experiments. Mating pairs were setup and carefully monitored each day until they gave birth. All animals were maintained in a specific-pathogen-free facility. Experiments were conducted according to the guidelines of the University of Minnesota Animal Care and Use Committee. Neonatal mice that were less than two days old were used for the studies with three exceptions (see Table I for complete list of mouse ages). Any mouse that became moribund was humanely euthanized throughout the study.
Table 1.
Effect of Vector, DNA Dose, Volume, Mouse Strain, and Age on Tumor Formation
| Experiment | Vector 1 | Vector 2 | Vector 3 | Vector 4 | Ratioa | Total DNA (ug) | Volume (uL) | Age (Hours) | # w/Tumor | Strain |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 13/15 (87%) | C57BL/6 |
| 2 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.5 | 2.0 | ≤48 | 4/4 (100%) | C57BL/6 |
| 3 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.5 | 1.0 | ≤48 | 3/4 (75%) | C57BL/6 |
| 4 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.25 | 1.0 | ≤48 | 4/5 (80%) | C57BL/6 |
| 5 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.25 | 0.5 | ≤48 | 3/6 (50%) | C57BL/6 |
| 6 | SB13/FfLuc | SV40-LgT | Empty Vector | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 0/4 (0%) | C57BL/6 |
| 7 | SB13/FfLuc | SV40-LgT | Empty Vector | - | 1:2:2 | 0.5 | 1.0 | 24–48 | 1/11 (9%) | C57BL/6 |
| 8 | SB13/FfLuc | Empty Vector | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 0/4 (0%) | C57BL/6 |
| 9 | SB13/FfLuc | Empty Vector | NRAS G12V | - | 1:2:2 | 0.5 | 1.0 | 24–48 | 0/8 (0%) | C57BL/6 |
| 10 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 7/7 (100%) | FVB/n |
| 11 | SB13/FfLuc | AKT | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 5/10 (50%) | FVB/n |
| 12 | SB13/FfLuc | SV40-LgT | EGFRvIII | - | 1:2:2 | 1.0 | 2.0 | ≤48 | 2/10 (20%) | FVB/n |
| 13 | SB13/FfLuc | AKT | NRAS G12V | shP53/GFP | 1:1:1:1 | 1.0 | 2.0 | ≤48 | 9/12 (75%) | FVB/n |
| 14 | SB13/FfLuc | EGFRvIII | NRAS G12V | shP53/GFP | 1:1:1:1 | 1.0 | 2.0 | 48–72 | 9/9 (100%) | FVB/n |
| 15 | SB13/FfLuc | EGFRvIII | NRAS G12V | Empty Vector | 1:1:1:1 | 1.0 | 2.0 | 24–48 | 0/6 (0%) | FVB/n |
| 16 | SB13/FfLuc | EGFRvIII | Empty Vector | shP53/GFP | 1:1:1:1 | 1.0 | 2.0 | 24–48 | 0/6 (0%) | FVB/n |
| 17 | SB13/FfLuc | Empty Vector | NRAS G12V | shP53/GFP | 1:1:1:1 | 1.0 | 2.0 | 24–48 | 3/5 (60%) | FVB/n |
| 18 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | 2160 | 0/5 (0%) | FVB/n |
| 19 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 1.0 | 2.0 | ≤ 96 | 3/6 (50%) | FVB/n |
| 20 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.25 | 1.0 | 24–48 | 5/5 (100%) | BALB/c |
| 21 | SB13/FfLuc | SV40-LgT | NRAS G12V | - | 1:2:2 | 0.125 | 1.0 | 24–48 | 4/4 (100%) | BALB/c |
with respect to vectors listed left to right
Plasmid Construction and Preparation
PT2/C-FLuc, pT/CMV-SV40-LgT, pT/CAGGS-NRASV12 and PGK-SB13 were created as previously described(26). PT2/C-Luc//PGK-SB13 was created by excising the PGK-SB transposase expression cassette from pPGK-SB13 as a Xmn I/Pme I fragment and ligating into pT2/C-Luc as a Xmn I/Pme I fragment. PKT2/PGK-Bsd:GFP CLP-Luc was a kind gift from Andy Wilbur (University of Minnesota, Minneapolis, MN, USA). PLXIN-EGFRvIII containing the human EGFRvIII cDNA was a kind gift from Dr. Michael J. Ciesielski (Roswell Park Cancer Institute, Buffalo, NY, USA). PT3.5/CMV-EGFRvIII was created by subcloning EGFRvIII from pLXIN-EGFRvIII into litmus 29 (New England Biolabs) as a Spe I fragment, followed by ligation into pT3.5/CMV-GFP as a Xho I/Age I fragment. MSCV-LTRmiR30-SV40 (27) contained a microRNA short hairpin against Trp53 and a second expression cassette encoding GFP; it was a kind gift from Dr. Scott Lowe (Cold Spring Harbor, NY, USA). The shP53 and GFP expression cassette-containing fragment was released from MSCV-LTRmiR30-SV40 as a Pvu II fragment and ligated into PT2/HB (28) as an EcoR V fragment to generate pT2/shP53/GFP4. The MSCV-AKT vector was a kind gift from Dr. Scott Lowe. MSCV-AKT was cut with EcoR I/ Nco I to release the AKT cDNA and ligated into Litmus 29, followed by final ligation into pKT2/CLP as a Nco I/Bgl II to generate pKT2/CLP-AKT. Plasmids were purified using a maxiprep kit (Invitrogen) and stored in 0.1X TE buffer (pH 8.0, from a 1X stock solution comprised of 10 mM Tris-Cl and 1mM EDTA).
PEI/DNA administration
In all oncogene experiments 25% mannitol (20 µl) was injected intraperitoneally immediately prior hypothermia anesthesia and PEI/DNA injection. Neonatal mice were then placed on ice for three minutes to induce anesthesia before being secured in a cooled, “neonatal rat” stereotaxic frame (Stoelting) maintained at 4–8°C by a dry ice/ethanol reservoir. A 10 µl syringe (Hamilton Company) fitted with a 30 gauge hypodermic needle (12.5° bevel, Hamilton Company) attached to a micropump (Stoelting) was used to inject plasmids at a flow rate of 0.7 µl/minute into the right lateral cerebral ventricle. Coordinates for injection were +1.5AP, 0.7ML, and −1.5DV from lambda. PEI/DNA complexes were prepared according to the manufacturer’s instructions to achieve N/P ratio of 7 (Polyplus Transfection) (29). One half to two microliters of PEI/DNA solution was administered at a maximum concentration of 0.5 µg/µl (higher concentrations resulted in precipitation). No incision was made for injection. The skull of a neonate was penetrated with the needle (sharp, beveled needle is critical) for all injections.
Tissue harvest, luciferase assays, immunofluorescent and immunohistochemical staining
Luciferase enzyme activity was assessed in vivo in mice three weeks of age and older using methods previously described (30). For in vitro luciferase assays animals were deeply anesthetized before transcardial perfusion with Phospho-buffered saline (PBS, pH 7.0–7.4). The brains were removed and homogenized in 400 µl of 1X tissue lysis buffer (Promega). Cell lysates and substrate solution provided in the luciferase tissue assay kit (Promega) were thawed and left at room temperature for 30 minutes. Luminescence was determined by mixing 100 µl lysate with 20 µl of substrate and immediately measured on a luminometer with a 15 second exposure time. Relative light units were normalized to mg of protein as determined by Bradford assay.
For histological analysis, animals were perfused with PBS followed with Z-fix (Anatech Ltd) or 4% paraformaldehyde. For immunofluorescence the brains were dissected and placed in 30% sucrose for 48 hours for cryoprotection. Brains were sectioned and immunofluorescent staining was carried out using rabbit anti-GFP (Molecular Probes). Images were acquired and processed as described (31). Immunohistochemistry (IHC) and H&E staining was carried out using standard clinical techniques on formalin-fixed, paraffin-embedded tissue (32). The EGFRvIII antibody was a kind gift from Dr. Darrel Bigner (Duke University, Durham, North Carolina).
Tissue Culture and Western blot
Western blot was conducted as described (26) using the following antibodies: mouse anti-LgT (Calbiochem), rabbit anti-NRAS (Santa Cruz), rabbit anti-p53 (Santa Cruz), mouse anti-β actin (Santa Cruz), and rabbit anti-ERK (Santa Cruz). Cell lines were derived from euthanized mice as previously described using TrypLE enzymatic digestion (33). Unless otherwise stated all tumor cells derived from mice were cultured in NSC media consisting of DMEM/F12, N2 and B27 supplements (1X), 1% penicillin/streptomycin (Invitrogen), and supplemented with 20 ng/ml EGF and FGF (Peprotech). Cytokines were added every 2–3 days and cells were passaged 1–2 times each week depending on density. GL261 glioma cells were cultured as described (34).
Southern Blot and Insertion Cloning
Southern blot hybridization for NRAS was performed as described (26). Transposon insertion sites were identified by a combination of linker-mediated PCR (35), using the pCR4-TOPOcloning vector and One-Shot TOP10 competent cells (Invitrogen) for shot-gun cloning, and pyrosequencing (454 Life Sciences, Roche) as previously described (36). Genomic sequences directly flanking the transposon were mapped using Ensemble (www.ensemble.org, NCBI m37 mouse assembly).
Statistical analysis
Survival was analyzed by log rank test as described (37). Graphing and statistical analysis was performed using Prism4 software (Graph Pad Software) as described (37).
Results
Characterization and optimization of SB-mediated gene transfer
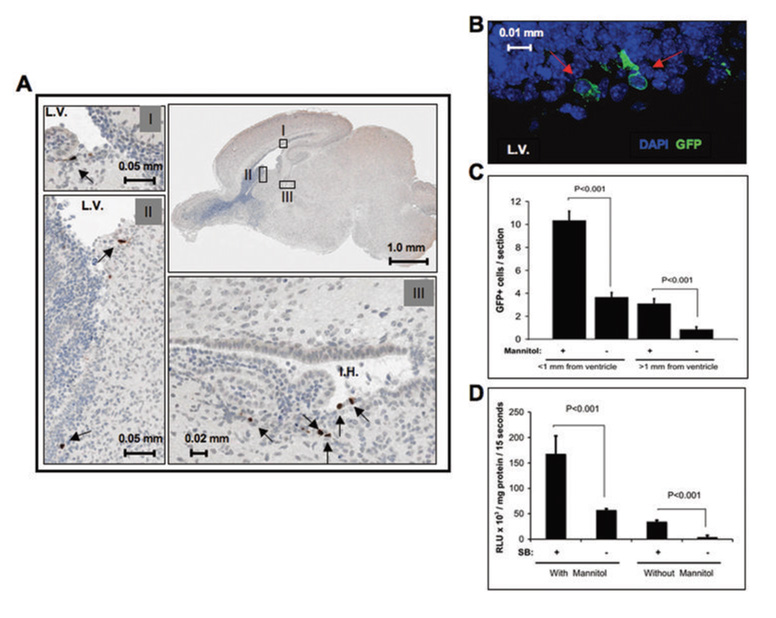
In order to determine the localization of transfected cells, PEI/DNA complexes encoding simian virus forty large T antigen (SV40-LgT) were injected into the lateral cerebral ventricle of neonatal mice. IHC was conducted to detect cells expressing SV40-LgT. Transfected cells were identified on all sides of the lateral ventricles, typically 1–4 cell diameters deep into the brain parenchyma (Fig. 1A). PEI/DNA complexes also diffused into the inferior horn of the lateral ventricle because numerous transfected cells were present in this region (Fig. 1A). Transfection of cells in the choroids plexus, third, and fourth ventricles was not observed (data not shown). Since intraperitoneal administration of mannitol is known to enhance viral transduction in the brain (38), we attempted to increase transfection efficiency by mannitol pre-treatment. PEI/DNA complexes encoding GFP were delivered into mice with and without mannitol injection prior to gene transfer. In accordance with SV40-LgT IHC, cells expressing GFP localized around the lateral ventricle wall (Fig 1B). GFP-positive cells were counted in serial sections to quantify the extent of transfection. Mannitol more than doubled the number of GFP-positive cells within 1 mm of the lateral ventricle and also increased transfection deeper into the brain parenchyma (Fig 1C). To determine if SB could enhance transgene expression in the brain, neonatal mice were intracerebrally injected with a FLuc transposon with or without transposase-encoding DNA. One month later brain lysates were assayed for FLuc activity as a measure of long-term expression. Luciferase activity was over seven times higher in brains injected with a plasmid encoding SB transposase relative to brains injected without SB-encoding DNA (Fig. 1D). Intraperitoneal administration of mannitol prior to co-transfection with SB-encoding DNA further increased FLuc activity three-fold (Fig. 1D). These results established co-delivery of SB transposase-encoding DNA and mannitol pre-treatment as the optimal parameters for long-term gene transfer.
Figure 1. Characterization of gene transfer.
(a) One microgram of pT/CMV-SV40-LgT plasmid DNA complexed in PEI was delivered at 0.5 µg/μl in a 2 µl volume into the right lateral ventricle of five neonatal (less than one day old) mice. IHC staining for SV40-LgT was conducted on brains harvested 48 hrs. after gene transfer. A representative sagittal section is shown. Grey boxes labeled with roman numerals represent 20X magnification of the corresponding black boxes shown at low power. Black arrows mark positive cells. The lateral ventricle (L.V.) and inferior horn (I.H.) of the lateral ventricle are labeled. (b) Similar experiment as in “A” except that a GFP vector was injected into mice that were or were not given an intraperitoneal injection of mannitol 1 hr. prior to gene delivery (n=4/group). Coronal sections were processed 48 hrs. later for GFP immunofluorescence. Cells reacting with anti-GFP antibody are marked by red arrows. (c) Quantification of the cells counted from “B”. Data represent the average GFP-positive cells counted per section +/− S.D (four brains were used, and four sections were counted from each brain). (d) Neonatal mice were injected with PT2/C-FLuc alone or in combination with PGK-SB13 at 1:30 ratio (N=4/group; 1µg total dose). Thirty days later brain lysates were assayed for luciferase activity (graph indicates average +/− SD).
Oncogene transfection resulted in development of spontaneous glioma
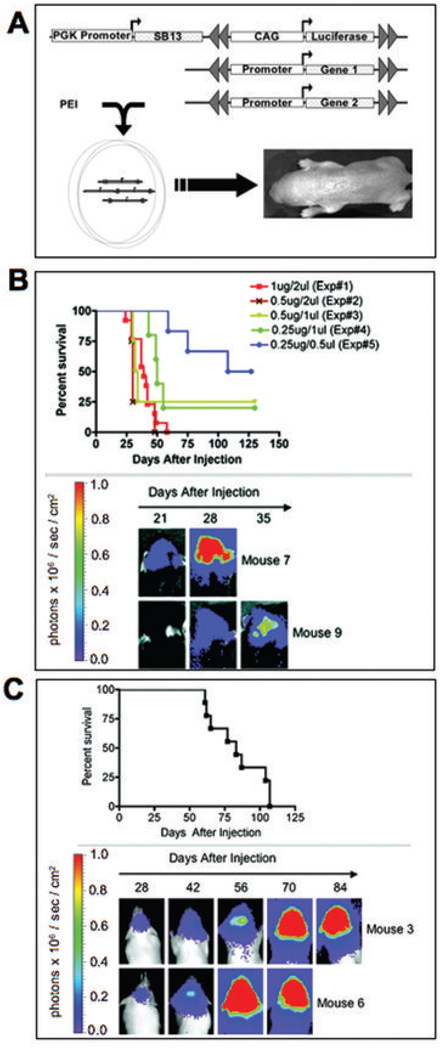
To promote spontaneous tumor formation we delivered transposons encoding SV40-LgT, a hyper-active human NRAS (26), and a transposon carrying a FLuc expression cassette on the same DNA molecule as the SB transposase gene into C57BL/6 mice (Fig. 2A). In experiment one (Table 1), tumors arose rapidly, visualized by increasing bioluminescence within three to four weeks (Fig. 2B). In general, well-demarcated tumors arose in the right (injected) cerebral hemisphere that exhibited infiltrative clusters of cells migrating away from the bulk tumor frequently surrounding blood capillaries (Fig. 3A). In some mice, apparent multifocal tumors arose near both lateral ventricles and macroscopic hydrocephalus was observed with tumors occupying the ventricular space (Supplementary Fig. 1 online). All tumors exhibited a high cell density with 2–5 mitotic cells often visible in a single microscopic field (Fig. 3A). Areas of necrosis were apparent in some of the tumors (Fig. 3B).
Figure 2. Oncogene transfection scheme, imaging, and survival.
(a) Schematic of co-transfection technique used to induce tumors by oncogene transfer. SB transposase expression is regulated by the phosphoglycerate kinase (PGK) promoter on the plasmid DNA backbone of the same vector harboring the FLuc transposon regulated by the CAG promoter. Double grey arrows indicate the inverted repeats of the transposon, marking the termini of the integrating DNA following transposition. (b) Neonatal mice were injected with SV40-LgT, NRAS, and FLuc vector loaded with SB-encoding DNA as detailed in Table I. Survival following each injection condition is shown above with representative bioluminescence images from experiment one below (note 2/15 mice in experiment 1 died from complications of hydrocephalus not tumor). (c) Survival and bioluminescent imaging of FVB/n mice from experiment 14 detailed in Table I
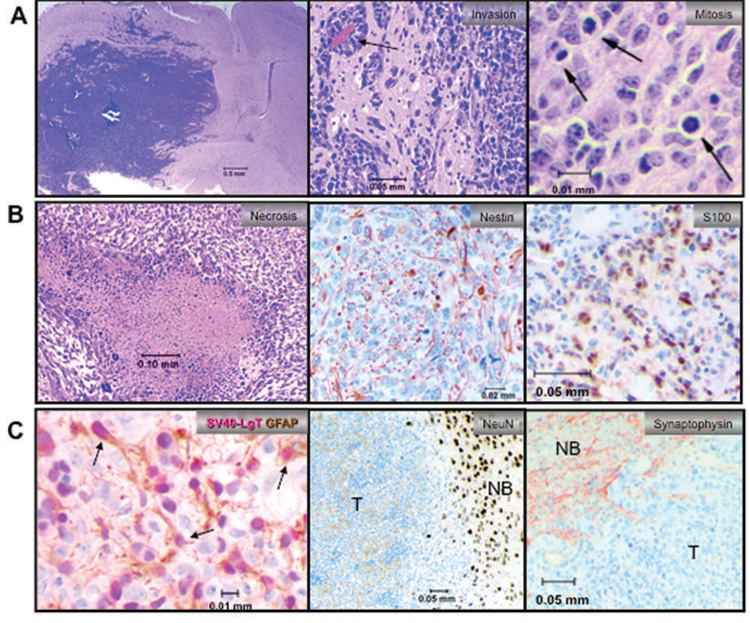
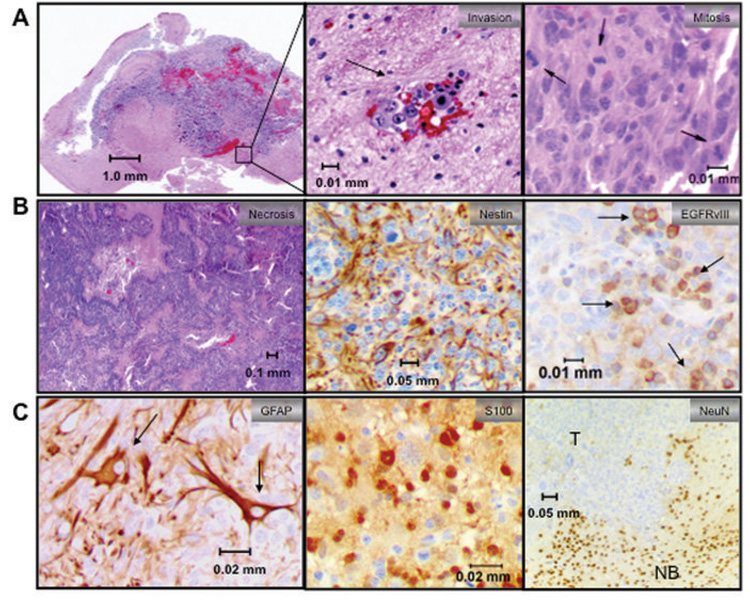
Figure 3. Characterization of NRAS/SV40-LgT-induced tumors.
(a) Left panel shows a representative tumor in the right hemisphere causing a mass effect. Middle panel shows tumor cells clustering around capillaries (black arrow). Right panel shows representative mitotic figures. (b) Representative images of necrosis, nestin, and S100 IHC are shown (c) Left panel is two-color IHC staining for GFAP in brown and SV40-LgT in pink (arrows mark some dual positive cells). Other panels show that the bulk tumor (T) is negative for neuronal markers Neun and synaptophysin by IHC whereas normal brain (NB) is strongly positive.
These tumors exhibited relatively homogenous expression of nestin (Fig. 3B), a marker for neural progenitor cells that is expressed in several types of brain tumors including gliomas. Immunopositivity for glial fibrillary acid protein (GFAP), a marker for astrocytes, was heterogeneous within individual tumors (supplementary Fig. 2 online). In order to determine whether the GFAP-positive cells were reactive astrocytes or tumor cells, two-color IHC was conducted staining for SV40-LgT and GFAP (Fig. 3C). Numerous tumor cells were immunopositive for GFAP and SV40LgT (Fig. 3C), ruling out reactive astrocytes as the only source of GFAP immunoreactivity. Consistent with an astrocytic phenotype, tumor cells also stained positive for S100. These tumors were negative for neuronal markers including Neun and synaptophysin, but the surrounding normal brain was positive as an internal control (Fig. 3C). In addition, tumors were negative for reticulin, ruling out a sarcoma phenotype (data not shown). Taken together the histological features of these murine tumors are most consistent with the diagnosis of a poorly differentiated astrocytoma corresponding to WHO grade III and IV. We refer to tumors induced in this murine mode as “GEM” glioma of various grades hereafter to acknowledge that such models are only an approximation of the human disease. In order to determine the frequency of each tumor grade, thirteen tumors that had been induced by NRAS/SV40-LgT were graded based on the presence of necrosis; 77% of exhibited minimal or microscopic necrosis (grade III) and 23% exhibited large regions of necrosis (grade IV; supplementary Table I).
The injection volume and concentration of PEI/DNA impacted penetrance and lethality of oncogene transfection. The highest injection volume (2 µl) tended to result in the development of hydrocephalus that was apparent by an enlarged skull, particularly in C57BL/6 mice, which are prone to this developmental defect (39). In experiment one, mice developed rapidly lethal tumors with 87% penetrance and median survival of 39 days (Fig. 2A). By reducing volume and PEI/DNA dose, hydrocephalus development was attenuated in C57BL/6 mice (experiments 2–5; Table I). Tumor initiation required the combination of NRAS and SV40-LgT. Tumors did not arise when the single genes were injected in combination with empty vector to control for total PEI/DNA dose (with the exception of one LgT-treated mouse, experiment 7 in Table I). In C57BL/6 mice the lowest injection volumes tended to decrease penetrance (Table I, experiments 4 and 5). In the BALB/c strain, as little as 0.125 µg of DNA in a 1 µl volume initiated tumors with 100% penetrance without hydrocephalus (experiment 21; Table I).
Characterization of cell lines derived from NRAS/SV40-LgT/Fluc tumors
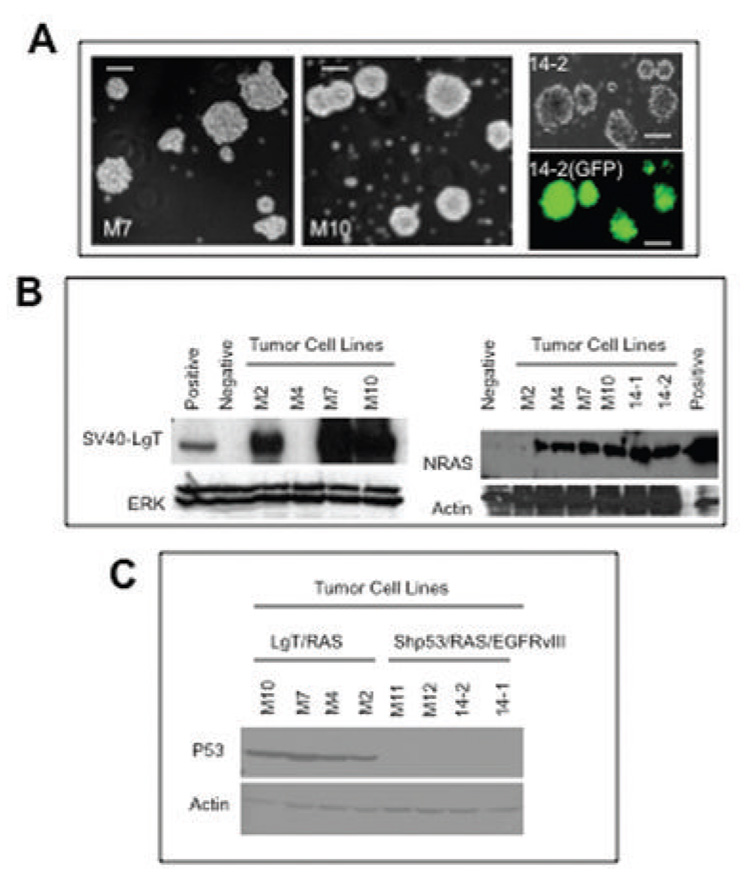
Four cell lines were established from the brains of C57BL/6 mice treated with NRAS/SV40-LgT that we designated M2, M4, M7, and M10. These mice were littermates sacrificed following in vivo imaging at 29 days after injection. Cells were cultured in serum free conditions shown to enrich for non-adherent neurospheres with an undifferentiated phenotype (6, 40). As expected, each cell line grew as non-adherent neurospheres (Fig. 4A). Three of the four cell lines expressed SV40-LgT and every line expressed NRAS at levels detectable by western blot (Fig. 4B).
Figure 4. Characterization of cell lines derived from SB-induced tumors.
(a) Left and middle panels show phase contrast image of M7 and M10 cells forming tumor spheres. Right panels show 14–2 cells in phase contrast and GFP fluorescent images (bar is equal to 100 µm). (b) GL261 glioma cells were transfected with a SV40-LgT- or NRAS-encoding plasmid as a positive control or a GFP-encoding plasmid as a negative control. Western blot of cell lysates from cell lines derived primary tumors using anti-SV40-LgT (left) or anti-NRAS (right) antibody. (c) Western blot for p53 using the indicated cell lysates.
Transposase-mediated insertions were cloned and mapped to the mouse genome. We cloned a total of 46 insertions from M2, M7, and M10 cells (supplementary Table II online). Twenty-eight separate insertions were recovered from M7 and fewer were isolated from M2 and M10 (supplementary Table II online). Four of nine insertions cloned from M10 mapped to chromosome 2. M7 and M2 had a more random pattern. Twenty-eight percent of the integrants mapped within introns of known or predicted genes (supplementary Table II online). To investigate the clonogenicity of these tumors individual subclones were isolated from M2, M7, and M10 cells that had been established from the entire tumor mass. Subclones were expanded into independent cell lines. Southern blots were conducted using a probe specific for NRAS vector sequence. Each of the four subclones derived from the parental cell lines had identical banding patterns (data not shown).
Tumor initiation was tested in transplantation experiments. Different numbers of M7 and M10 cells were injected into the brain parenchyma of SCID recipient mice. All mice injected with 10,000 M7 cells developed tumors and as few as 1,000 cells initiated tumors in 75% of mice (supplementary Fig. 3 online). Tumors that arose were traceable by bioluminescent imaging similarly to the primary tumor from which M7 was derived. The histological phenotype of the primary tumors shown in Fig. 3 was indistinguishable from the tumors that arose upon transplantation of M7 including areas of necrosis (supplementary Fig. 3 online). Transplantation of 10,000 M10 cells initiated tumors in 66% of mice with a median survival of 35 days, significantly delayed compared to the same dose of M7 cells with a median survival of 20 days (supplementary Fig. 3 online; P=0.02).
Development of alternative MG models with human oncogenes
We next sought to determine how flexible the requirements for tumor initiation via SB-mediated transfection could be. Some of the most commonly dysregulated pathways in human MG are high-level RAS and AKT activation via signaling from receptor tyrosine kinases (RTKs), Trp53 loss, and mutation of EGFR rendering it constitutively active (EGFRvIII) (41). We delivered a microRNA-based short hairpin RNA vector that reduces p53 expression (shp53) (27) in combination with NRAS (26), AKT, and EGFRvIII in seven different combinations (Table I). The combination of NRAS, EGFRvIII, and shP53 was the most robust, inducing MG with 100% penetrance in experiment 14 (Table I).
All mice treated with NRAS/shP53/EGFRvIII exhibited increasing bioluminescence that correlated with survival (Fig. 2C; median survival 83 days). These tumors were mitotically active and highly invasive, exhibiting single cell infiltrates invading normal brain away from the bulk tumor (Fig. 5A). Hemorrhaging and pseudopalisading necrosis was apparent in some tumors (Fig. 5A–B). Nearly the entire tumor mass expressed nestin. However, tumor cells were heterogeneous in appearance including the presence of giant cells and mixed immunoreactivity of GFAP (supplementary Fig. 4 online). Similarly to the human disease, expression of EGFRvIII was also patchy (Fig. 5B). Despite this heterogeneity a significant fraction of tumor cells expressed glial markers GFAP and S100 but not neuronal markers Neun or synaptophysin (Fig 5; supplementary Fig. 4 online). The expression of astrocytic markers, hemorrhaging, whole brain infiltration, and pseudopalisading necrosis closely resembles the most common forms of human GBM (1). However, approximately 60% of these tumors lacked appreciable necrosis and were thus considered grade III GEM astrocytoma (supplementary Table I online). The penetrance of tumor initiation was attenuated when one of the three oncogenes (e.g., NRAS/shP53/EGFRvIII) was removed from the cocktail. Mice injected with shP53/EGFRvIII and NRAS/EGFRvIII did not develop tumors, whereas 60% of mice injected with NRAS/shP53 developed GEM grade III or IV gliomas (experiments 15–17 in Table I; supplementary Table I online).
Figure 5. Characterization of the NRAS/shP53/EGFRvIII model.
(a) Left panel shows large tumor in the right hemisphere infiltrating the left hemisphere. Middle panel is 20X magnification of region marked by the black box in “A” showing single cell infiltrates invading normal brain tissue. Left panel shows representative mitotic figures (arrows). (b) Representative images of pseudopalisading necrosis, nestin, and EGFRvIII IHC are shown (black arrows mark a few EGFRvIII positive cells). (c) Representative images of GFAP, S100, and NeuN staining are shown where “T” marks the bulk tumor mass and “NB” marks normal brain.
Cell lines were established from mice injected with NRAS/shP53/EGFRvIII when they developed neurological symptoms. We designated the lines derived from mice in experiment 14 as 14–1 and 14–2, and two additional lines not shown in Table I as M11 and M12. Because the shP53 vector contains a GFP expression cassette, the expression of GFP was investigated using fluorescent microscopy. The vast majority tumor spheres in any microscopic field exhibited green fluorescence (Fig. 4A), confirming presence of shP53/GFP vector. Both 14–1 and 14–2 cells expressed NRAS by western blot (Fig. 4B; M11 and M12 were not tested). In addition, western blot analysis revealed that all cell lines derived from tumors induced with NRAS/shP53/EGFRvIII lacked detectable p53 protein (Fig. 4C). These results confirm that the tumor cell lines were genetically engineered by in vivo co-transfection using four separate plasmid DNA molecules.
Discussion
Despite decades of intensive research the prognosis for MG patients has only improved marginally (42). Arguably, unsatisfactory progress may be attributable to the paucity of animal models that are biologically accurate yet convenient enough to use in preclinical trials and flexible enough for basic research. A model that utilizes mutations relevant to human MG and can be flexibly modified to mimic the genetic heterogeneity and biology of human MG would be valuable. This SB-induced somatic model appears to meet these criteria, producing transplantable tumor initiating cells, histological and genetic heterogeneity similarly to human MG. Whether this model will be better at predicting clinical responses relative to established models remains to be determined.
There are several key advantages of the SB-induced model that make it novel and potentially useful for a variety of applications. First, the SB system can integrate transposons up to 10 kb in size (28) and 80% of human cDNAs are less than 7 kb (43). Therefore, after including a promoter and poly adenylation signal for gene expression, SB is suitable to deliver 70–80% of all human cDNAs. In contrast, the RCAS model is limited to 2.6 kb (14) and most retroviruses greater than 7.0 kb in size are difficult to grow at high titer. The SB-induced model could be used to rapidly validate MG candidate genes discovered by comparative genomic approaches or forward genetic screens. Second, straightforward approaches to measure glioma viability using bioluminescence in spontaneous models have been lacking. In the RCAS model, FLuc was expressed only in dividing cells (16) or when the Gli pathway is activated (44). While these approaches are elegant and provide important information, they require breeding doubly transgenic mice, the majority of the tumor cells within a glioma are not dividing, and the activity of the Gli pathway is not a direct measurement of tumor viability. In contrast, the SB system can induce tumors immediately in many strains of mice where FLuc expression is regulated by a ubiquitous CAG promoter, making it more flexible and convenient for measuring viability of the bulk tumor mass. Third, when the appropriate combinations of oncogenes are used 100% of animals develop tumors. This rate of tumor formation is equivalent or better than existing GEM models and is therefore for very appealing for preclinical trials. In addition, we have noted that animals that fail to develop Fluc expression do not develop tumors, and can therefore be excluded before beginning any preclinical trial. Forth, the SB-induced model offers unprecedented speed and flexibility because it can be utilized in existing GEM, rather than requiring creation of new GEM to interrogate new biological questions. While our initial studies have focused on mice, we speculate that the SB-induced model could be applied to rats and perhaps larger animals because it is based on somatic cell gene transfer into wild type animals. We have preliminarily determined effective transfection of the rat brain using SB (data not shown).
Molecular analyses demonstrated that 3/4 of the cell lines derived from SV40-LgT/NRAS-treated mice expressed the expected oncogenes. We could not detect expression of SV40-LgT in M4 cells by western blot (Fig. 4B). Fifty thousand M4 cells were not tumor initiating (data not shown). The mouse from which M4 was derived had barely detectable luciferase signal relative to M2, M7, and M10 at the time of sacrifice (data not shown). Thus, we were unable to isolate the relatively few tumorogenic clone(s) from the brain used to derive M4 cells. Because the combination of SV40-LgT and NRAS was required to induce a tumor it is likely that the brain from which M4 was derived contained cells expressing SV40-LgT and NRAS. EGFRvIII expression was detected in the primary tumor induced by NRAS/shP53/GFP/EGFRvIII (Fig. 5B). NRAS and GFP were also expressed but p53 was not in the cell lines derived from these mice (Fig. 4A–C). All together, our results demonstrate that up to five transgenes (including FLuc) were stably expressed in a significant fraction (but not all) of the tumor mass. Therefore, the SB-induced model could be useful to produce genetically engineered cell lines in many different mouse strains.
It is possible that insertional mutagenesis played a role in the heterogeneity of SB-induced tumors, as 28% of insertions mapped to introns (one in an exon) and others could activate transcription of nearby genes (supplementary Table II online). The observation that no common gene was hit twice argues against this possibility. In addition NRAS or SV40-LgT gene transfer alone did not result in detectable tumor (Table I). However, more insertions will have to cloned and characterized to determine what extent insertional mutagenesis may play in this model.
The clonality and cell of origin for these tumors are not entirely clear. Southern blot revealed that single clones derived from the cell lines exhibited identical banding patterns (data now shown), indicating tumors were monoclonal or that a single clone overtook the culture via selective outgrowth. In experiment ten we analyzed tumors histologically before overt symptoms developed at 28 days. This experiment revealed that tumors arise in lateral ventricle and invade the parenchyma laterally (supplementary Fig. 5 online). In one animal it appeared independent tumors were present in the lateral ventricles, and migratory tumor cells were seen in the olfactory bulb (supplementary Fig. 6 online). We speculate that a subset of tumors in this model may be oligoclonal, arising simultaneously and sometimes migrating from the subventricular zone (SVZ) to the olfactory bulb similarly to normal neural stem cells (45). IHC data indicated that both ependymal cells, subependymal cells, and cells greater than 1 mm from the ventricle wall were transfected (Fig. 1). While we determined that the tumor cell lines derived from the SB model expressed nestin, CD133, and CD15 (data not shown), it is not clear whether expression of these stem cell markers is a consequence of dedifferentiation or transformation of putative neural progenitor cells. This is an active area of investigation in our laboratory.
Human MG evolves with the host immune system, a different circumstance than that modeled by injecting glioma cells into the brain of adult mice. MG patients often develop tolerance to tumor antigens facilitated by an elevated fraction of regulatory T cells (Tregs) (47). Nonetheless, spontaneous anti-glioma immune responses have been documented following successful treatment with radiation and chemotherapy (48). In the current study tumors developed in immunocompetent mice presumably tolerized to foreign antigens used to drive MG formation and Treg frequency was not assessed. Future studies will address these features in this SB-induced model. We have previously shown that neonatal mice are tolerized to challenge with human neoantigen delivered within 24 hours of birth (49). We made observations that indicate mice in the current study were tolerant or ignorant to the neoantigens we delivered using SB: the penetrance of tumor development dropped in mice older than two days despite equivalent transfection efficiency and we could not induce tumors with SV40-LgT/NRAS in adult mice (experiments 18 and 19; Table I), and M7 cell transplants were rejected in 2/5 immunocompetent C57BL/6 mice (data not shown), but not in SCID mice (supplementary Fig. 3 online). The SB-induced somatic tumors could be initiated in GEM for immunology studies, creating “humanized” models that express human tumor antigens and human major histocompatibility complex class I (50). Likewise, human RTK genes could drive tumor growth so that small molecule drugs or humanized antibodies could be tested on the human RTK to diminish artifacts with species specificity. This SB-induced somatic model offers a new tool to understand the biology of MG and realize effective targeted therapy.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the NIH/NINDS 1R21-NS055738-01A2 (JRO), Randy Shaver Foundation (JRO), Minnesota Medical Foundation (JRO), NIH/NIDA T32 DA022616 (SD, WCL), NIH/NCI R01CA113636-01A1 (DAL), Minnesota Department of Employment and Economic Development SPAP-05-0013-P-FY06 (DAL). We thank Aaron Sarver for bioinformatics support in analyzing pyrosequencing data.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis FG, Kupelian V, Freels S, McCarthy B, Surawicz T. Prevalence estimates for primary brain tumors in the United States by behavior and major histology groups. Neuro-oncol. 2001;3(3):152–158. doi: 10.1093/neuonc/3.3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiesner SM, Freese A, Ohlfest JR. Emerging concepts in glioma biology: implications for clinical protocols and rational treatment strategies. Neurosurg Focus. 2005;19(4):E3. doi: 10.3171/foc.2005.19.4.4. [DOI] [PubMed] [Google Scholar]
- 4.Ren ZP, Olofsson T, Qu M, et al. Molecular genetic analysis of p53 intratumoral heterogeneity in human astrocytic brain tumors. J Neuropathol Exp Neurol. 2007;66(10):944–954. doi: 10.1097/nen.0b013e318156bc05. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y, Ng HK, Ding M, Zhang SF, Pang JC, Lo KW. Molecular analysis of microdissected de novo glioblastomas and paired astrocytic tumors. J Neuropathol Exp Neurol. 1999;58(2):120–128. doi: 10.1097/00005072-199902000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Apetoh L, Ghiringhelli F, Tesniere A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature medicine. 2007;13(9):1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 8.Weiss WA, Israel M, Cobbs C, et al. Neuropathology of genetically engineered mice: consensus report and recommendations from an international forum. Oncogene. 2002;21(49):7453–7463. doi: 10.1038/sj.onc.1205936. [DOI] [PubMed] [Google Scholar]
- 9.Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26(1):109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 10.Xiao A, Yin C, Yang C, Di Cristofano A, Pandolfi PP, Van Dyke T. Somatic induction of Pten loss in a preclinical astrocytoma model reveals major roles in disease progression and avenues for target discovery and validation. Cancer research. 2005;65(12):5172–5180. doi: 10.1158/0008-5472.CAN-04-3902. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer cell. 2005;8(2):119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer research. 1998;58(23):5275–5279. [PubMed] [Google Scholar]
- 14.Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12(23):3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci U S A. 1998;95(3):1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhrbom L, Nerio E, Holland EC. Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nature medicine. 2004;10(11):1257–1260. doi: 10.1038/nm1120. [DOI] [PubMed] [Google Scholar]
- 17.Momota H, Holland EC. Bioluminescence technology for imaging cell proliferation. Current opinion in biotechnology. 2005;16(6):681–686. doi: 10.1016/j.copbio.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Reilly KM, Tuskan RG, Christy E, et al. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci U S A. 2004;101(35):13008–13013. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawes JJ, Tuskan RG, Reilly KM. Nf1 expression is dependent on strain background: implications for tumor suppressor haploinsufficiency studies. Neurogenetics. 2007;8(2):121–130. doi: 10.1007/s10048-006-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1996;7(16):1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 21.Ohlfest JR, Demorest ZL, Motooka Y, et al. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther. 2005;12(5):778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- 22.Hirko AC, Buethe DD, Meyer EM, Hughes JA. Plasmid delivery in the rat brain. Biosci Rep. 2002;22(2):297–308. doi: 10.1023/a:1020194807665. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Yadava P, Hughes J. Polyethylenimine strategies for plasmid delivery to brain-derived cells. Methods. 2004;33(2):144–150. doi: 10.1016/j.ymeth.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 25.Ivics Z, Izsvak Z. Transposons for gene therapy! Curr Gene Ther. 2006;6(5):593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- 26.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci U S A. 2005;102(47):17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature genetics. 2005;37(11):1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 28.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8(1):108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 29.Ohlfest JR, Lobitz PD, Perkinson SG, Largaespada DA. Integration and long-term expression in xenografted human glioblastoma cells using a plasmid-based transposon system. Mol Ther. 2004;10(2):260–268. doi: 10.1016/j.ymthe.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Ohlfest JR, Demorest ZL, Motooka Y, et al. Combinatorial Antiangiogenic Gene Therapy by Nonviral Gene Transfer Using the Sleeping Beauty Transposon Causes Tumor Regression and Improves Survival in Mice Bearing Intracranial Human Glioblastoma. Mol Ther. 2005 doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- 31.Wu A, Oh S, Wiesner SM, et al. Persistence of CD133(+) Cells in Human and Mouse Glioma Cell Lines: Detailed Characterization of GL261 Glioma Cells with Cancer Stem Cell-Like Properties. Stem Cells Dev. 2008;17(1):173–184. doi: 10.1089/scd.2007.0133. [DOI] [PubMed] [Google Scholar]
- 32.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer research. 1997;57(18):4130–4140. [PubMed] [Google Scholar]
- 33.Panchision DM, Chen HL, Pistollato F, Papini D, Ni HT, Hawley TS. Optimized flow cytometric analysis of CNS tissue reveals novel functional relationships between CD133, CD15 and CD24 expressing cells. Stem Cells. 2007 doi: 10.1634/stemcells.2006-0260. [DOI] [PubMed] [Google Scholar]
- 34.Wu A, Oh S, Ericson K, et al. Transposon-based interferon gamma gene transfer overcomes limitations of episomal plasmid for immunogene therapy of glioblastoma. Cancer Gene Ther. 2007;14(6):550–560. doi: 10.1038/sj.cgt.7701045. [DOI] [PubMed] [Google Scholar]
- 35.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436(7048):272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 36.Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nature medicine. 2006;12(7):852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 37.Wu A, Oh S, Gharagozlou S, et al. In vivo vaccination with tumor cell lysate plus CpG oligodeoxynucleotides eradicates murine glioblastoma. J Immunother (1997) 2007;30(8):789–797. doi: 10.1097/CJI.0b013e318155a0f6. [DOI] [PubMed] [Google Scholar]
- 38.Ghodsi A, Stein C, Derksen T, Martins I, Anderson RD, Davidson BL. Systemic hyperosmolality improves beta-glucuronidase distribution and pathology in murine MPS VII brain following intraventricular gene transfer. Exp Neurol. 1999;160(1):109–116. doi: 10.1006/exnr.1999.7205. [DOI] [PubMed] [Google Scholar]
- 39. http://jaxmice.jax.org/strain/000664.html. viewed on 4/2/2008.
- 40.Gunther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2007 doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 41.Fenstermaker RA, Ciesielski MJ. Deletion and tandem duplication of exons 2 – 7 in the epidermal growth factor receptor gene of a human malignant glioma. Oncogene. 2000;19(39):4542–4548. doi: 10.1038/sj.onc.1203802. [DOI] [PubMed] [Google Scholar]
- 42.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 43.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 44.Becher OJ, Hambardzumyan D, Fomchenko EI, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer research. 2008;68(7):2241–2249. doi: 10.1158/0008-5472.CAN-07-6350. [DOI] [PubMed] [Google Scholar]
- 45.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 46.Beier D, Hau P, Proescholdt M, et al. CD133+ and CD133− Glioblastoma-Derived Cancer Stem Cells Show Differential Growth Characteristics and Molecular Profiles. Cancer research. 2007;67(9):4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 47.Fecci PE, Mitchell DA, Whitesides JF, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer research. 2006;66(6):3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 48.Ueda R, Low KL, Zhu X, et al. Spontaneous immune responses against glioma-associated antigens in a long term survivor with malignant glioma. Journal of translational medicine. 2007;5:68. doi: 10.1186/1479-5876-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohlfest JR, Frandsen JL, Fritz S, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105(7):2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 50.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. The Journal of experimental medicine. 1997;185(12):2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.