Abstract
Aneuploidy, an incorrect chromosome number, is the leading cause of miscarriages and mental retardation in humans and is a hallmark of cancer. We examined the effects of aneuploidy on primary mouse cells by generating a series of cell lines that carry an extra copy of one of four mouse chromosomes. In all four trisomic lines proliferation was impaired and metabolic properties were altered. Immortalization, the acquisition of the ability to proliferate indefinitely, was also affected by the presence of an additional copy of certain chromosomes. Our data indicate that aneuploidy decreases not only organismal but also cellular fitness and elicits traits that are shared between different aneuploid cells.
Numerical alterations in an organism's karyotype, a condition known as aneuploidy, is associated with developmental abnormalities in all species examined to date. Studies in budding yeast (1), fission yeast (2, 3), Drosophila (4), maize (5), rice (6) and mice (7, 8) showed that aneuploidy interferes with organismal fitness and development. In humans as well, aneuploidy is detrimental, representing the major cause of mental retardation and miscarriages (9, 10). However, aneuploidy is also a characteristic of the disease of uncontrolled proliferation, cancer. This raises the question: If aneuploidy is so deleterious, why are most solid tumors aneuploid? We thus examined the consequences of aneuploidy on cell proliferation and physiology by generating four primary mouse cell lines that carry an additional chromosome. Aneuploidy was detrimental at the cellular level, causing a slowing of cell proliferation and changes in cellular metabolism. We speculate that tumor development requires the acquisition of aneuploidy-tolerating mutations and propose that the mechanisms that elicit the traits shared by aneuploid cells are ideal targets for cancer therapeutics.
Generation of mouse embryonic fibroblasts (MEFs) trisomic for chromosome 1, 13 16 or 19
To determine the effects of an additional chromosome on murine cell physiology we generated mouse embryonic fibroblast (MEF) lines that carried an additional chromosome (henceforth trisomic MEFs). We used a breeding scheme to obtain trisomic (Ts) embryos (Fig. S1) (10). Mice homozygous for a Robertsonian translocation (for example, a fusion between chromosomes 6 and 16; strain A) were crossed to a strain homozygous for a second Robertsonian translocation (for example, between chromosomes 16 and 17; strain B). From this cross, male offspring were selected that carried both Robertsonian translocations (compound heterozygotes) and mated to wild-type mice lacking any Robertsonian translocation. Between 7 to 40% of the resulting progeny [the exact percentage depended on the strain background, the stage of embryogenesis analyzed, and identity of the translocation chromosome (8)] were trisomic for the chromosome common to the two Robertsonian translocations due to a meiotic non-disjunction event in the male germline.
With the exception of mice trisomic for Chromosome (Chr) 19 (Ts19), of which a small percentage of embryos develop to term and survive for a short period of time, trisomic embryos die in utero. However, many of these embryos develop past embryonic day 10.5 allowing for the generation of MEF lines (7). We used mice carrying different combinations of Robertsonian translocations to generate embryos trisomic for Chromosome 1, 13, 16, or 19. We chose these four chromosomes because they cover a large portion of the size and coding spectrum of mouse chromosomes (Chr1: 197Mbp, 1228 genes; Chr13: 120Mbp, 843 genes; Chr16: 98Mbp, 678 genes; Chr19: 61Mbp, 734) (11).
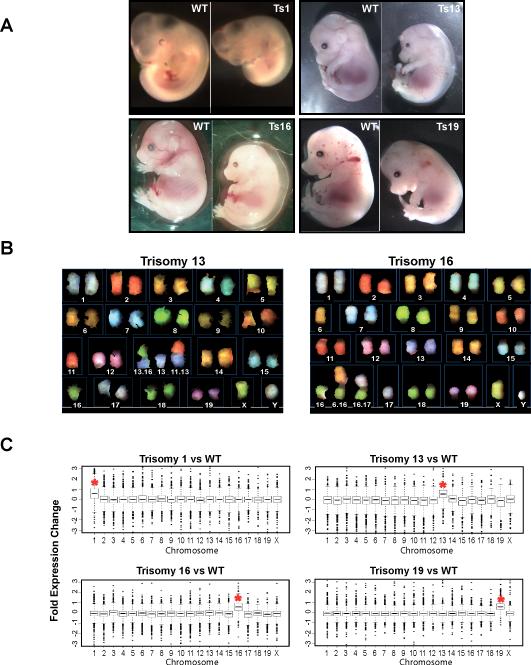
Initially, trisomic embryos were identified by their distinctive morphology. They developed more slowly than their euploid littermates and many exhibited nuchal edema and other developmental abnormalities (Fig. 1A) (7). To verify that the embryos were indeed trisomic for a particular chromosome, we counted the number of chromosome arms in preparations of spread metaphase chromosomes from early passage (≤2) MEF cultures generated from the trisomic embryos (12). We also used Spectral Karyotype Analysis (SKY), which identifies each chromosome by a unique fluorescent color, to confirm that the cell lines generated were trisomic for a single, specific chromosome, and that other changes in chromosomal composition had not occurred, at least during the early stages of cell culture (Fig. 1B).
Fig. 1. Generation of trisomic embryos and mouse embryonic fibroblast (MEF) cell lines.
(A) Trisomic (Ts) embryos were recovered by timed matings. The trisomy 1 (Ts1) embryo was recovered at 10.5 days postcoitum (dpc). Ts13, Ts16 and Ts19 embryos were recovered at 14.5 or 15.5dpc. In all instances Ts embryos were identified by their developmental abnormalities and reduced size (7).
(B) Examples of Spectral karyotype (SKY) analysis of metaphase spreads prepared from early passage (≤p3) Chr13 and Chr16 trisomic MEFs. Chromosomes and Robertsonian translocations are identified.
(C) Gene expression pattern of aneuploid cell lines and lines from euploid littermate controls (12). Transcripts were binned by chromosome and the average gene expression/total chromosome is shown. The asterisk indicates the identity of the trisomic chromosome. The increase in gene expression was highly significant (p≤ 1x10−74, all trisomies, Student's t-Test).
Gene expression from the additional chromosomes is proportional to gene copy number
The presence and consequence of an additional chromosome in MEFs was further determined by a genome-wide transcript expression analysis. Total RNA was isolated from passage 2 cultures (12). Overall, gene expression changed according to gene copy number, with expression of genes present on Chr16 increasing by, on average, 1.52-fold in cells trisomic for this chromosome (Fig. 1C, Table S1; n=3 independent cell lines). The expression of genes on Chr13 increased, on average, 1.46-fold in Ts13 cell lines (Fig. 1C, Table S1; n=4 independent cell lines), the expression of genes on Chr1 increased an average of 1.55-fold in a Ts1 cell line (Fig. 1C, Table S1; n=1 cell line), and the expression of genes on Chr19 increased, on average, 1.51-fold in a single Ts19 cell line (Fig. 1C, Table S1; p≤10−74 for all cell lines, Student's t-Test). The approximate 1.5-fold increase in gene expression of genes present on the trisomic chromosome indicates that the genes present on the additional chromosome are transcribed, which is consistent with expression profiles obtained from patients with Down Syndrome (13). Comparison of the expression patterns of the different trisomic cell lines did not reveal genes that showed increased or decreased expression in all four different trisomic MEFs (Tables S1 and S2) suggesting that a gene expression pattern common to all aneuploid cell lines does not exist. We conclude that the majority of the genes present on the additional chromosome are expressed. Thus, dosage compensation at the transcriptional level does not occur in these cells.
Proliferation defects of aneuploid cells
We next examined the ability of trisomic MEFs to proliferate in culture. We used four independent cell lines trisomic for either Chr13, Chr16 and Chr19 and three independent cell lines trisomic for Chr1. These trisomic cell lines were compared to cell lines derived from euploid littermates, which, due to the breeding scheme, carried a single Robertsonian translocation.
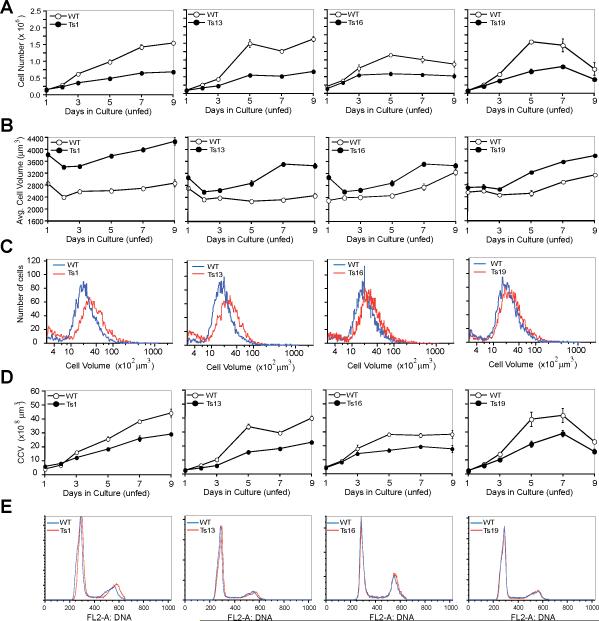
MEFs, kept for a short time in culture, (we used MEFs at passage 3 to ensure that both the euploid and trisomic cells were karyotypically consistent with those of the embryo) were seeded on multiple plates and the number of cells present in the wells was counted for 7 or 9 days. In these accumulation assays, the medium was changed every other day (Figs. S2A and S3A) or cells were kept in the same medium for the entire experiment (Fig. 2A; Fig. S3B) (12). In both fed and unfed euploid cultures, cell number increased during the first 5 to 7 days and then remained constant thereafter. The trisomic cell lines behaved similarly to the wild-type cells over the first two days of the experiment. However, after the initial two days, proliferation of the trisomic cells decreased compared to that of euploid controls. The decrease in proliferative capacity was severe in Ts1 and Ts13 cells, but less dramatic and more variable though still statistically significant in Ts16 and Ts19 cells (Figs. 2A and 3D; Figs. S2A, S3A, S3B, and S4A). Thus, the presence of an additional chromosome inhibits cell proliferation in culture. This reduced proliferation appears to be more pronounced as the size of the additional chromosome increases.
Fig. 2. Proliferation defects in trisomic MEFs.
Wild type (open circles) and Ts cells (closed circles) were plated and counted daily for cell number increase (12). Error bars are +/− SD. The data for each column come from the same cell line.
(A) Growth of early passage (p3) trisomic cells under “unfed” (medium was not changed) conditions.
(B, C) Average cell volume of cells under growth conditions where the medium was not changed (unfed; B) and the distribution of cell volumes in the culture at day 5 of the accumulation assay (C). Note that the low amounts of small size particles in (C) indicate that cells are not undergoing lysis.
(D) Analysis of the cumulative cell volume (CCV) (number of cells x average cell volume) during the proliferation assay in A. Error bars are +/− SD.
(E) DNA content analysis of asynchronous wild-type and trisomic cells
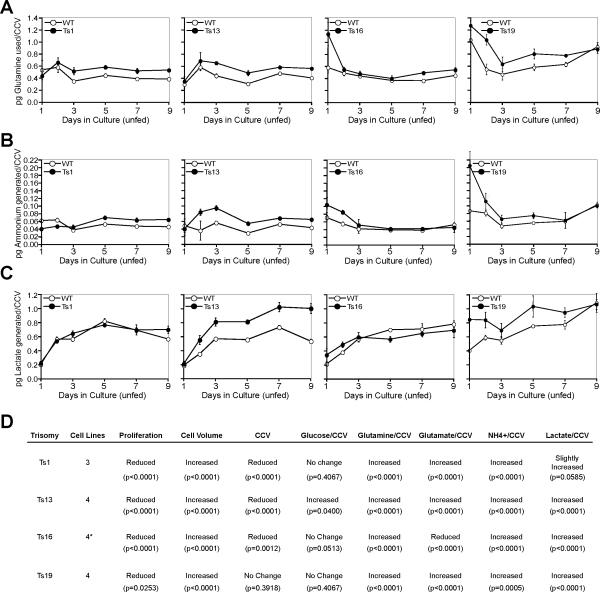
Fig. 3. Cellular metabolism in trisomic MEFs.
(A – C) Tissue culture supernatants of proliferation experiments were subjected to metabolic analyses (12) and the amount of glutamine used (A) and ammonium (B) and lactate (C) generated per cumulative cell volume was determined at the indicated times. The data for each column come from the same cell line. Error bars are +/− SD. (D) The table summarizes the changes in cell proliferation, cell and cumulative cell volume a well as glucose and glutamine uptake and production of glutamate, ammonium and lactate. These p-values are shown for measurements of the proliferation assays. P-values were determined through a 2-way nested ANOVA with standard statistical packages for values obtained for days 3, 5 and 7 (Table S4). Values below p=0.05 were interpreted to mean that the values obtained were either significantly increased or reduced. P-values > 0.06 were interpreted to mean no difference between Ts line and wild-type. The asterisk denotes that cell number, cell volume, and cumulative cell volume was determined for 4 trisomy 16 cell lines, whereas the metabolic analyses were performed with three Ts16 lines.
The trisomic cell lines analyzed carried two Robertsonian translocations (Fig. 1B; Fig S1). To test the possibility that the Robertsonian translocations rather than the presence of an extra chromosome caused the proliferation defect, we analyzed euploid cell lines that harbored 0, 1, or 2 Robertsonian translocations. The presence of Robertsonian chromosomes did not affect cell proliferation regardless of whether the medium was changed or not (Fig. S5A). We conclude that the presence of an additional chromosome, not the chromosomal fusion, reduced cell proliferation.
Cell volume is increased in trisomic cells
During the establishment of MEF cultures we observed that the average size of trisomic cells was increased (12). This was not caused by an increase in the breadth of the size distribution, but was due to a shift in the distribution of the cell size toward a larger average size (Figs. 2B, 2C, and 3D; Figs. S2B, S2C, S3C, S3D, and S4B). The increase in cell volume was readily detectable by passage 3, the beginning of the proliferation experiment, in all trisomic lines, and persisted and sometimes even increased during the course of the experiment (Fig. 2B; Figs. S2B and S3C). As observed in the cell proliferation analysis, the increase in cell volume was more pronounced in cells carrying an extra copy of the larger chromosomes (Ts1 and Ts13 cells) and was less dramatic and variable but nevertheless statistically significant in cells carrying an extra copy of the smaller chromosomes 16 or 19 (Figs. 2C and 3D; Figs. S2B, S3C, and S4B). Analysis of euploid cells with 0, 1 or 2 Robertsonian translocations showed that this increase in cell volume was not due to the presence of fusion chromosomes but was due to the presence of the additional chromosome (Fig. S5B).
Because proliferation of primary MEFs is inhibited when the cells come into close contact with one another (14), the increased size of trisomic MEFs raised the possibility that the lower cell number observed in these cultures resulted from earlier contact inhibition rather than from a decreased ability to proliferate. To test this possibility, we calculated the cumulative cell volume (CCV) by multiplying the cell number by the average cell volume. If a larger cell size and thus earlier contact inhibition was responsible for the decreased cell number in trisomic cultures, the CCV should be the same in wild-type and trisomic cell lines. This was not the case. Trisomic cell lines produced less CCV than euploid controls. This defect was pronounced in cell cultures trisomic for Chr 1 or Chr13, more subtle and variable but nevertheless statistically significant in cell cultures trisomic for Chr16, and not detectable in cells trisomic for Chr 19 (Figs. 2D and 3D; Fig. S2D, S3E, and S4C). Our results indicate that the cumulative cell volume of cultures trisomic for chromosome 1, 13 or 16 is less than that of euploid controls. Analysis of euploid cells with 0, 1 or 2 Robertsonian translocations showed that this decrease in CCV was not due to the presence of fusion chromosomes but was due to the presence of the additional chromosome (Fig. S5C). We conclude that the reduced cell accumulation in trisomic cultures is due to proliferation defects. These defects are more severe in cells carrying an extra copy of larger chromosomes.
To examine whether the cell proliferation defect observed in trisomic cells arose from delays in a specific cell cycle stage, we compared the DNA content between asynchronously growing trisomic and euploid MEFs (12). The flow cytometric profile was similar in trisomic cells and euploid controls examined, (Fig. 2E; Fig. S3F). Neither cell lysis, as judged by the presence of large amounts of cellular debris in the cell volume determination, (Fig. 2C; Figs. S2C and S3D), nor senescence-associated β-galactosidase activity (Fig. S6) was increased in trisomic cell lines. Cell proliferation was also impaired in primary cells from humans with Down Syndrome (Trisomy 21) but a specific cell cycle defect was not observed either (15, 16). It is possible that progression through the cell cycle is slowed overall in trisomic mouse and human cells. However, we favor the idea that specific cell cycle defects exist, but are too subtle to be detected in asynchronously growing cells. Although our results did not reveal a specific cell cycle defect, they clearly show that aneuploidy hampers rather than promotes cell proliferation. Thus, during tumorigenesis the aneuploid state of a cell per se would impair rather than accelerate the process.
Altered metabolic properties of aneuploid cells
Many metabolic pathways are altered in tumor cells (17). The trisomic MEFs we generated allowed us to examine whether the aneuploid state could contribute to these metabolic changes (12). We first analyzed the use of glucose, a carbon source of tissue culture cells, and of glutamine, another carbon source as well as primary nitrogen source. To measure the amount of glucose and glutamine used by trisomic MEFs, we grew cells over nine days without changing the medium and then measured the amount of glucose and glutamine per CCV remaining in the medium (12). Glucose consumption was slightly increased in cells trisomic for chromosome 13 but was not affected in other trisomic cell lines (Fig. 3D; Figs. S7A and S7C). In contrast, glutamine consumption was increased in all trisomic cells lines. It was higher in cells trisomic for chromosome 1 and 13 and slightly, though statistically significantly, increased in cells carrying an extra copy of chromosome 16 or 19 (Fig. 3A and 3D; Figs. S7C and S8A).
We also examined the production of the metabolites ammonium, glutamate and lactate per CCV. Ammonium and glutamate are produced by the degradation of glutamine in tissue culture cells. Additionally, ammonium is produced as a result of the breakdown of amino acids due to higher rates of autophagy or perturbations in amino acid metabolism. We observed an increase in the production of ammonium in all trisomic cell lines (Figs. 3B and D; Figs. S7C and S8B). Glutamate production was increased in Ts1, 13 and 19 cells but reduced in Ts16 cells (Fig. 3D; Figs. S7B, S7C, and S8C), indicating that production of not all metabolites is increased in all trisomic cells. Lactate, is produced when pyruvate accumulates in cells as a result of an increase in glycolysis, defects in mitochondrial function, disruption of pyruvate import into the mitochondria, or increased activity of lactate dehydrogenase. Lactate production was slightly, though statistically significantly, increased in Ts13, Ts16, and Ts19 cell cultures and was approaching significance in Ts1 cultures (Figs. 3C and 3D; Fig. S8D). The changes in metabolism observed in aneuploid cells were specific to the presence of an additional chromosome and not to the Robertsonian fusion event. Analysis of euploid cells carrying 0, 1 or 2 Robertsonian translocations showed that the changes in metabolism were not due to the presence of fusion chromosomes in cells, as all the characteristics were indistinguishable between these cell lines (Fig. S5).
We conclude that primary aneuploid cells display alterations in glutamine use and the production of ammonium and lactate and speculate that these phenotypes may reflect a general alteration in energy production in the aneuploid cells. An increase in lactate production was first described nearly 100 years ago by Otto Warburg in Flexner-Jobling's rat carcinomas (18). It is now clear that many aspects of cellular metabolism are altered in tumor cells. Our results raise the possibility that one (but by no means the only) cause of the metabolic alterations observed in tumor cells is their aneuploid state.
Effects of trisomy on immortalization
Aneuploidy is a characteristic of many tumors and has been proposed to play a key role in promoting tumorigenesis (19). Consistent with this idea is the observation that the occurrence of acute lymphoblastic leukemia and acute megakaryoblastic leukemia is greatly increased in Down Syndrome patients (20). However, the incidence of many solid tumors in these individuals is only half of that in the normal population, raising the possibility that aneuploidy also restricts the formation of certain tumors (21, 22). Studies of mouse mutants that result in an increased frequency of aneuploidy also revealed mixed results. A mouse model in which chromosome mis-segregation was induced by inactivation of a component of the chromosome segregation machinery, CENP-E, indicated that aneuploidy acts in an oncogenic manner in some cell types but inhibits tumorigenesis in others (23). Random aneuploidy caused by transient overexpression of Mad2 in the mouse appears to initiate tumor formation only in certain cell types (24). A mouse model expressing a hypomorphic allele of the spindle assembly checkpoint protein, BubR1, displays progressive aneuploidy and exhibits an accelerated aging phenotype but without increased incidence of tumorigenesis (25). Finally, segmental trisomy reduces tumor number in the colon cancer APCMin mouse model (26). Thus, it is unclear whether aneuploidy inhibits or promotes tumorigenesis or does both. The primary trisomic cell lines we generated allowed us to begin to address this question and test the possibility that the identity of the additional chromosome determines whether aneuploidy promotes or inhibits tumor formation. We did this by examining the effects of specific additional chromosomes on immortalization induced by serial passage in vitro. Although it is clear that in vitro immortalization does not recapitulate all aspects of tumorigenesis, it is, in most cases, accompanied by two important characteristics of many solid tumors: (1) loss of p53 tumor suppressor pathway function and (2) aneuploidy (27).
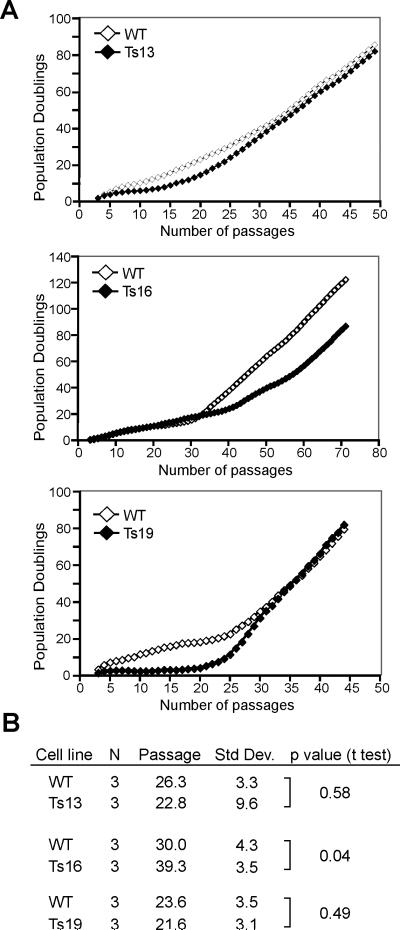
MEFs can be serially passaged and after a period of reduced proliferation, these cells will spontaneously overcome this period of reduced growth (27). This process of serial passaging until the culture fails, or until a subpopulation acquires the ability to grow indefinitely (which is usually caused by loss of p53 function) is referred to as a 3T3 protocol (27). We cultured four Ts16, three Ts13 and Ts19 cell lines and one Ts1 line in parallel with littermate euploid controls through serial passages to analyze the number of passages required for trisomic MEFs to generate immortalized cells (12). To determine the passage at which immortalization occurred, we fit the population doublings for each culture to a double-linear fit model (12); the point at which the two lines intersect represents the passage by which immortalization had occurred. Immortalization was delayed in cell lines trisomic for Chr16. One line failed to immortalize and 3 lines showed a significant delay in the process as judged by passage number (Figs. 4A and 4B). In the trisomic cell lines that spontaneously immortalized, immortalization required, on average, 39 (+/− 4, SD) passages compared to 30 (+/− 4, SD) passages in matched euploid cultures (Figs. 4A and 4B). Thus, the presence of an extra copy of Chr16 hampers spontaneous immortalization. The one cell line trisomic for Chr1 failed to immortalize (Fig. S9), raising the possibility that an extra copy of Chr1 also antagonizes immortalization.
Fig. 4. Rates of spontaneous immortalization of primary aneuploid MEFs.
(A) Cells were serially passed in a 3T3 immortalization assay (12) and the number of population doublings are shown as a function of number of passages.
(B) The mean number of passages (passage) until immortalization was calculated as described (12). The standard deviations are shown in the column to the right. P-values are given in the last column. Note that the average number of passages until immortalization for the Ts16 cell lines does not include the Ts16 cell line that failed to immortalize. Note that for Ts19 two of the three immortalization assays were performed in independent experiments with cells obtained from the same embryo.
In contrast to cell cultures trisomic for Chromosome 16 or 1, the number of passages necessary to achieve spontaneous immortalization was similar in control and Ts13 cell lines with 26 (+/− 3, SD) passages in Ts13 compared to 23 (+/− 9, SD) passages in euploid cultures (Figs. 4A and 4B). The number of passages was also indistinguishable from the euploid controls for the Ts19 cell lines analyzed. Ts19 cell lines required 22 (+/− 3, SD) passages, while the euploid counterparts immortalized at 24 (+/− 4, SD) passages (Figs. 4A and 4B). These results indicate that although proliferation is slower in cells trisomic for Chromosome 13 or 19, immortalization occurs after a similar number of passages compared to wild-type. In fact, when the time of immortalization is described as a function of population doublings, Ts13 cells immortalize earlier than wild-type controls. On average only 12 (+/− 5, SD) doublings were necessary to immortalize Ts13 cell lines compared to 27 (+/− 8, SD) doublings in euploid controls (p≤ 0.05, Student's t-Test). Our results indicate that aneuploidy affects the rate of immortalization in MEFs and this effect depends on the identity of the extra chromosome. These findings imply that the immortalization barrier caused by the proliferation defect due to aneuploidy (with perhaps the exception of that in Ts1) can eventually be overcome. The difference in the efficiency with which various trisomic lines overcome the proliferation barrier indicates that the underlying mechanism might differ in the individual aneuploid cell lines. However, once immortalized, trisomic cells do not consistently differ from immortalized euploid cells in their chromosome number. All immortalized trisomic and euploid cell lines were near tetraploid (Table S3), suggesting that once immortalization occurred the degree of aneuploidy does not differ between euploid and trisomic cell lines.
Discussion
Our analysis of MEFs each containing a different additional chromosome revealed that in addition to chromosome specific traits, the four trisomic MEFs share characteristics such as a cell proliferation delay and an altered metabolism. MEFs carrying hypomorphic mutations in the spindle checkpoint component BubR1 frequently carry one or two extra chromosomes and their proliferation is also impaired (25) indicating that at least the defect in cell proliferation is shared among different types of aneuploidies in the mouse. Primary foreskin fibroblasts of individuals with Down Syndrome also exhibited a proliferation delay and an increase in cell volume (16, 28), suggesting that aneuploidy may also hamper proliferation in human cells. In budding yeast, the proliferation defects of aneuploid cells is caused by imbalances in intracellular protein composition due to expression of genes on the additional chromosome (1). Because the genes present on the additional chromosome are also transcribed in the trisomic MEFs and thus are likely also translated, the same could be true in mouse cells.
Most solid tumors are aneuploid. Our results and that of others indicate that aneuploidy suppresses rather than enhances tumorigenesis. We found that the presence of an extra chromosome hampered cell proliferation. There is also evidence to suggest that at least human Ts21 cells do not proliferate as well as euploid cells either (28). Also, the percentage of cells in S phase in solid tumors, which are mostly aneuploid, varies between 2 − 8%, whereas a normal renewing epithelium such as the intestine exhibits a DNA replication index of approximately 16% (29). Furthermore, individuals carrying an extra copy of chromosome 21 have a 50% lower probability of developing solid tumors than individuals with the correct chromosome number (21, 22). Segmental trisomy in the mouse has been shown to reduce incidence of neoplasia in the sensitized APCMin genetic background (26). Additionally, mouse models in which low-level aneuploidy was induced through interference with the chromosome segregation machinery prevented tumor formation in many tissues and caused tumor formation only relatively late in others (23-25).
A few findings, however, argue for a cancer-promoting role of aneuploidy. Loss of heterozygosity, which can arise from chromosome loss or aneuploidy, is detected in atypical ductal hyperplasias, which can be precursors of breast cancer (30) and in small (2mm diameter) adenomas, which are thought to represent early stage colon cancers (31-33). Finally, even though tumors form late in mice carrying a low-level aneuploidy-inducing mutation, they do occur with an increased frequency in some tissues.
How can we reconcile these results? We propose that aneuploidy per se is a barrier towards tumorigenesis, but the very events that cause aneuploid cells to proliferate slowly, the cellular imbalances caused by aneuploidy and the stresses it is associated with, might promote tumorigenesis in a small fraction of aneuploid cells (1). The stresses associated with cellular imbalances could lead to an increase in mutation rate, in gene amplification, and/or in genomic instability. Precedents exist for all these scenarios in bacteria, yeast and tissue culture cells (34-38). Aneuploidy-tolerating and proliferation-promoting mutations could then eventually lead to the selection of tumor cells with high proliferative capacity. Furthermore, aneuploidy would also shield the evolving tumor from lethal mutations. Thus, in a rather counter-intuitive manner as has been suggested for chemical carcinogens (39, 40), the proliferation inhibiting imbalances of aneuploidy may, under some circumstances, promote tumorigenesis.
Irrespective of whether or not aneuploidy can promote tumorigenesis, it is clear that aneuploidy causes a proliferative disadvantage in budding yeast (1), S. pombe (2, 3), primary mouse cells (this study) and human cells (28). This property of aneuploidy functions as a barrier towards transformation and this disadvantage must be overcome during tumorigenesis. Identifying mutations that can overcome the proliferation-inhibiting effects of aneuploidy, could provide new pathways to exploit in cancer treatment. Given that most solid tumors are aneuploid, the cellular consequences of aneuploidy could also provide novel targets in cancer therapy. Characterizing the phenotypes associated with aneuploidy in human cells, as well as identifying small molecules that specifically target aneuploid cells may provide new avenues in the treatment of cancer. (41)
Supplementary Material
References
- 1.Torres EM, et al. Science. 2007 Aug 17;317:916. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 2.Niwa O, Tange Y, Kurabayashi A. Yeast. 2006 Oct 15;23:937. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- 3.Niwa O, Yanagida M. Current Genetics. 1985;9:463. doi: 10.1007/BF00420224. [DOI] [PubMed] [Google Scholar]
- 4.Lindsley DL, et al. Genetics. 1972 May;71:157. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClintock B. Genetics. 1929 Mar;14:180. doi: 10.1093/genetics/14.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh K, Multani DS, Khush GS. Genetics. 1996 May;143:517. doi: 10.1093/genetics/143.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyban AP, Baranov VS. Cytogenetics of mammalian embryonic development. Clarendon Press; Oxford University Press; Oxford, New York: 1987. p. x.p. 362. Oxford science publications. [Google Scholar]
- 8.Gropp A, Winking H, Herbst EW, Claussen CP. J Exp Zool. 1983 Nov;228:253. doi: 10.1002/jez.1402280210. [DOI] [PubMed] [Google Scholar]
- 9.Pai GS, Lewandowski RC, Borgaonkar DS. Handbook of chromosomal syndromes. J. Wiley; New York: 2003. p. xiii.p. 361. [Google Scholar]
- 10.Gropp A, Kolbus U, Giers D. Cytogenet Cell Genet. 1975;14:42. doi: 10.1159/000130318. [DOI] [PubMed] [Google Scholar]
- 11.Chromosome size and coding gene statistics obtained from NCBI m37 mouse assembly. http://www.ensembl.org/Mus_musculus/index.html.
- 12.See Supporting Online Material.
- 13.Mao R, Zielke CL, Zielke HR, Pevsner J. Genomics. 2003 May;81:457. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 14.Abercrombie M. Exp Cell Res. 1961;(Suppl 8):188. doi: 10.1016/0014-4827(61)90348-2. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen K, Marcus M, Gropp A. Hereditas. 1985;102:77. doi: 10.1111/j.1601-5223.1985.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 16.Rosner M, et al. J Neural Transm Suppl. 2003:51. doi: 10.1007/978-3-7091-6721-2_4. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Febs J. 2007 Mar;274:1393. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O, Posener K, Negelein E. Biochem Z. 1924;152:309. [Google Scholar]
- 19.Boveri T. Verhandlungen der physikalisch-medizinischen Gesellschaft zu Würzburg. Neu Folge. 1902;35:67. [Google Scholar]
- 20.Satge D, et al. Am J Med Genet. 1998 Jul 7;78:207. [PubMed] [Google Scholar]
- 21.Hasle H, Clemmensen IH, Mikkelsen M. Lancet. 2000 Jan 15;355:165. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 22.Satge D, Sasco AJ, Lacour B. Int J Cancer. 2003 Aug 20;106:297. doi: 10.1002/ijc.11212. [DOI] [PubMed] [Google Scholar]
- 23.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Cancer Cell. 2007 Jan;11:25. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Sotillo R, et al. Cancer Cell. 2007 Jan;11:9. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker DJ, et al. Nat Genet. 2004 Jul;36:744. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 26.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Nature. 2008 Jan 3;451:73. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 27.Todaro GJ, Green H. J Cell Biol. 1963 May;17:299. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal DJ, McCoy EE. J Cell Physiol. 1974 Feb;83:85. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- 29.Tannock I, Hill RP. The Basic science of oncology. Pergamon Press; New York: 1987. p. viii.p. 398. [Google Scholar]
- 30.Larson PS, et al. J Pathol. 2006 Jul;209:307. doi: 10.1002/path.1973. [DOI] [PubMed] [Google Scholar]
- 31.Bomme L, et al. Cancer Genet Cytogenet. 1998 Oct 1;106:66. doi: 10.1016/s0165-4608(98)00047-8. [DOI] [PubMed] [Google Scholar]
- 32.Bomme L, et al. Int J Cancer. 2001 Jun 15;92:816. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- 33.Shih IM, et al. Proc Natl Acad Sci U S A. 2001 Feb 27;98:2640. doi: 10.1073/pnas.051629398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edlund T, Normark S. Nature. 1981 Jul 16;292:269. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- 35.Ponder RG, Fonville NC, Rosenberg SM. Mol Cell. 2005 Sep 16;19:791. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 36.Selmecki A, Forche A, Berman J. Science. 2006 Jul 21;313:367. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung HM, Yeamans G, Ross CA, Yasbin RE. J Bacteriol. 2003 Apr;185:2153. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mihaylova VT, et al. Mol Cell Biol. 2003 May;23:3265. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farber E. Biochem Pharmacol. 1990 Jun 15;39:1837. doi: 10.1016/0006-2952(90)90599-g. [DOI] [PubMed] [Google Scholar]
- 40.Haddow A. Acta Unio Intern Contra Cancrum. 1938;3:342. [Google Scholar]
- 41.We thank E. Vazile in the Koch Institute Microscopy and Imaging Facility for help with SKY analysis, M. Luo in the MIT BioMicro Center for help with microarray analysis, L. Chan for assistance with the double linear fit models for immortalization kinetics, A. Regev and M. Guttman for advice and assistance with statistical analysis, and M. Dunham, M. Hemann, J. Lees, F. Solomon and members of the Amon lab for critical reading of this manuscript. Supported by the Howard Hughes Medical Institute , a David Koch Research Award, a grant from the Curt W. and Kathy Marble Cancer Research Fund and a David Koch Graduate Fellowship (VRP). All microarray data are provided in final processed form in Table S1 and as raw data through the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) under accession number GSE12501.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.