Abstract
Localized induction of DNA damage is a valuable tool for studying cellular DNA damage responses. In recent decades, methods have been developed to generate DNA damage using radiation of various types, including photons and charged particles. Here we describe a simple ultrasoft X-ray multi-microbeam system for high dose-rate, localized induction of DNA strand breaks in cells at spatially and geometrically adjustable sites. Our system can be combined with fixed- and live-cell microscopy to study responses of cells to DNA damage.
Keywords: Soft X-rays, Microbeam irradiation, DNA damage, DNA double-strand breaks, Live-cell microscopy
Introduction
Today various microbeam irradiation techniques are widely used, but localized induction of DNA damage in cells posed a great challenge when Professor Christoph Cremer and colleagues developed a microbeam instrument for studying the spatial organization of chromosomes inside the cell nucleus (Cremer et al. 1981). In their experiments, a small area of the nucleus was exposed to the focused beam of a UV-C laser. Using cytogenetic analysis, the authors identified a subgroup of cells that showed a small number of chromosomes with heavy damage surrounded by the majority of chromosomes that were unaffected by the micro-irradiation. This pioneering study revealed that chromosomes occupy distinct territories in the interphase cell nucleus, an observation that has contributed in a fundamental way to our understanding of nuclear architecture.
Since these early experiments, microbeam irradiation of living cells has become a powerful and standard method for studying the induction and processing of DNA lesions. Localized irradiation activates damage signaling and repair responses at sites where the nucleus is exposed to the microbeam and initiates a complex choreography of events, including protein binding and parting, modification of chromatin, and DNA processing. Microbeam irradiation techniques have enabled analysis of the dynamics of fluorescently tagged repair proteins at damage sites and have helped to define the temporal and spatial organization of the DNA damage response (DDR) (Bekker-Jensen et al. 2006; Essers et al. 2006; Feuerhahn and Egly 2008). Results obtained using microbeam irradiation have also provided important insights into molecular pathways connecting DNA repair to cell death, genomic instability, and malignant cell transformation (Hill et al. 2006).
By taking advantage of the many different types of photon and charged-particle sources presently available, and the characteristic spectra of DNA lesions they induce, an increasing variety of DNA repair processes can now be studied with microbeams. Advanced irradiation systems incorporate focused or collimated photon and/or charged particle beams (Czub et al. 2006; Endo et al. 2006; Folkard et al. 2001, 2005; Funayama et al. 2008; Garty et al. 2006; Gerardi 2006; Greif et al. 2006; Hamada et al. 2006; Hill et al. 2006; Nelms et al. 1998a, b; Schettino et al. 2002; Sowa et al. 2005; Stap et al. 2008; Sun et al. 2006; Tanno et al. 2006; Tartier et al. 2003). Recently, some of these microbeam instruments have been combined with confocal or wide-field fluorescence microscopes for observation of DDR in living cells (Chang et al. 2006; Folkard et al. 2003; Hauptner et al. 2004; Jakob et al. 2005; Kruhlak et al. 2006; Moné et al. 2001; Sowa Resat and Morgan 2004; Walter et al. 2003). The radiation effects of the different photon and particle sources can be summarized as follows: UV-A photons produce singlet oxygen species that initiate free-radical-driven reactions leading to oxidative damage to DNA and chromatin (Dizdaroglu 1992; Marrot and Meunier 2008) whereas UV-A exposure of pre-sensitized cells is more effective in producing DNA double-strand breaks (Limoli and Ward 1994). UV-B photons predominantly produce dimers of pyrimidine bases located on the same strand, leading to distortion of DNA structure, halting of transcription and replication (Marrot and Meunier 2008, 2004). UV-C photons induce DNA oxidation and, to a lesser extent, pyrimidine dimers, but not strand breaks (Perdiz et al. 2000; Rodrigo et al. 2000). X-ray photons, alpha particles, and accelerated electrons or ions transfer more energy at impact than UV photons. When absorbed in the cell nuclei, these high-energy radiations produce multiple ionizations that generate single- and double-strand DNA breaks (DSBs) and some DNA base damage (Preston 2005).
Here we describe a vertically oriented ultrasoft X-ray multi-microbeam (MMB) system designed to induce up to 10 DSBs per second at a number of spatially and geometrically adjustable sites within a large number of cell nuclei simultaneously. We opted for DSB induction by ultrasoft X-rays because poorly characterized chromatin damage generated as a by-product of the Hoechst/BrdU/UV-A laser microirradiation, used frequently for DSB induction, could affect our results (Dinant et al. 2007; Williams et al. 2007). The aim of this study is to apply MMB irradiation to quantitatively study accumulation of repair-related proteins in live cells and in cells fixed after irradiation. MMB irradiation also allows studying the kinetics of one or more fluorescent repair proteins at a number of damaged areas in a single cell nucleus. We expect that simultaneous analysis of different fluorescence signals in the same nucleus after MMB irradiation can provide detailed quantitative information about the dynamics and potential interactions of proteins at the damaged DNA.
Experiments described here show that the MMB system produces a uniform pattern of DNA-damaged areas in the cell nuclei and that the accumulation of repair proteins at the damaged areas can be quantitatively analyzed using fixed- and live-cell microscopy techniques.
Materials and methods
Setup of the multi-microbeam (MMB) soft X-ray system
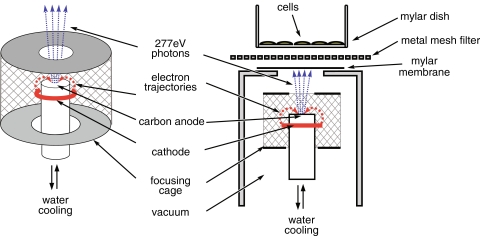
Our experimental setup consists of an X-ray source contained in a vacuum chamber (Fig. 1), evacuated by a turbo-molecular pump and an in-series rotary pump, achieving a vacuum of <10−5 mbar, as measured by Bayert Alpert ionization gauge. The samples are irradiated in ambient environment via a vacuum-sealed window with a diameter of 14 mm, formed by a 2 μm-thick Mylar membrane, supported by a stainless-steel grid. A 14-μm thick, photolithographically manufactured nickel mesh filter (Melo et al. 2008) with openings of between 2.2 and 3.5 μm in diameter, placed on top of the window, was used as a multi-microaperture system, providing ≈5 × 106 microbeams/cm2. The soft X-ray source (Fig. 1) is based on an obsolete e-beam evaporation system of vacuum generators. In contrast to traditional sources, this system has cylindrical symmetry. Electrons emitted by the circular cathode are focused at the top of the cooled anode (carbon target) by the surrounding cylindrical electrode (focusing cage). The advantage is that the size of the focal spot can be chosen by adjusting the distance of the top orifice of the electrode to the top of the anode. A spot size <0.5 mm can easily be achieved, but we increased the size to about 4 mm to prevent evaporation of the anode material and increase the photon-emitting surface. We conducted our experiments with 277 eV, 4.4 nm carbon K radiation (Agarwal and Sparrow 1981) using a graphite anode. To prevent system overheating, we connected the target to the cooled copper anode by conductive glue. Heat radiation from the cathode is also optically shielded by the top part of the focusing cage.
Fig. 1.
Schematic representation of the multi-microbeam irradiation setup
Our samples were irradiated using an anode voltage of 3 keV and a cathode emission current of 8 mA, which can be increased to ~20 mA. Approximately 90% of the emissions consisted of carbon K radiation (~277 eV), and the residual part was Bremsstrahlung with a maximum intensity at ~1.6 keV, as determined from the pulse height distribution produced by a classical proportional counter.
Cell culture and irradiation
Wild-type and 53BP1-GFP-expressing U2OS cells (Bekker-Jensen et al. 2005) were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% FCS at 37°C in an atmosphere containing 10% CO2. Then, 24 h before irradiation, approximately 106 cells were plated in custom-made glass rings with Mylar bottoms as described before (Stap et al. 2008). Cells were irradiated by placing the dishes on top of the nickel mesh filter for the required period of time.
Immunocytochemistry and microscopy
After irradiation, cells were fixed and stained for indicated proteins as described before (Stap et al. 2008) using the following antibodies: rabbit anti-RAD51 (Essers et al. 2002), mouse anti-γH2AX (05-636, Milipore), rabbit anti-MDC1 (A300-051A, Bethyl Laboratories), goat anti-mouse-Cy3 (115-165-166), and goat anti-rabbit-FITC (111-095-144) (Jackson Immunoresearch). For live-cell analysis, the mylar membrane with irradiated cells was placed on top of a glass cover-slip, mounted into a push-to-seal custom-made imaging chamber in CO2-independent medium (Gibco BRL) and positioned on the stage of an inverted Leica SP2 confocal microscope equipped with a heated stage. Fluorescence recovery after photobleaching (FRAP) analysis was performed using the 488-nm laser line. Immunocytochemically stained cells were imaged using a Leica DMRA wide-field fluorescence microscope, and wide-field 3-D images were reconstructed using Huygens Pro (Scientific Volume Imaging) and processed using ImageJ and Photoshop CS3 (Adobe Systems). Data processing was performed using Excel (Microsoft) and Prism 4 (GraphPad Software). Scanning electron microscopy (SEM) images of the nickel mesh filters were obtained using an SEM 525 (Philips).
Results
Geometrically and spatially tunable MMB irradiation elicits normal DNA damage response
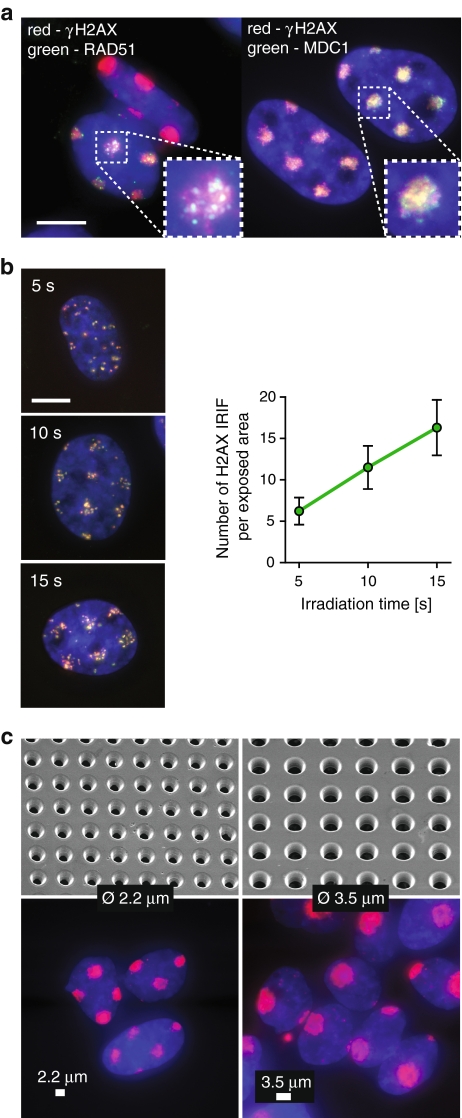
Many DSB repair-related proteins accumulate in the vicinity of the damaged DNA, forming so-called ionizing radiation-induced foci (IRIF) at damage-containing chromatin within minutes after exposure of cells to IR (Lukas et al. 2005). Among the earliest events triggered by DSBs is phosphorylation of histone H2AX by the ATM kinase (Foster and Downs 2005). Phosphorylated H2AX (γH2AX) was present at confined areas of U2OS cell nuclei at 30 min after irradiation by MMB (Fig. 2a). MMB irradiation also induced accumulation of the damage-mediator MDC1 (Stewart et al. 2003) and repair-protein RAD51 (Tashiro et al. 2000) at the exposed areas (Fig. 2a). These results indicate that MMB irradiation elicits a normal DDR.
Fig. 2.
Induction of DNA double-strand breaks by multi-microbeam irradiation. a MMB irradiation induces DSB responses. U2OS cells were irradiated for 6 min through a metal mesh filter with openings of 2.2 μm in diameter, fixed 30 min later, and stained for DNA (blue), γH2AX (red), and Rad51 (green, left panel) or MDC1 (green, right panel). Inset shows magnification of a single irradiated area. Scale bar 10 μm. b The number of DSBs induced by MMB irradiation depends linearly on irradiation time. Left panels show U2OS cells irradiated for 5, 10, or 15 s through a metal mesh filter with openings of 2.2 μm in diameter, fixed 5 min later, and stained for DNA (blue), γH2AX (red), and MDC1 (green). Right panel shows numbers of γH2AX foci representing individual DSBs per exposed area in cells from b. c The geometry of exposed areas can be controlled by applying metal mesh filters of various parameters. Cells were irradiated for 6 min using filters with openings of 2.2 μm (left) or 3.5 μm in diameter (right), fixed 5 min later, and stained for DNA (blue) and γH2AX (red). Upper panels show mesh filters used, imaged by a scanning electron microscope. Lower panels show stained cells irradiated using the respective filters. Scale bar 2.2 μm (left panel) and 3.5 μm (right panel)
We then measured the dose-rate of the MMB by counting the number of individual γH2AX IRIF in exposed areas after different irradiation periods (Fig. 2b). Exposure time of between 5 and 15 s was linearly correlated with the numbers of γH2AX IRIF (Fig. 2b), allowing estimation of irradiation time for experiments requiring induction of large numbers of DSBs, when scoring of individual IRIF is impossible.
Our irradiation setup provides a possibility to control the geometry and distribution of irradiated areas by using metal mesh filters of various parameters (Fig. 2c). Indeed, when we applied mesh filters with opening diameters of 2.2 and 3.5 μm, we detected γH2AX presence at confined circular areas of the expected diameter (Fig. 2c). Importantly, few γH2AX IRIF were present outside of these areas. We conclude that MMB allows precise control over irradiation dose, distribution, and geometry of the exposed areas.
MMB irradiation is suitable for analysis of DDR in living cells
Live-cell analysis can provide valuable information about the cellular response to DSBs. UV laser microirradiation of presensitized cells is a frequently used method for DSB induction in combination with live-cell microscopy. However, the broad spectrum of DNA damage induced by this approach includes large numbers of UV-specific lesions, and the deposited dose is difficult to estimate. DNA damage induced by ultrasoft X-rays is well characterized and does not include UV-specific lesions (Gobert et al. 2004; Goodhead et al. 1981; Griffin et al. 1998; Hawkins 2006; Hill et al. 2001; Nikjoo et al. 1999; Yokoya et al. 1999; de Lara et al. 2001).
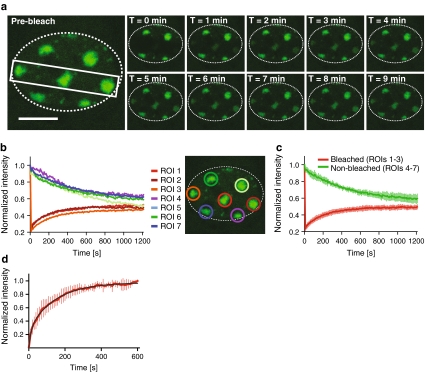
Microscopical observation of MMB-irradiated U2OS cells expressing GFP-tagged 53BP1, one of the early DSB markers (Bekker-Jensen et al. 2005), revealed accumulation of this protein at damaged chromatin consistent with the geometry of the mesh filter used (Fig. 3a). We then applied the technique of fluorescence recovery after photobleaching (FRAP) (van Royen et al. 2009) to visualize the dynamics of 53BP1-GFP at damaged chromatin. We bleached the 53BP1-GFP fluorescence at three out of seven irradiated areas in a single cell. Subsequent imaging revealed recovery of the fluorescence at the bleached regions of interest (ROIs) and concomitant loss of fluorescence at the nonbleached ROIs (Fig. 3b, c), confirming the dynamic character of the interaction of 53BP1 with damaged chromatin. Next, we quantified the dynamics of 53BP1 by estimating its residence time at damaged chromatin (Fig. 3d). To achieve this, we irradiated cells using a metal mesh filter that produces an average of one irradiated circular area of 5 μm in diameter per cell. Then, 20 min after irradiation, we bleached the GFP signal at the damage-containing chromatin areas by exposing them to five pulses of the 488-nm laser set to 100% emission. Subsequently, we imaged the cells for 10 min at 10-s intervals. We then measured the average GFP fluorescence intensities at the bleached regions and normalized them as described earlier (Bekker-Jensen et al. 2005). To obtain residence time of 53BP1 at damaged chromatin, we fitted the obtained data to the following equation (Bekker-Jensen et al. 2005):
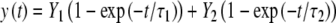 |
Fig. 3.
Dynamics of 53BP1-GFP at areas of DNA damage induced by the MMB system. a U2OS cells expressing 53BP1-GFP were irradiated for 6 min through a metal mesh filter with openings of 2.2 μm in diameter and mounted under a confocal microscope. The indicated area of the selected cell was then bleached, and the cell was imaged for 20 min at intervals of 10 s. The gallery shows images captured at indicated times after bleaching. The perimeter of the cell nucleus is indicated by the dotted line. Scale bar 10 μm. b Quantification of the fluorescence intensities measured at the different ROIs in the cell (a) at indicated times after bleaching. The fluorescence intensities at each ROI were normalized to the intensities at the respective ROI before bleaching. c Quantification of average fluorescence intensities at bleached (1–3) and non-bleached (4–7) regions at indicated times after bleaching. Error bars indicate standard deviations. d Quantification of recovery of 53BP1-GFP intensity at damaged chromatin areas after photobleaching. The graph represents normalized average intensity of the bleached areas obtained from measurement of 10 cells. The black curve represents nonlinear fit obtained as described earlier (Bekker-Jensen et al. 2005). Error bars represent standard deviation
Residence time of the damage-associated fraction of 53BP1-GFP (τ1) was 145 ± 5 s, more than 3.5 times longer than that reported before for 53BP1-GFP at chromatin after UVA laser microirradiation in BrdU presensitized cells (Bekker-Jensen et al. 2005). Whether this discrepancy reflects physiological differences in protein dynamics at UV- and IR-irradiated chromatin requires further investigation.
Discussion
We constructed a vertical ultrasoft X-ray multi-microbeam source for simultaneous, localized irradiation of large numbers of cell nuclei. The purpose of this instrument is to induce large amounts of DNA damage by ionizing radiation at several sites in the cell nucleus. This allows detection of repair-related proteins that accumulate at the damage sites at low concentrations. Using custom-made metal mesh filters, we were able to obtain a uniform pattern of DNA damage-containing areas. We could observe the irradiated samples under a live-cell confocal microscope and perform FRAP analysis at the individual exposed sites. This procedure will allow evaluation of the dynamics and interactions of several fluorescently labeled repair proteins simultaneously in a single cell as an alternative to UV laser microirradiation.
The MMB ultrasoft X-ray irradiation system is still in the initial phase of development. We intend to incorporate the MMB into an upright confocal fluorescence microscopy system equipped with a water-dipping objective. This will allow real-time observation of fast DDR events, such as accumulation of the Ku or DNA-PKcs proteins (Mari et al. 2006; Uematsu et al. 2007). However, for this purpose, a higher dose-rate is required as accumulation of these factors reaches the maximum within the first minute after laser microirradiation. Our current setup offers rates of up to 10 DSB per second, which can be further increased by shortening the distance between the source and cells (currently approximately 14 cm) and by raising the anode voltage to 5 keV (currently 3 keV).
Ionizing radiation is widely accepted as the reference inducer of single- and double-strand breaks in studies focused on DSB repair. In contrast, UV or multi-photon laser microirradiation is difficult to control and reproduce between different experimental setups and generates large amounts of UV-specific lesions and possibly other types of DNA damage (Dinant et al. 2007; Kong et al. 2009). Therefore, the relatively well-defined spectrum of DNA damage induced by ultrasoft X-rays generated by our setup presents a direct advantage over laser microirradiation techniques. Recently, we used the MMB to confirm the recruitment of the HP1 protein to DSB-containing chromatin, which was first observed using UV laser microirradiation (Luijsterburg et al. 2009).
The metal mesh filters that form the multi-microaperture structure provide broad control over the spatial distribution of induced DNA damage. For instance, sparsely distributed openings could be used to study “bystander effects” in neighbors of cells exposed to irradiation. Currently, an area of about 150 mm2, containing approximately 2 × 105 cells, can be irradiated at the same instant. However, some experiments require irradiation of a limited area of the sample so that several irradiation conditions can be tested in one dish. This can be easily achieved by using metal mesh filters with openings restricted to a small area. In the near future, we will upgrade the MMB system to make it compatible with α-particle and UV radiation sources and soft X-ray sources of other photon energies.
Acknowledgments
We thank Dr. Claudia Lukas for providing the p53BP1-GFP expressing cell line, and Prof. Fred Bijkerk, Ing. Rob Kemper, Ing. Kees Grootkarzijn and The Institute for Atomic and Molecular Physics (AMOLF) for facilitating instrument development. We also thank Prof. Eddie Barendsen for helpful discussions and Dr. Menno Visser (Stork VECO BV) for kindly providing a metal mesh filter. This work was supported by grants from the Dutch Cancer Society (KWF) and the Maurits and Anna de Kock foundation.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
This article has been submitted as a contribution to the Festschrift entitled “Uncovering cellular sub-structures by light microscopy” in honour of Professor Cremer’s 65th birthday.
C. van Oven and P. M. Krawczyk contributed equally to this work.
Contributor Information
Jan Verhoeven, Phone: +31-34-6241543, Email: verhoeven@physics.leidenuniv.nl.
Jacob A. Aten, Phone: +31-20-5664757, Email: j.a.aten@amc.uva.nl
References
- Agarwal BK, Sparrow JH (1981) Line intensities in the soft X-ray region. J Phys F Met Phys 11:1303–1309. doi:10.1088/0305-4608/11/6/014 [DOI]
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J (2005) Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol 170:201–211. doi:10.1083/jcb.200503043 [DOI] [PMC free article] [PubMed]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J et al (2006) Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173:195–206. doi:10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed]
- Chang S, Zhang J, Bordelon D, Schreiber E, Cox A, Zhou O (2006) Development of a nanotechnology based low-LET multi-microbeam array single cell irradiation system. Radiat Prot Dosimetry 122:323–326. doi:10.1093/rpd/ncl508 [DOI] [PubMed]
- Cremer C, Cremer T, Zorn C, Zimmer J (1981) Induction of chromosome shattering by ultraviolet irradiation and caffeine: comparison of whole-cell and partial-cell irradiation. Mutat Res 84:331–348. doi:10.1016/0027-5107(81)90202-5 [DOI] [PubMed]
- Czub J, Banas D, Braziewicz J, Choinski J, Jaskóla M, Korman A et al (2006) An irradiation facility with a horizontal beam for radiobiological studies. Radiat Prot Dosimetry 122:207–209. doi:10.1093/rpd/ncl518 [DOI] [PubMed]
- de Lara CM, Hill MA, Jenner TJ, Papworth D, O’Neill P (2001) Dependence of the yield of DNA double-strand breaks in Chinese hamster V79-4 cells on the photon energy of ultrasoft X rays. Radiat Res 155:440–448 [DOI] [PubMed]
- Dinant C, de Jager M, Essers J, van Cappellen WA, Kanaar R, Houtsmuller AB et al (2007) Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J Cell Sci 120:2731–2740. doi:10.1242/jcs.004523 [DOI] [PubMed]
- Dizdaroglu M (1992) Oxidative damage to DNA in mammalian chromatin. Mutat Res 275:331–342 [DOI] [PubMed]
- Endo S, Hoshi M, Takada J, Takatsuji T, Ejima Y, Saigusa S et al (2006) Development, beam characterization and chromosomal effectiveness of X-rays of RBC characteristic X-ray generator. J Radiat Res (Tokyo) 47:103–112. doi:10.1269/jrr.47.103 [DOI] [PubMed]
- Essers J, Hendriks RW, Wesoly J, Beerens CEMT, Smit B, Hoeijmakers JHJ et al (2002) Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair (Amst) 1:779–793. doi:10.1016/S1568-7864(02)00110-6 [DOI] [PubMed]
- Essers J, Vermeulen W, Houtsmuller AB (2006) DNA damage repair: anytime, anywhere? Curr Opin Cell Biol 18:240–246. doi:10.1016/j.ceb.2006.03.004 [DOI] [PubMed]
- Feuerhahn S, Egly J (2008) Tools to study DNA repair: what’s in the box? Trends Genet 24:467–474. doi:10.1016/j.tig.2008.07.003 [DOI] [PubMed]
- Folkard M, Prise KM, Vojnovic B, Gilchrist S, Schettino G, Belyakov OV et al (2001) The impact of microbeams in radiation biology. Nucl Instrum Methods Phys Res B 181:426–430. doi:10.1016/S0168-583X(01)00463-3 [DOI]
- Folkard M, Vojnovic B, Gilchrist S, Prise KM, Michael BD (2003) The design and application of ion microbeams for irradiating living cells and tissues. Nucl Instrum Methods Phys Res B 210:302–307. doi:10.1016/S0168-583X(03)01043-7 [DOI]
- Folkard M, Prise K, Schettino G, Shao C, Gilchrist S, Vojnovic B (2005) New insights into the cellular response to radiation using microbeams. Nucl Instrum Methods Phys Res B 231:189–194. doi:10.1016/j.nimb.2005.01.055 [DOI]
- Foster ER, Downs JA (2005) Histone H2A phosphorylation in DNA double-strand break repair. FEBS J 272:3231–3240. doi:10.1111/j.1742-4658.2005.04741.x [DOI] [PubMed]
- Funayama T, Wada S, Yokota Y, Fukamoto K, Sakashita T, Taguchi M et al (2008) Heavy-ion microbeam system at JAEA-Takasaki for microbeam biology. J Radiat Res (Tokyo) 49:71–82. doi:10.1269/jrr.07085 [DOI] [PubMed]
- Garty G, Ross GJ, Bigelow AW, Randers-Pehrson G, Brenner DJ (2006) Testing the stand-alone microbeam at Columbia University. Radiat Prot Dosimetry 122:292–296. doi:10.1093/rpd/ncl454 [DOI] [PubMed]
- Gerardi S (2006) A comparative review of charged particle microbeam facilities. Radiat Prot Dosimetry 122:285–291. doi:10.1093/rpd/ncl444 [DOI] [PubMed]
- Gobert FN, Lamoureux M, Hervé du Penhoat MA, Ricoul M, Boissière A, Touati A et al (2004) Chromosome aberrations and cell inactivation induced in mammalian cells by ultrasoft X-rays: correlation with the core ionizations in DNA. Int J Radiat Biol 80:135–145. doi:10.1080/09553000310001654710 [DOI] [PubMed]
- Goodhead DT, Thacker J, Cox R (1981) Is selective absorption of ultrasoft X-rays biologically important in mammalian cells? Phys Med Biol 26:1115–1127. doi:10.1088/0031-9155/26/6/010 [DOI] [PubMed]
- Greif K, Beverung W, Langner F, Frankenberg D, Gellhaus A, Banaz-Yasar F (2006) The PTB microbeam: a versatile instrument for radiobiological research. Radiat Prot Dosimetry 122:313–315. doi:10.1093/rpd/ncl436 [DOI] [PubMed]
- Griffin CS, Hill MA, Papworth DG, Townsend KM, Savage JR, Goodhead DT (1998) Effectiveness of 0.28 keV carbon K ultrasoft X-rays at producing simple and complex chromosome exchanges in human fibroblasts in vitro detected using FISH. Int J Radiat Biol 73:591–598. doi:10.1080/095530098141834 [DOI] [PubMed]
- Hamada N, Schettino G, Kashino G, Vaid M, Suzuki K, Kodama S et al (2006) Histone H2AX phosphorylation in normal human cells irradiated with focused ultrasoft X rays: evidence for chromatin movement during repair. Radiat Res 166:31–38. doi:10.1667/RR3577.1 [DOI] [PubMed]
- Hauptner A, Dietzel S, Drexler GA, Reichart P, Krücken R, Cremer T et al (2004) Microirradiation of cells with energetic heavy ions. Radiat Environ Biophys 42:237–245. doi:10.1007/s00411-003-0222-7 [DOI] [PubMed]
- Hawkins RB (2006) Mammalian cell killing by ultrasoft X rays and high-energy radiation: an extension of the MK model. Radiat Res 166:431–442. doi:10.1667/RR3594.1 [DOI] [PubMed]
- Hill MA, Stevens DL, Stuart Townsend KM, Goodhead DT (2001) Comments on the recently reported low biological effectiveness of ultrasoft X rays. Radiat Res 155:503–510. doi:10.1667/0033-7587(2001)155[0503:COTRRL]2.0.CO;2 [DOI] [PubMed]
- Hill MA, Stevens DL, Kadhim M, Blake-James M, Mill AJ, Goodhead DT (2006) Experimental techniques for studying bystander effects in vitro by high and low-LET ionising radiation. Radiat Prot Dosimetry 122:260–265. doi:10.1093/rpd/ncl429 [DOI] [PubMed]
- Jakob B, Rudolph JH, Gueven N, Lavin MF, Taucher-Scholz G (2005) Live cell imaging of heavy-ion-induced radiation responses by beamline microscopy. Radiat Res 163:681–690. doi:10.1667/RR3374 [DOI] [PubMed]
- Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ et al (2009) Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. doi:10.1093/nar/gkp221 [DOI] [PMC free article] [PubMed]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG et al (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172:823–834. doi:10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed]
- Limoli CL, Ward JF (1994) Response of bromodeoxyuridine-substituted Chinese hamster cells to UVA light exposure in the presence of Hoechst dye #33258: survival and DNA repair studies. Radiat Res 138:312–319. doi:10.2307/3578678 [DOI] [PubMed]
- Luijsterburg M, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S et al (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185:577–586. doi:10.1083/jcb.200810035 [DOI] [PMC free article] [PubMed]
- Lukas C, Bartek J, Lukas J (2005) Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma 114:146–154. doi:10.1007/s00412-005-0011-y [DOI] [PubMed]
- Mari P, Florea BI, Persengiev SP, Verkaik NS, Brüggenwirth HT, Modesti M et al (2006) Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA 103:18597–18602. doi:10.1073/pnas.0609061103 [DOI] [PMC free article] [PubMed]
- Marrot L, Meunier J (2008) Skin DNA photodamage and its biological consequences. J Am Acad Dermatol 58:S139–S148. doi:10.1016/j.jaad.2007.12.007 [DOI] [PubMed]
- Melo A, Kornberg M, Kaufmann P, Piazzetta M, Bortolucci E, Zakia M et al (2008) Metal mesh resonant filters for terahertz frequencies. Appl Opt 47:6064–6069. doi:10.1364/AO.47.006064 [DOI] [PubMed]
- Moné MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ et al (2001) Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep 2:1013–1017. doi:10.1093/embo-reports/kve224 [DOI] [PMC free article] [PubMed]
- Nelms BE, Mackie TR, MacKay JF, Hill CK, DeLuca PMJ, Lindstrom MJ et al (1998a) A comparison of cytotoxicity after whole- or partial-cell irradiation with synchrotron-produced ultrasoft X rays. Radiat Res 150:521–527. doi:10.2307/3579868 [DOI] [PubMed]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH (1998b) In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590–592. doi:10.1126/science.280.5363.590 [DOI] [PubMed]
- Nikjoo H, O’Neill P, Terrissol M, Goodhead DT (1999) Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat Environ Biophys 38:31–38. doi:10.1007/s004110050135 [DOI] [PubMed]
- Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E (2000) Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J Biol Chem 275:26732–26742 [DOI] [PubMed]
- Preston RJ (2005) Radiation biology: concepts for radiation protection. Health Phys 88:545–556. doi:10.1097/00004032-200506000-00003 [DOI] [PubMed]
- Rodrigo G, Roumagnac S, Wold MS, Salles B, Calsou P (2000) DNA replication but not nucleotide excision repair is required for UVC-induced replication protein A phosphorylation in mammalian cells. Mol Cell Biol 20:2696–2705. doi:10.1128/MCB.20.8.2696-2705.2000 [DOI] [PMC free article] [PubMed]
- Schettino G, Folkard M, Prise KM, Vojnovic B, Michael BD (2002) Upgrading of the Gray Laboratory soft X ray microprobe and V79 survival measurements following irradiation of one or all cells with a CK X ray beam of different size. Radiat Prot Dosimetry 99:287–288 [DOI] [PubMed]
- Sowa Resat M, Morgan WF (2004) Microbeam developments and applications: a low linear energy transfer perspective. Cancer Metastasis Rev 23:323–331. doi:10.1023/B:CANC.0000031770.96320.1e [DOI] [PubMed]
- Sowa MB, Murphy MK, Miller JH, McDonald JC, Strom DJ, Kimmel GA (2005) A variable-energy electron microbeam: a unique modality for targeted low-LET radiation. Radiat Res 164:695–700. doi:10.1667/RR3463.1 [DOI] [PubMed]
- Stap J, Krawczyk PM, Van Oven CH, Barendsen GW, Essers J, Kanaar R et al (2008) Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nat Methods 5:261–266. doi:10.1038/nmeth.f.206 [DOI] [PubMed]
- Stewart GS, Wang B, Bignell CR, Taylor AMR, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966. doi:10.1038/nature01446 [DOI] [PubMed]
- Sun GM, Kim EH, Song KB, Jang M (2006) Beam characterisation of the KIRAMS electron microbeam system. Radiat Prot Dosimetry 121:84–91. doi:10.1093/rpd/ncl009 [DOI] [PubMed]
- Tanno Y, Kobayashi K, Tatsuka M, Gotoh E, Takakura K (2006) Mitotic arrest caused by an X-ray microbeam in a single cell expressing EGFP-aurora kinase B. Radiat Prot Dosimetry 122:301–306. doi:10.1093/rpd/ncl512 [DOI] [PubMed]
- Tartier L, Spenlehauer C, Newman HC, Folkard M, Prise KM, Michael BD et al (2003) Local DNA damage by proton microbeam irradiation induces poly(ADP-ribose) synthesis in mammalian cells. Mutagenesis 18:411–416. doi:10.1093/mutage/geg015 [DOI] [PubMed]
- Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T (2000) Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol 150:283–291. doi:10.1083/jcb.150.2.283 [DOI] [PMC free article] [PubMed]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G et al (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177:219–229. doi:10.1083/jcb.200608077 [DOI] [PMC free article] [PubMed]
- van Royen ME, Farla P, Mattern KA, Geverts B, Trapman J, Houtsmuller AB (2009) Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells. Methods Mol Biol 464:363–385 [DOI] [PubMed]
- Walter J, Cremer T, Miyagawa K, Tashiro S (2003) A new system for laser-UVA-microirradiation of living cells. J Microsc 209:71–75. doi:10.1046/j.1365-2818.2003.01117.x [DOI] [PubMed]
- Williams ES, Stap J, Essers J, Ponnaiya B, Luijsterburg MS, Krawczyk PM et al (2007) DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat Genet 39:696–698. doi:10.1038/ng0607-696 (author reply 698–699) [DOI] [PubMed]
- Yokoya A, Watanabe R, Hara T (1999) Single- and double-strand breaks in solid pBR322 DNA induced by ultrasoft X-rays at photon energies of 388, 435 and 573 eV. J Radiat Res (Tokyo) 40:145–158. doi:10.1269/jrr.40.145 [DOI] [PubMed]