Abstract
Background
Chromosomal deletions and duplications, which result in halving or doubling of copy number in a block of genes, are an important source of variation between individuals. Phenotypic effects of copy number variation are commonly observed but, to our knowledge, effects on sensitivity to volatile anesthetics have not been assessed in any organism.
Methods
The potency with which halothane depresses the righting reflex of fruit flies was measured in congenic Drosophila strains, each of which was heterozygous for a deletion of average size ~ 400 kb. Over 200 strains were examined, thereby scanning~half of the fly genome.
Results
Although the vast majority of deletion heterozygotes were indistinguishable from the control, eight had significantly altered sensitivity to halothane. Genetic tests supported the hypothesis that the change in anesthetic sensitivity was due to reduction in copy number and not adventitious mutations in the strains. Among the eight outliers, the difference in halothane potency ranged from a 25% increase to a 15% decrease. When these lines were tested with enflurane, isoflurane and sevoflurane, changes of similar magnitude but distinctive patterns were found.
Conclusions
Variation in gene copy number has a significant impact on anesthetic sensitivity in Drosophila melanogaster. The level of transcription of a few genes must thus be limiting for a normal response to volatiles. Since coupling between gene copy and gene expression is universal and since the components of the fly’s nervous system are highly conserved, our work provides a rationale for investigating the clinical impact of copy number variation.
Introduction
The study of genetic variation in the human population has benefited greatly from the application of sophisticated molecular biological techniques. The most recent advance is the demonstration that, in addition to variable number tandem repeats and single nucleotide polymorphisms, the chromosomes of individuals differ from each other by deletion or duplication of blocks of DNA that range in size from a few kb to >1Mb. 1 Such rearrangements, which can affect virtually any segment of the genome, are surprisingly common: the first human to have his genome completely sequenced was found to harbor over 60 such blocks. 2 By definition, deletions and duplications produce a 50% change in the copy number of the affected set of genes. Since gene dosage typically regulates the level of gene expression, copy number variation (CNV) is a priori likely to have significant effects on human biology, especially on complicated processes in which a two-fold change in gene expression could be disruptive. Indeed, a handful of human diseases are now known to be associated with duplicated or deleted segments of the genome 3 and increasing attention is being paid to the possibility that CNV underlies others. A catalog of the genes affected by CNV in the human population has not yet been assembled. However, even in one individual the list of genes affected by CNV includes an ion channel (KCNN3) and a vesicle trafficking protein (SYT8), both of which are expressed in adult brain. 2 Given the likelihood that the population of surgical patients will harbor copy number variants of many genes that affect neural function, the potential impact of CNV on anesthesia needs to be addressed.
One way to begin such an assessment is via a model organism in which gene copies can be manipulated systematically. Past work from our lab and others have shown that the fruit fly, Drosophila melanogaster, responds to anesthetics at concentrations comparable to those used in the clinic. 4,5 Moreover, mutations that influence anesthetic endpoints in the fly often map to well-conserved genes that have significant roles in the functioning of the nervous system. 6,7 However, because previous studies used strong mutagens, they may have been biased toward isolation of mutations that severely disrupt gene function. Thus, past work with Drosophila does not provide a clear guide to the effects on anesthesia that would be expected from the subtle changes in gene expression that accompany CNV. To address this uncertainty, we tested a collection of over 200 fly strains, each of which has a 50% reduction in the copy number of a substantial block of genes. The results provide the first estimation of the frequency with which such variants influence anesthetic sensitivity and the magnitude of this influence.
Materials and Methods
Drosophila Stocks, Crosses and Transformants
Flies were grown on cornmeal/molasses agar at 25°C and 50% humidity under a 12:12 light/dark cycle using standard techniques. 8 Over 220 members of the DrosDel collection of deletion stocks 9 were obtained from the Bloomington Drosophila Stock Center at Indiana University. Each of these lines was not only fully characterized at the molecular level but had been constructed so as to maintain the genetic background of the corresponding parental stock. The mean (±sd) and median size of the deleted region in the strains that we tested were, respectively, 441±236 kb and 429 kb; taken together the deletions affected ~50% of the fly genome. For the initial screen of this set, the parental DrosDel line and the deletion stocks derived from it were individually crossed to a derivative of the Canton-S line into which had been introduced a recessive allele, har38, of the narrow abdomen (na) gene. 10 The female offspring that were subsequently tested were thus heterozygous both for a particular deletion and for a strong allele of na; this was done with three aims in mind. The first was to place the deletion chromosome in trans to a standard chromosome rather than the multiply-inverted, multiply-marked balancer chromosomes 8 present in the original stock. This permitted valid comparisons to be made between deletions that mapped to different chromosomes and thus came with different balancers. The second aim was to provide the tested lines with a wild-type copy of the white (w) gene, since the DrosDel parental and deletion lines all bear an allele of this gene (w1118) that is known to alter anesthetic sensitivity. 11,12 The final aim was to enable the possibility of finding genes that have a specific interaction with na. When homozygous, mutations in this gene confer strong effects on anesthesia sensitivity, but heterozygotes are indistinguishable from wild-type. 10,13,14 We reasoned that, if na depended on a component that was present in limiting amounts, a strain that was heterozygous both for na and the gene encoding such a putative component might be uniquely altered in anesthetic sensitivity. Although this possibility was examined whenever an anesthetic phenotype was suspected, we did not find such an enhancer locus: heterozygosity for na was not essential for the effects we found. Accordingly, the final analysis of “outlier” DrosDel deletions was done after crossing them to the parental Canton-S line rather than to the na mutant derivative of it. Similarly, selected deletion lines (also obtained from the Bloomington Stock Center) from the Exelixis collection 15 were evaluated, relative to the Exelixis parental line, for the presence of dosage-sensitive loci by crossing to Canton-S and testing non-balancer female offspring.
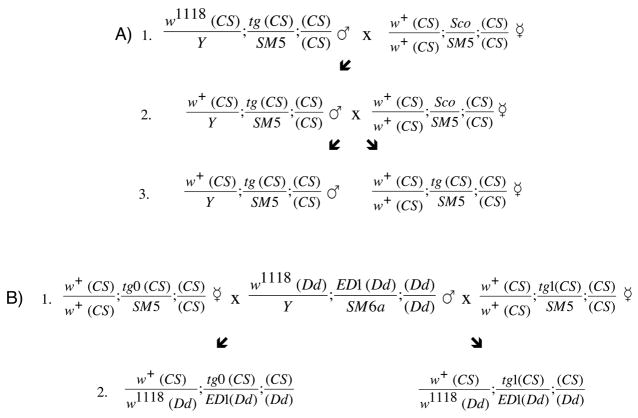
To evaluate the ability of transgenes to overcome the effect of a particular heterozygous deletion, we employed a commercial service (Genetic Services, Inc., Sudbury, MA) to introduce by standard techniques 16 the DNA segments described below (Plasmid Constructs) into the genome of a w1118 derivative of our Canton-S line. 11 For each plasmid construct we isolated several transformants, at least one on each of the two autosomes. The white eye color of the parental line permitted facile detection of individuals that inherited the transgene, which carries a miniaturized version of white and partially restores red eye color. However, since miniwhite lacks many features of the natural gene, it cannot be relied upon to overcome the anesthesia defects associated with the w1118 mutation. 11,12 Thus, using the crossing scheme outlined in Figure 1A, for each transformant line we replaced the X chromosome (bearing the w1118 allele) with that from wild-type Canton-S. The resulting lines were then crossed to the deletion line and the desired offspring were tested (Figure 1B). The experiments presented in the text were carried out with transformants bearing transgenic inserts on chromosome II, but comparable results were also obtained with transformants bearing inserts on chromosome III (data not shown).
Fig. 1.
Genetic Manipulations with Transgenes. (A) Replacing the X chromosome of the original transformant lines with a wild-type X chromosome. Each line diagrams a cross between males (♂) and virgin females (☿) of the indicated genotype. Chromosomes that derive from the Canton-S line are indicated as (CS); individual genetic elements that distinguish these chromosomes are indicated (w=white;tg=transgene). The crossing scheme relies on the use of a balancer, a chromosome in which the normal complement of genes has been severely rearranged so as to prevent recombination with a homologous chromosome. 8 In the example shown, the SM5 balancer carries a dominant marker that yields curly wings; since this marker is a recessive lethal, it is maintained as a heterozygote with a chromosome that carries a dominant bristle marker, Sco (detailed descriptions of these markers and the balancer can be found at FlyBase 41). In each of the two crosses shown, selection for the Curly phenotype and against the Sco phenotype insures inheritance of the transgene. (B) Testing rescue of a CNV phenotype by transgenes. Males that are heterozygous for the ED1 deletion (made in the DrosDel (Dd) genetic background) 9 are crossed (line 1) to females bearing a transgene. The latter are the offspring from a stock made by crossing males and females from line A3. In the examples shown, the transgene either carries no genomic DNA (tg0) or carries a partial segment of the genomic region under the ED1 deletion (tg1). The female offspring from these crosses (line 2) are assayed for halothane sensitivity in the distribution test; comparison of the resulting EC50 values indicates the extent of rescue. Another control (not shown) is generated by using males from the DrosDel parental line instead of ED1 males.
Plasmid Constructs
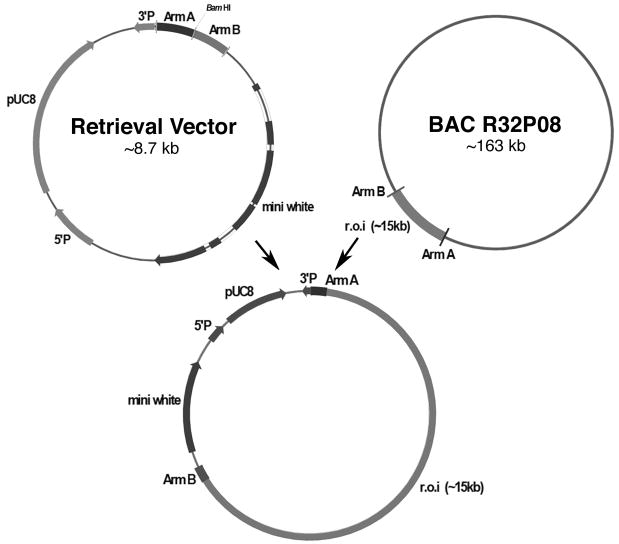
The vector used for our studies was CaSpeR4 (GenBank Accession X81645), a plasmid engineered to facilitate the creation of transgenic lines of Drosophila. 16 The key feature of this vector is a pair of transposon ends that, with the aid of a source of transposase that is coinjected with the vector into fertilized eggs, direct the insertion into the Drosophila genome of the segment of DNA that lies between the ends. To introduce 15–20 kb segments of the Drosophila genome into CaSpeR4, we used a recently described recombineering procedure. 17 First, using polymerase chain reaction we amplified a ~500 bp fragment from each end of the desired insert. These were cloned using conventional techniques to create a derivative of CaSpeR4 in which the two “homology arms” were adjacent to each other and separated by a unique BamHI restriction site (Figure 2). This so-called retrieval vector was then linearized at the unique site and transformed into a derivative of the recombination-proficient E.coli strain SW102. 18 To make this derivative, we introduced into SW102 a bacterial artificial chromosome, chosen from the Berkeley collection 19 because it bears a ~ 160 kb segment of the Drosophila genome that encompasses all the material deleted in the DrosDel ED1 line. ‡ Within the transformed bacteria, recombination between homologous sequences initiate a repair process in which the retrieval vector is restored to a circular form that incorporates the segment between the homology arms from BACR32P8 (Figure 2). The ensuing plasmids were examined by restriction digestion and by polymerase chain reaction amplification of Drosophila genes from the region of interest; correct constructs were amplified and purified by conventional techniques.
Fig. 2.
Creation of rescue constructs by recombineering. At the top left is shown a vector used to retrieve sequences from the bacterial artificial chromosome (BAC) shown at the top right. Some key features of the vector are marked. These include the two ends (5′P and 3′P) of a P transposable element 16 and a pair of “homology arms”, ~500 bp segments (marked A and B) whose sequences come from the two ends of a region of interest (marked r.o.i.) within the BAC. Upon introduction by electroporation of a BamHI-linearized vector into a BAC-containing strain that has been engineered to overexpress recombination proteins, gap repair by homologous recombination promotes the transfer to the vector of the entire segment of the BAC that lies between the arms. Two attractive features of this recombineering (recombination-mediated genetic engineering) procedure 42 is that it does not depend on fortuitously placed restriction sites (any sequence from the BAC can be amplified and cloned as a homology arm) and that it employs the natural replication machinery of E. coli to achieve the transfer of genetic information and thus is much less prone to introduction of errors than polymerase chain reaction-based methods.
Anesthetics
Halothane was obtained from Halocarbon Laboratories, River Edge, NJ, sevoflurane and isoflurane from Abbott Laboratories, North Chicago, IL, and enflurane from Baxter Healthcare Corp, Deerfield, IL. These agents were volatilized using Gas Washing Bottles (Fisher Scientific, Pittsburgh, PA) to bubble air through liquid anesthetic; the resulting stream was mixed with air that had been humidified in the same way. The concentration of anesthetic delivered to the glove box (total flow ~10L/min) was monitored using a MIRAN M1ACVF-A single beam infrared spectrometer (Invensys Process Systems, Plano, TX). The spectrometer was calibrated with standards made by injecting precise quantities of liquid anesthetic into a large vessel of known volume and, after volatilization was complete, pumping the mixture through the spectrometer. This calibration was repeated at 6 month intervals, with little or no adjustment required.
Tests of Anesthesia Sensitivity
Flies that were three to seven days old were collected and sorted in groups of 10 12 under carbon dioxide anesthesia and allowed to recover for one to two days. Without further recourse to anesthesia, they were then transferred from food vials to testing vials (50 ml Falcon tubes perforated with tiny holes to permit gas exchange but retain flies), which were then placed in a glove box. Following equilibration with a fixed concentration of volatile agent for 30–70 min, the flies were examined for their ability to escape the conical bottom of the testing vial following three brief mechanical shocks. This assay of a fly’s righting and climbing reflex, also known as the distribution test, 10,11 was quantified as the minimum number of flies found at the bottom during a one minute trial. Typically, each vial of flies was tested sequentially at increasing concentrations of drug, with an additional equilibration period of 30–70 min for each new concentration. This procedure was adopted for the following reasons. First, flies do not feed during the assay and, although the anesthetic is delivered in humidified air, are in danger of becoming dehydrated and/or starved after several hours in the glove box. Second, the exchange of gases in insects like fruit flies is primarily passive, via diffusion through blunt-ended trachea that start at the cuticle and ramify throughout the internal organs. Washout to the baseline is thus predicted to be asymptotic and, at least as judged by observation of behavior, appears to be quite slow. Thus, to maximize the amount of useful information obtained within a fixed time window, we tested flies at incrementally higher concentrations (stair-stepping) without interposing no-anesthesia intervals between them. This procedure is reliable and reproducible; moreover in the few cases tested, it yields EC50 values that are similar to those obtained with a procedure in which no fly is tested at more than one concentration. 10,12
The initial screen of deletion heterozygotes was done with groups of 20–25 lines, 3 vials of flies per line, which were tested together with 10–15vials of the control heterozygote. In total, 220 DrosDel strains were tested: 39carried deletions of material from the X chromosome, 42 from the 2L autosomal arm, 35 from 2R, 53 from 3L, and 51 from 3R. The complete list of tested strains is available upon request. As a compromise between throughput and detection, each line was tested only at three concentrations of halothane. For most of the survey these were 0.30%, 0.375% and 0.45%, concentrations that induced ~ 10to 80 %failure of reactive climbing in control flies. For each line tested, a crude estimate of potency (EC50 value) was derived by a logit analysis. 20 We focused on those deletion heterozygotes that gave an initial estimate of EC50 that differed by ≥10% from the value deduced from the contemporaneous dataset for the control heterozygote. This criterion reflected our assessment of the uncertainty in the calculated potency values. To begin this assessment, we treated individual datasets from the control heterozygote as if they were obtained both from control and a deletion line; i.e., we compared a dataset from the control heterozygote to itself. The 95% confidence limits on the resulting potency ratio typically were 0.97 to 1.03. Consistent with this estimate is the fact that, when all 18 EC50 values determined for the control heterozygote throughout the course of this study were compared to each other, the coefficient of variation around the mean value of 0.385% halothane was 0.057. Although these estimates might have encouraged focusing on deletion heterozygotes that differed from control by ± 5%, as noted above (Testsof Anesthesia Sensitivity, 2nd paragraph) each deletion heterozygote was tested with 4–5 times fewer flies than the control. This fact, which reflected the need to manage the manpower cost of the project, meant that the proportion of flies that failed the distribution test was less precisely determined for the deletion than the control heterozygotes. Accordingly, we expanded the “gray zone”, in which we ignored potential differences between lines, to include EC50 values that differed from control by ±10%.
Lines that passed this initial test were retested, using fresh offspring from the cross. This eliminated about half of the initial candidates and focused our attention on 12 of the 220 lines. These were subjected to a final evaluation, in which one set of vials (comprising 24 to 48 animals) was tested at four concentrations of halothane (0.10%, 0.20%, 0.30%, and 0.35%) and a second set of the same genotype was tested at four higher concentrations (0.40%, 0.50%, 0.60%, and 0.70%). The eight lines that differed from control in this evaluation were subsequently evaluated in separate rounds of testing for sensitivity to enflurane, isoflurane, and sevoflurane.
Statistical Analysis
All statistical comparisons were made on the basis of a logit analysis 20 of 8-point concentration-response curves using a commercial software package (SPSS Inc, Chicago, IL). The input for such analyses were experiments performed in parallel on a group of deletion heterozygotes and the relevant control heterozygote. The output consisted of maximum likelihood estimates for two parameters that describe concentration-response curves. The first parameter characterized the slope or steepness of the curves; here, a single value was calculated that best fit all the lines tested. The second parameter provided distinct estimates of potency for each line. These were presented in two forms: a) EC50 values and 95% confidence limits for each strain; and b) potency ratios (EC50 for strain x/EC50 for strain y) and 95% confidence limits for each pair of strains. The shift in EC50 was calculated from the control heterozygote/deletion heterozygote potency ratio, R, as 100 × [(1/R)−1]. The 95% confidence limits on this shift were similarly calculated from the corresponding limits provided for R. Significant differences were assigned to those deletion heterozygotes for which the confidence limits on the shift in EC50 from that of the control heterozygote were entirely above or below zero. For the rescue construct strains, significant differences between relevant pairs were assigned if the confidence limits on their EC50 values did not overlap.
Results
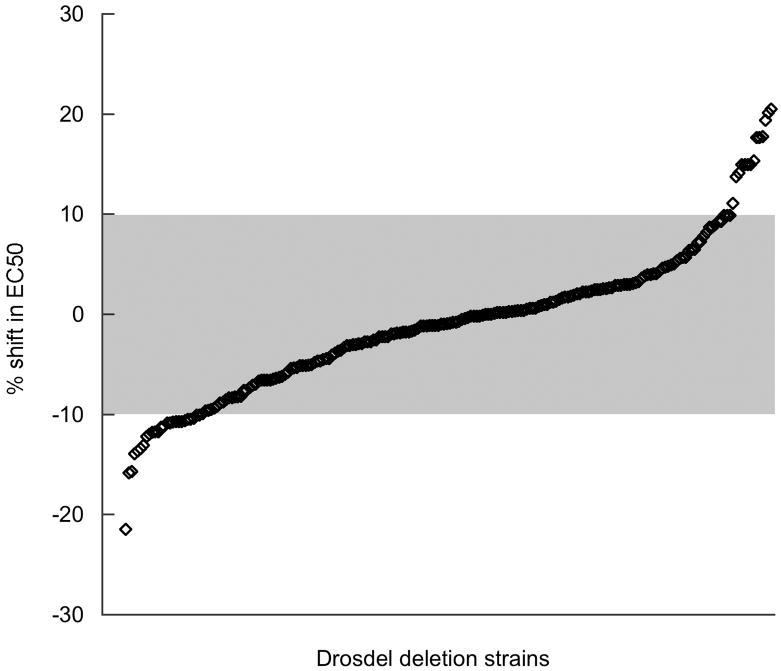
Our study was made possible by the existence of a collection of Drosophila strains bearing chromosomal deletions. 9 Many of these variants were crossed to a laboratory stock to generate a large set of deletion heterozygotes, each distinguished by a single contiguous block of genes in which copy number was reduced from two to one. We exposed each of these strains to a few concentrations of halothane and measured the effect of the drug on the ability of flies to right themselves and climb after a brief mechanical shock. 10 From this we estimated an EC50 value and compared it to the comparable value for a control strain. As seen in the overview shown in Figure 3, the vast majority of strains have EC50 values that fall into the range that can not be reliably distinguished from the control: if copy number variation in these strains produces an effect on anesthesia, it is too small to be detected by our methods. We conclude that, at least in this model organism, subtle changes in gene expression are not a ubiquitous source of variability in anesthetic responsiveness.
Fig. 3.
A survey of the effect of copy number variation on halothane potency. EC50 values for 220 deletion heterozygotes were estimated as described in Materials and Methods. Each strain was assigned a rank based on the percentage difference between its EC50 value and that determined in parallel for the control strain. The figure presents these differences as a function of rank number. Note that very few of the strains can be reliably distinguished from control line because the shift in their EC50 falls within the region of experimental uncertainty (gray shading).
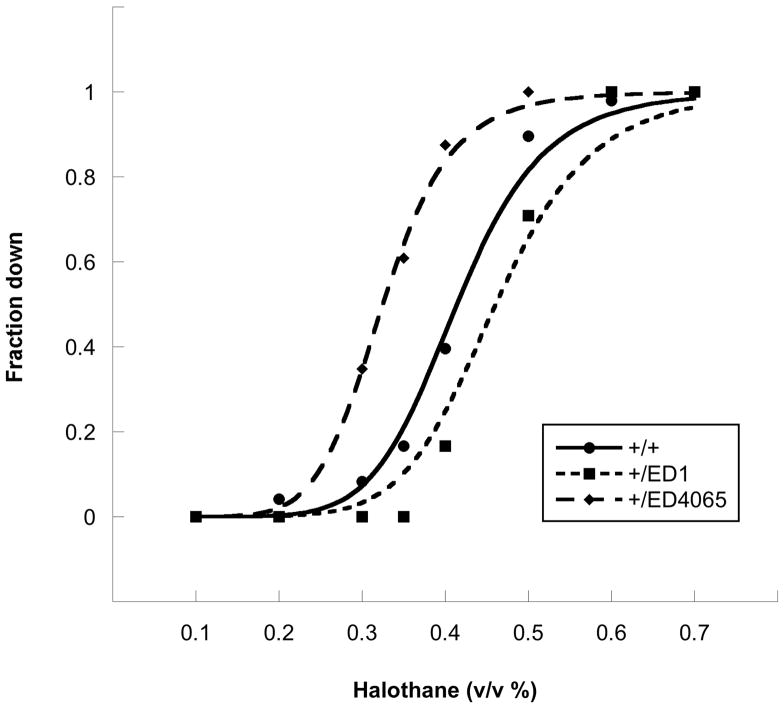
It is also evident from Figure 3 that, against the featureless background formed by most of the strains, a few deletion heterozygotes appear to show significant effects of CNV on the response to halothane. We retested these “outliers”; those that held up were then studied over a wide range of concentrations. As typified by the examples shown in Figure 4, this proved that several strains have bona fide alterations in halothane EC50. From the detailed examination of the outliers that we have studied most carefully (Table 1), a few facts stand out. First, none of the effects are large. Although we have examined deletions that span over half of the fly genome, the biggest shift in EC50 seen is ~25%. Second, relative resistance to halothane is detected with similar frequency to halothane hypersensitivity. The rough parity between increased and decreased sensitivity argues that CNV is not just making flies sick, since this would be expected to favor hypersensitivity. Third, as described in Materials and Methods, the initial screen used strains that were heterozygous not only for a deletion but also for a strong allele of a gene (na) that influences anesthetic sensitivity in flies. However, in no case did we find a deletion whose effect was the result of a genetic interaction with the na gene, and the data in Table 1 therefore come from strains that were simply heterozygous for the indicated deletion. If there are genes that interact with na, more sensitive methods will have to be used to find them. Fourth, since all the arms of the fly genome were well represented in the set of tested deletions, even if one assigns a single count to pairs of very tightly linked hits, the distribution appears to be non-random. Although we have no clear explanation for this phenomenon, we wonder if the centromere-proximal portion of 2R (the locus of six of the seven hits in this arm) is less able than other regions to compensate for reduced gene dosage. 21
Fig. 4.
Representative concentration-response curves for the halothane-induced loss of the righting/climbing reflex. The data for the control strain (+/+) and two deletion heterozygotes (+/ED1 and +/ED4065) were fitted to the logistic function, resulting in a common slope coefficient of 7.9 and EC50 values, respectively, of, 0.40, 0.46, and 0.32.
Table 1.
Summary information on deletions with unequivocal effects on halothane sensitivity
| Strain Designation | Location of deletion (Chromosome arm:genomic coordinate) | Deletion size (bp) | Δ potency (95% CL) |
|---|---|---|---|
| +/ED4065 | 2R:20290189 | 540173 | −19.7% (−13.0 to −26.9) |
| +/ED5495 | 3R:5996223 | 716259 | −25.0% (−15.6 to −35.9) |
| +/ED2247 | 2R:7487611 | 388614 | −16.7% (−10.2 to −23.6) |
| +/ED2308 | 2R:8667875 | 216614 | −16.6% (−10.1 to −23.5) |
| +/ED1725 | 2R:3501429 | 542121 | +9.9% (+4.9 to +15.4) |
| +/ED1715 | 2R:3214456 | 589972 | +9.2% (+4.2 to +14.5) |
| +/ED1 | 2R:12914232 | 70595 | +16.1% (+10.6 to +22.5) |
| +/ED2751 | 2R:12744676 | 240132 | +15.6% (+10.1 to +21.8) |
The designated name indicates that the tested strains are heterozygotes between a strain without a deletion (+) and a strain (ED#) from the DrosDel collection carrying a deletion with the indicated number. The start point of each deletion is given with reference to the coordinate system of Drosophila Genome Release 5.1 (GenBank Accession AE013599.4 and AE014297.2). The change in potency of the deletion heterozygotes relative to the control heterozygote (+/parental ED line with no deletion) is deduced from eight-point concentration-response curves. These values, together with their 95% confidence limits, are calculated as described in Materials and Methods from the potency ratios derived from a logit analysis of the data.
Taken at face value, the data shown in Table 1 and Figure 4 suggests that CNV has a definite influence on anesthesia. However, an alternative hypothesis is that the anesthetic phenotype of the listed strains is not due to the deletions they contain but to adventitious mutations that they harbor. Such mutations might have been introduced during the original construction of the deletion line 9 or might have arisen in the deletion line during subsequent passages. 22 We have used two strategies to confront this possibility. The first is based on the rationale that, if a deleted region is responsible for the anesthetic phenotype, then putting an extra copy of that region at some other location in the genome should reverse the phenotype. Indeed, a transgene bearing a portion of the region missing in ED1 fully reverses the halothane resistance associated with this deletion (Table 2). The larger size of the other deletions in Table 1 encumbered the rescue strategy and prompted the use of a complementary approach: if the original effect is actually due to removal of one or more genes from the indicated region, the effect should be recapitulated by a different deletion that also removes this critical block. Accordingly, for several of the regions implicated in Table 1, we acquired and tested independently derived strains 15 that contain deletions with at least one endpoint in the region of interest. Figure 5 provides an example of this approach by comparing the effect of a deletion from the original collection (ED2308) with the effect of independently derived deletions that remove DNA from the same chromosomal region. A comparable level of halothane hypersensitivity is observed in each case; a similar outcome has been obtained following examination of deletions from within the region of reduced copy number in strains ED4065/+ and ED2247/+ (Dongyu Guo, PH. D., Research Fellow, Laboratory of Molecular Biology, National Institute of Mental Health, Bethesda, MD, USA, unpublished observations). Although some of the deletions of Table 1 have not been examined with either strategy, the success of our initial trials indicates that adventitious mutations do not seriously confound the conclusion that variation in gene copy number produces significant changes in halothane sensitivity.
Table 2.
Transgenic rescue of the anesthesia phenotype in a deletion heterozygote
| Row | Deletion carried by parent 1 | Content of transgene carried by parent 2 | Halothane EC50, v/v% (95% CL) |
|---|---|---|---|
| 1 | None | None | 0.388 (0.378–0.399) |
| 2 | ED1 | None | 0.434 (0.423–0.446) |
| 3 | ED1 | 2R: 12945571–12967489 | 0.426 (0.415–0.437) |
| 4 | ED1 | 2R: 12973364–12988824 | 0.373 (0.364–0.384) |
Each row of the table contains information about a line of flies made by crossing a strain from the DrosDel collection (either the parental stock or the ED1 deletion derived from it) to a transgenic derivative of the Canton-S strain (for a diagram of the crossing scheme, see Figure 1). The transgene either consisted of an empty vector (rows 1 and 2) or (in rows 3 & 4) the same vector modified by the addition of a segment of the region deleted in ED1 (see Figure 2 for an outline of this construction). The potency with which halothane depressed the climbing ability of the offspring from these crosses was determined from 8-point concentration-response curves and is expressed as an EC50 value. Comparison of the EC50 values in rows 2 and 4 shows that an extra copy of a ~15 kb region can restore the sensitivity of an ED1 heterozygote to a level that is indistinguishable from that of wild-type flies (row 1). The specificity of the rescue is demonstrated by the failure of a different segment from the ED1 region to produce this effect (compare rows 2 and 3).
Fig. 5.
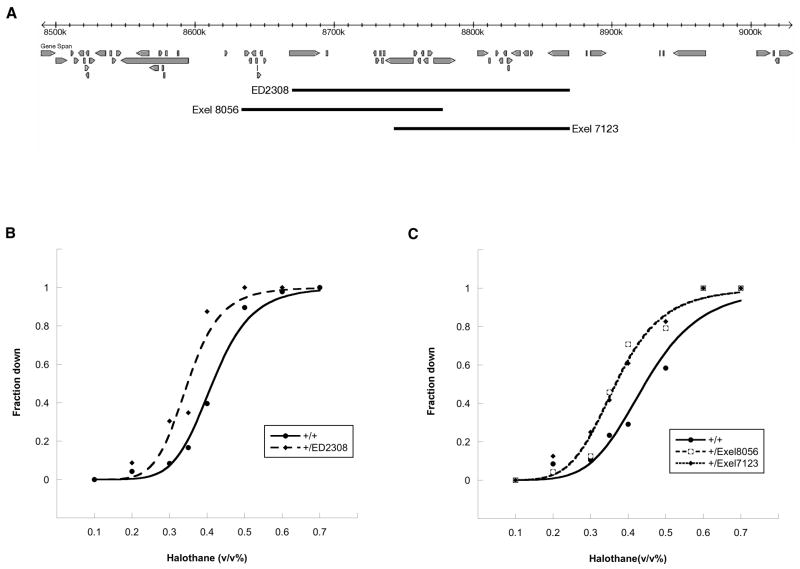
Hypersensitivity to halothane associated with multiple deletions involving one region of the genome. (A) An overview of a 540 kb segment of chromosome arm 2R. Beneath the scale ruler is shown the rough position of genes predicted= to lie in this region, the extent of the DrosDel ED2308 deletion described in Table 1, and the extent of two independently derived deletions from the Exelixis collection.(B, C) Concentration-response curves. The tested lines were generated by crossing the Canton-S wild-type strain to the indicated deletion strain or, in the case of the control heterozygotes, to the parental line from which the deletion was derived. Halothane sensitivity was assessed as described in Materials and Methods. Compared to the corresponding control line (+/+), there is a leftward shift in the EC50 value of the DrosDel deletion heterozygote (B) and both of the heterozygotes made with deletions from the Exelixis collection (C). The magnitude of this shift for +/ED2308, +/Exel8056, and +/Exel7123 is 16.6%, 17.7%, and 17.3%, respectively.
Diverse volatiles often display differential effects on molecular and cellular systems. 23–25 Reflecting this complexity, previous studies have found that the effect of a point mutation on sensitivity to one anesthetic is not a good predictor of its effect on other volatiles. 26,27 To see if the subtler effects of CNV also display agent-specificity, we tested the strains described in Table 1 for sensitivity to anesthetics other than halothane. On the one hand, some of the dosage-sensitive regions, e.g., that which is deleted in ED4065, affect all volatile agents equivalently (Table 3). This could imply the presence of genes within the deleted segments that contribute to processes affected by all agents. However, although gross climbing ability of these strains in the absence of anesthetics appears normal, it is hard to rule out the possibility that uniform hypersensitivity is merely the result of subtle defects in baseline locomotor performance that are enhanced under the stress of anesthesia. On the other hand, some CNVs show clear evidence of agent-specificity and thus by definition cannot have suffered a change in baseline performance. For example, the segment deleted in ED2247 seems important for sensitivity to halothane and enflurane but not isoflurane and sevoflurane (Table 3). In these cases it appears there are genes whose expression is limiting for the operation of neural circuits that are disparately affected by different volatile agents. A powerful way to find such circuits is to identify the critical genes within the deleted segments and then to determine where they must be expressed to influence anesthesia sensitivity.
Table 3.
Spectrum of anesthetic effects in deletion heterozygotes that show an altered sensitivity to halothane.
| Strain/Genotype | Enflurane | Isoflurane | Sevoflurane |
|---|---|---|---|
| +/ED4065 | −27.0% (−20.1 to −34.2) | −28.7% (−22.7 to −35.3) | −20.3% (−16.0 to −24.9) |
| +/ED5495 | −13.2% (−7.8 to −19.7) | −13.1% (−9.5 to −16.9) | −22.1% (−18.0 to −26.6) |
| +/ED2247 | −14.7 (−9.1 to −21.7) | +0.3% (−4.7 to +6.0) | +0.7% (−2.9 to +4.7) |
| +/ED2308 | −10.1% (−5.1 to −15.3) | −1.7% (−4.3 to +0.9) | −7.0% (−3.6 to −10.4) |
| +/ED1725 | +2.9% (−2.2 to +8.5) | +2.7% (0.0 to +5.6) | 0.0% (−3.8 to +3.6) |
| +/ED1715 | +1.3% (−3.8 to + 6.8) | +2.4% (−0.3 to +5.3) | −3.2% (−6.7 to +0.4) |
| +/ED1 | −2.8% (−7.7 to +2.2) | +1.0% (−1.6 to +3.7) | −0.9% (−4.5 to +2.9) |
| +/ED2751 | −14.4% (−9.2 to −20.0) | +1.0% (−1.6 to +3.7) | −2.6% (−6.1 to +1.0) |
The strains described in Table 1 were tested for sensitivity of the righting/climbing reflex to enflurane, isoflurane and sevoflurane. The protocol for testing was exactly as described in Materials and Methods for construction of concentration-response curves for halothane except that the following sets of concentrations were used. For enflurane: 0.10%, 0.20%, 0.30%, and 0.35% plus 0.40%, 0.50%, 0.60%, and 0.70%. For isoflurane: 0.10%, 0.20%, 0.30%, 0.40%, and 0.43% plus 0.45%, 0.48%, 0.50%, and 0.60%. For sevoflurane: 0.10%, 0.30%, 0.50%, 0.60% and 0.75% plus 0.65%, 0.70%, 0.78%, and 0.90%. For each of these agents the % change in potency from the control heterozygote and the 95% confidence limits on this value, calculated as in Table 1, is given.
Discussion
In this work we have used genetically engineered strains of Drosophila to mimic the kind of variation in gene copy number that is found in the human population. Our results show that a small fraction of such variants do have a significant impact on sensitivity to anesthetics and thus focus attention on this recently appreciated type of polymorphism.
Copy number variants are thought to arise by “unequal crossing-over”, i.e., recombination between perfect or near-perfect repeats that are located 10–1000 kb apart. 28 Since repeated DNA is a common genomic feature and homologous recombination is part of the normal process of meiosis, it is expected that CNVs will be generated with considerable frequency. One might have imagined that selection pressures would severely limit the survival of such variants, so the surprise from recent studies of these aberrations is their prevalence. A recent hybridization study designed to examine this issue stringently for 30 individuals found ~1000 instances of CNV, 29 consistent with the frequency found in the two individuals whose entire genome has been scrutinized. 2,30 Although spontaneous CNVs have also been found in Drosophila, 31,32 the deletions used in our study were made by expression of a foreign site-specific recombinase so as to promote unequal crossing-over between selected repeats that had been artificially placed in the genome. 9 Nevertheless, in terms of size and ubiquity, the deletions we used mimic those typical of CNVs found in nature.
The conventional rationale for expecting a close relationship between copy number of a gene and its expression is straightforward: a) gene expression is often limited by the amount of message per cell and b) for a given concentration of active RNA polymerase in a cell, transcription is often limited by the amount of template. 33 The classic evidence supporting this rationale was the discovery that amplification of the dihydrofolate reductase (DHFR) gene in cultured mouse cells is accompanied by a corresponding increase in DHFR message and, ultimately, enzymatic activity. 34 Similarly, amplification of an esterase gene in mosquitoes is associated with a corresponding increase in expression. 35 Despite the abundant historical evidence for correlation between copy number and gene expression, it must be acknowledged that the coupling is not perfect. For example, in a recent study of gene expression phenotypes, for some 90 human genes that displayed CNV, the correlation coefficients relating expression level and copy number were generally low. 36 The weak association may be due in part to the methodology employed in such a large-scale study, but it could also reflect the operation of compensatory mechanisms that blunt the effect of a two-fold change in copy number on message level. 21 Nevertheless, the association of CNV with disease states 3 proves that compensation is not uniformly effective and reduction in gene copy number can produce significant changes in human health. Similarly, classic studies of segmental aneuploidyin Drosophila provide many examples of a tight coupling between the expression of an enzyme and the copy number of its encoding gene. 37 Thus, it seems reasonable to infer that the effects on anesthesia that we have observed upon reduction in the copy number of certain regions of the genome is the result of reduced expression of genes that lie within that region.
Much work will need to be done in order to identify the critical genes that lie within the haploin sufficient regions. However, for a few such regions the process has begun and has resulted in a narrowed focus within the deleted segment. This advance is a side benefit of the tests we employed to confirm that the observed anesthesia effects were indeed due to reduced copy number of genes in the relevant segment. For example, the two deletions used to challenge the ED2308 region remove partially overlapping segments of the genome (Figure 5). Although we cannot rule out more complex possibilities, the simplest interpretation of this result is that a critical gene whose expression is responsible for the dosage-sensitive effects on halothane sensitivity lies in the relatively small region of overlap between these two deletions. An even sharper focus is provided by the successful transgenic experiment used to query the ED1 region. Specifically, the data of Table 2 clearly indicate that the critical gene responsible for the CNV effect of the ED1 deletion is one of the three that are carried by the rescue construct. Modification of the rescuing transgene to eliminate each of these genes in turn offers a straightforward, albeit laborious, method to refine this hypothesis.
Another issue addressed by our dissection of haploin sufficient regions concerns the magnitude of the observed changes in anesthetic sensitivity. We have noted that CNV effects are not large. Could this be due to a limitation inherent in our methods, e.g., the testing of flies at monotonically increasing concentrations? This is clearly not the case because, when studying null mutants rather than heterozygotes, the same methods have detected larger shifts in sensitivity 10,12 (J. L. Campbell, J. Q. Gu, and H.A. Nash, manuscript in preparation). Moreover, even though the CNV effects are modest, they are not trivial. Because of the steep slope of anesthetic concentration-response curves, even modest changes in potency can produce wholesale changes in population behavior. 38 Nevertheless, the magnitude of the CNV effect suggests that anesthesia is well-buffered against subtle changes in gene expression. Before accepting this interpretation, one should also consider the possibility that the true effect of CNV is obscured because deletions may remove genes with opposite influences on anesthesia. According to this scenario, the observed phenotype of a given deletion reflects a haphazard balance between the conflicting effects of reducing copy number of two antagonistic genes. If this were the case, a larger effect might be seen when a particular region of interest is reanalyzed with deletions that only remove part of it. However, such an enhancement was seen neither with the deletions used to subdivide ED2308 (Figure 5) nor with those that subdivided other intervals from Table 1 (Dongyu Guo, personal communication). Similarly, Table 2 shows that adding back the genes from the segment of the ED1 region that confers resistance to halothane does not unmask a hypersensitivity to the drug caused by reduced copy number of genes in the remainder of this region. Although these observations cannot conclusively eliminate the possibility that effects of copy number reduction on anesthesia are blunted by simultaneous manipulation of pairs of genes with opposite effects, our experience suggests that this scenario is uncommon.
At the outset of this work, we could not make a confident prediction either about the frequency with which deletion heterozygotes would show effects on anesthesia or about the magnitude of such effects. On the one hand, we worried that compensatory mechanisms 21 might commonly obscure the effect of reduction in copy number on gene expression. Moreover, even if compensation was not prevalent, we wondered about redundancy in the neural pathways affected by anesthetics. Taken together, these factors made it seem possible that none of the 220 deletion heterozygotes would generate a reliable phenotype, an outcome that would lead us to the conclusion that CNV had little or no impact on anesthesia. On the other hand, we were aware that Drosophilists have long used changes in copy number of one gene to enhance effects of a mutation in another gene. 39 And, since anesthetics are known to undermine the function of many gene products, we suspected that each of these effects could be subject to enhancement by a separate change in copy number. Moreover, the behavioral endpoint used for our study is obviously the result of many complex locomotor actions, each of which must depend on the functioning of many genes. Reduced copy number of these genes could have effects on locomotor ability too subtle to detect in the absence of anesthesia, but the drug might make them evident. Thus, it also seemed possible that the majority of deletion heterozygotes would have a dramatic anesthetic phenotype, leading us to conclude that CNV had a major impact on anesthesia. Our results inform us that the actual effects of CNV fall in between these extremes. Changes in copy number thus take their place alongside point mutations and transposon insertions as part of the spectrum of genetic aberrations that significantly influence anesthesia in flies. 4,6 Current efforts are exploring how CNV influences the effectiveness of various drugs in patient populations. 40 Our experience with Drosophila suggests that it would be worthwhile to include volatile anesthetics in such studies.
Acknowledgments
Supported by the Intramural Program of the National Institute of Mental Health. None of the authors has any financial interest in this work.
The authors thank Joy Qun Gu for performing the assays of Table 2, Robert Scott for help with the figures and Dongyu Guo for his permission to cite unpublished observations and for his comments on the manuscript. This research was supported by the Intramural Program of the NIMH.
Footnotes
http://www.fruitfly.org/data/sequence/release3/BACR32P08; last visited 12/29/08.
(http://flybase.org/cgi-bin/gbrowse/dmel/?name=2R:8490000..9029346); last visited 12/29/08.
References
- 1.Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–61. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 2.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J. Genomics. DNA duplications and deletions help determine health. Science. 2007;317:1315–7. doi: 10.1126/science.317.5843.1315. [DOI] [PubMed] [Google Scholar]
- 4.Gamo S. Studies on target genes of general anesthetics. Curr Drug Targets. 2002;3:31–41. doi: 10.2174/1389450023348118. [DOI] [PubMed] [Google Scholar]
- 5.Nash HA. In Vivo Genetics of Anesthetic Action. Brit J Anesth. 2002;89:143–55. doi: 10.1093/bja/aef159. [DOI] [PubMed] [Google Scholar]
- 6.Humphrey JA, Hamming KS, Thacker CM, Scott RL, Sedensky MM, Snutch TP, Morgan PG, Nash HA. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr Biol. 2007;17:624–9. doi: 10.1016/j.cub.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Takase M, Gamo S. Relationship between general anesthesia and memory in Drosophila involving the cAMP/PKA pathways and adhesion-related molecules. Curr Med Chem. 2007;14:1479–88. doi: 10.2174/092986707780831140. [DOI] [PubMed] [Google Scholar]
- 8.Stocker H, Gallant P. Getting started : an overview on raising and handling Drosophila. Methods Mol Biol. 2008;420:27–44. doi: 10.1007/978-1-59745-583-1_2. [DOI] [PubMed] [Google Scholar]
- 9.Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, O’Kane CJ, Huen D, Sharma P, Asztalos Z, Baisch H, Schulze J, Kube M, Kittlaus K, Reuter G, Maroy P, Szidonya J, Rasmuson-Lestander A, Ekstrom K, Dickson B, Hugentobler C, Stocker H, Hafen E, Lepesant JA, Pflugfelder G, Heisenberg M, Mechler B, Serras F, Corominas M, Schneuwly S, Preat T, Roote J, Russell S. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan Z, Scott RL, Nash HA. A New Assay for the Genetic Study of General Anesthesia in Drosophila melanogaster: Use in Analysis of Mutations in the 12E Region. J. neurogenet. 2000;14:25–42. doi: 10.3109/01677060009083475. [DOI] [PubMed] [Google Scholar]
- 11.Campbell JL, Nash HA. Volatile general anesthetics reveal a neurobiological role for the white and brown genes of Drosophila melanogaster. J Neurobiol. 2001;49:339–49. doi: 10.1002/neu.10009. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y, Nash HA. Visual mutations reveal opposing effects of illumination on arousal in Drosophila. Genetics. 2008;178:2413–6. doi: 10.1534/genetics.107.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12:2152–8. doi: 10.1016/s0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- 14.Alone DP, Scott RL, Nash HA. An ion channel that influences anesthesia sensitivity: Designing a genetic test for assessing a candidate anesthetic target. International Congress Series. 2005;1283:119–25. [Google Scholar]
- 15.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–92. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 16.Bachmann A, Knust E. The use of P-element transposons to generate transgenic flies. Methods MolBiol. 2008;420:61–77. doi: 10.1007/978-1-59745-583-1_4. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, Tabor PE, Bailey MR, DeShazo DR, Catanese J, Mammoser A, Osoegawa K, de Jong PJ, Celniker SE, Gibbs RA, Rubin GM, Scherer SE. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–4. doi: 10.1126/science.287.5461.2271. [DOI] [PubMed] [Google Scholar]
- 20.Waud DR. On biological assays involving quantal responses. J Pharm Exper Ther. 1972;183:577–607. [PubMed] [Google Scholar]
- 21.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–7. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Haag-Liautard C, Dorris M, Maside X, Macaskill S, Halligan DL, Houle D, Charlesworth B, Keightley PD. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–5. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 23.Matta JA, Cornett PM, Miyares RL, Abe K, Sahibzada N, Ahern GP. General anesthetics activate a nociceptive ion channel to enhance pain and inflammation. Proc Natl Acad Sci U S A. 2008;105:8784–9. doi: 10.1073/pnas.0711038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama T, Penheiter AR, Penheiter SG, Chini EN, Thompson M, Warner DO, Jones KA. Differential effects of volatile anesthetics on M3 muscarinic receptor coupling to the Galphaq heterotrimeric G protein. Anesthesiology. 2006;105:313–24. doi: 10.1097/00000542-200608000-00014. [DOI] [PubMed] [Google Scholar]
- 25.Pittson S, Himmel AM, MacIver MB. Multiple synaptic and membrane sites of anesthetic action in the CA1 region of rat hippocampal slices. BMC Neurosci. 2004;5:52. doi: 10.1186/1471-2202-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell DB, Nash HA. Use of Drosophila mutants to distinguish among volatile general anesthetics. Proc. Natl. Acad. Sci USA. 1994;91:2135–9. doi: 10.1073/pnas.91.6.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamo S, Tomida J, Dodo K, Keyakidani D, Matakatsu H, Yamamoto D, Tanaka Y. Calreticulin Mediates Anesthetic Sensitivity in Drosophila melanogaster. Anesthesiology. 2003;99:867–75. doi: 10.1097/00000542-200310000-00019. [DOI] [PubMed] [Google Scholar]
- 28.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–21. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 29.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, Park HS, Kim JI, Seo JS, Yakhini Z, Laderman S, Bruhn L, Lee C. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–95. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 31.Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:19920–5. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–31. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths AJF, Gelbart WM, Miller JH, Lewontin RC. In: Modern Genetic Analysis. Tenney S, editor. New York: W.H. Freeman, Inc; 1999. pp. 235–70. [Google Scholar]
- 34.Alt FW, Kellems RE, Bertino JR, Schimke RT. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978;253:1357–70. [PubMed] [Google Scholar]
- 35.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–53. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 36.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien SJ, Gethmann RC. Segmental aneuploidy as a probe for structural genes in Drosophila: mitochondrial membrane enzymes. Genetics. 1973;75:155–67. doi: 10.1093/genetics/75.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franks NP, Honore E. The TREK K2P channels and their role in general anaesthesia and neuroprotection. Trends Pharmacol Sci. 2004;25:601–8. doi: 10.1016/j.tips.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Hawley RS, Gilliland WD. Sometimes the result is not the answer: the truths and the lies that come from using the complementation test. Genetics. 2006;174:5–15. doi: 10.1534/genetics.106.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouahchi K, Lindeman N, Lee C. Copy number variants and pharmacogenomics. Pharmacogenomics. 2006;7:25–9. doi: 10.2217/14622416.7.1.25. [DOI] [PubMed] [Google Scholar]
- 41.Drysdale R. FlyBase : a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 42.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–99. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]