Abstract
AIM: To explore the effects of the nucleoside analogues β-L-D4A and β-LPA on hepatitis B virus (HBV) promoters.
METHODS: Four HBV promoters were amplified by polymerase chain reaction (PCR) and subcloned into the expression vector pEGFP-1. The four recombinants controlled by HBV promoters were confirmed by restriction analysis and sequencing. Human hepatoma HepG2 cells transfected with the recombinant plasmids were treated with various concentrations of β-L-D4A and β-LPA. Then, enhanced green fluorescent protein (EGFP)-positive cells were detected by fluorescence microscopy and using a fluorescence activated cell sorter (FACS).
RESULTS: Four HBV promoters were separately obtained and successfully cloned into pEGFP-1. Expression of EGFP under the control of the surface promoter (Sp) and the X promoter (Xp) was inhibited by β-L-D4A in a dose-dependent manner, while expression of EGFP under the control of the core promoter (Cp) and Xp was inhibited by β-LPA in a dose-dependent manner.
CONCLUSION: The two novel nucleoside analogues investigated here can inhibit the activities of HBV promoters in a dose-dependent manner. These findings may explain the mechanisms of action by which these two novel compounds inhibit HBV DNA replication.
Keywords: Hepatitis B virus, Nucleoside analogue, Hepatitis B virus promoter, Enhanced green fluorescent protein, Fluorescence activated cell sorter
INTRODUCTION
Hepatitis B virus (HBV) is the leading cause of chronic hepatitis in the world[1]. According to the World Health Organization, over 350-million people (5% of the world population) are chronically infected with HBV. Although safe and effective vaccination for HBV is available in developing countries[2–4], there is still no effective treatment for the millions of chronically infected individuals[5]. Consequently, long-term infection with chronic HBV could lead to cirrhosis and hepatocellular carcinoma[6,7]. In light of these facts, it is evident the discovery and development of novel antiviral agents for the treatment of HBV is an extremely important undertaking.
The number of formally approved anti-HBV drugs is limited. The necessity for new compounds acting on a variety of molecular targets within the viral replicative cycle is crucial. Thus, it remains important to discover new antiviral drugs, and to investigate new potential targets such as uncoating, transcription, packaging, excretion, or synthesis of cccDNA[8,9].
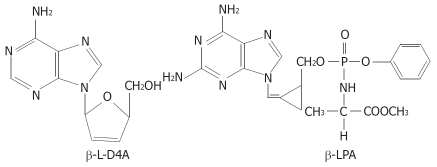
In our previous work[10–13], we synthesized two novel nucleoside analogues, β-L-D4A and β-LPA (Figure 1), and studied their inhibitory actions against HBV as well as their cytotoxicities. β-L-D4A and β-LPA possess potent inhibitory effects on the replication of HBV (EC50 = 0.2 and 0.01 μmol/L, respectively) with little cytotoxicity (IC50 = 200 and 50 μmol/L, respectively) or mitochondrial toxicity. Their TI values are 1000 and 5000, respectively. Therefore, they are expected to be developed as new clinical anti-HBV drugs. Our previous work also showed these two compounds possessed significant anti-HBV effects at the transcription level by inhibiting the production of HBV RNA. Therefore, we supposed the two compounds might act by inhibiting the activities of HBV promoters.
Figure 1.
Structures of β-L-D4A and β-LPA.
Complementary DNA for the Aequorea victoria green fluorescent protein (GFP) produces a fluorescent product when expressed in prokaryotic or eukaryotic cells. Because exogenous substrates and cofactors are not required for this fluorescence, GFP expression can be used to monitor gene expression and protein localization in living organisms[14–16]. pEGFP-1 is a promoterless EGFP vector, which can be used to monitor transcription from different promoters and promoter/enhancer combinations inserted into the Multiple Cloning Site located upstream of the EGFP coding sequence[17,18]. Hence, we chose pEGFP-1 as an expression vector to monitor the activities of HBV promoters. In this study, we explored the effects of our two novel nucleoside analogues on HBV promoters to uncover the mechanism underlying their anti-HBV effects.
MATERIALS AND METHODS
Materials
β-L-D4A and β-LPA were synthesized by ourselves with the help of the Pharmaceutic College of Wuhan University, and identified by infrared, mass spectra. and nuclear-magnetic resonance. Lamivudine was provided by Professor Cheng YC (School of Medicine, Yale University, New Haven, CT, USA). These compounds were dissolved in phosphate-buffered saline (PBS). The expression vector pEGFP-1 was purchased from BD ClonTech. Plasmid p3.6II was a kind gift from Prof. Wang Yuan (Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences). E. coli (Dh5α) and human hepatoma HepG2 cells were preserved by our institute. Fetal bovine serum (FBS) and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Hyclone Corp. All other reagents used were of analytical grade.
Polymerase chain reaction (PCR)
The desired four fragments were HBV preS gene promoter (preSp), Sp, Cp (including enhancer II) and Xp (including enhancerI)[19–23]. PCR was employed to amplify the four fragments from p3.6II, a plasmid containing the HBV complete genome (adr subtype). Specific primers were designed by us and synthesized at Sangong Company (Shanghai, China). The primers used are listed in Table 1. The lower-case nucleotides indicate the recognition sites for restriction endonucleases Acc65 I and Age I (Fermentas, USA). PCR products were separated by agarose gel electrophoresis. Fragments of interest were withdrawn and directly ligated into pGEM-T vector (Promega, USA). Positive clones were then screened by virtue of a blue/white screening system and sequenced after small-scale extraction. The four positive plasmids were cleaved by Acc65Iand AgeI, and fragments containing the HBV promoters were purified.
Table 1.
Primer sequences used for PCR
| Target | Primer sequences | Product size (bp) |
| preSp | F5’-GACAAAggtaccAAACCATATTATCC-3’ | 229 |
| R5’-GAGGAccggtAACAGAAAGATTCGT-3’ | ||
| Sp | F5’-GATTGgtaccTCAACCCCAACAAG-3’ | 458 |
| R5’-GAAAAAccggtCCTGTAACACGAG-3’ | ||
| Cp | F5’-CCACCggtaccTGCCCAAGGTCTTA-3’ | 302 |
| R5’-TCCAAAccggtTATACGGGTCAATG-3’ | ||
| Xp | F5’-GTATggtaccGAATTGTGGGTCTTTTG-3’ | 445 |
| R5’-ACGTAAACAccggtCGTCCCGCGC-3’ |
Construction of EGFP expression vectors controlled by HBV promoters
The expression vector pEGFP-1 was digested by Acc65Iand AgeIand vector fragments were collected. Then, the fragments containing HBV promoters were mixed with vector fragments at a ratio of 5 to 1, and these fragments were ligated using T4 DNA ligase (Promega, USA) at 16°C overnight. The ligated products were used to transform E. coli (Dh5α). Plasmids extracted from E. coli were analyzed by restriction enzymes and sequencing, and finally, the four promoter-controlled EGFP expression vectors pEGFP-Sp, pEGFP-preSp, pEGFP-Cp and pEGFP-Xp were produced.
Expression assays
HepG2 cells were incubated in DMEM medium with 10% (vol/vol) FBS at 37°C in a moist atmosphere containing 5% CO2/95% air. The cells were inoculated at a density of 3 × 105/mL per well in 24-well tissue culture plates. Twenty-four hours after plating, the four expression vectors were transfected into HepG2 cells (1 μg DNA per well) on different plates using Lipofectamine2000 (Invitrogen), according to manufacturer’s instructions (one well was left as a negative control group, which was needed as reference for FACS). After a 6 h incubation, the transfected cells were treated with various concentrations of β-L-D4A (0.08 μmol/L in 4 wells, 0.4 μmol/L in 4 wells, 2 μmol/L in 4 wells, 10 μmol/L in 4 wells). Four wells of cells were treated with lamivudine at 1 μmol/L as a comparative group, and 3 wells were not treated with any drug as a positive control group (that is, a blank control). The cells were grown in the presence of drugs for 42 h. EGFP-positive cells were detected using an inverted fluorescence microscope (ZEISS, Axiovert40) with an excitation wavelength of 488 nm. Then, all cells were digested with 0.25% trypsin; digestion was terminated by FBS and the cells were re-suspended in 500 μL of PBS per well before being subjected to flow cytometry (Becton Dickinson) analysis. The same experiments were performed with β-LPA.
Statistical analysis
Data are expressed as mean ± SD. Each experiment was repeated at least three times. Differences were considered statistically significant when P < 0.05, as analyzed by one way analysis of variance and Tukey’s post-hoc test. The analysis was conducted using the SPSS12.0.
RESULTS
Validation of cloning
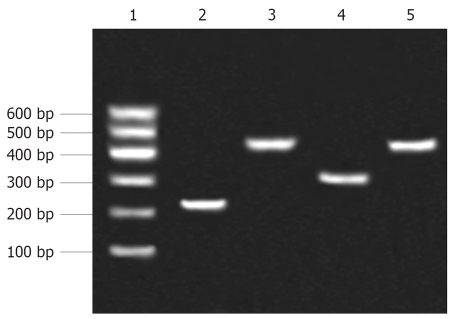
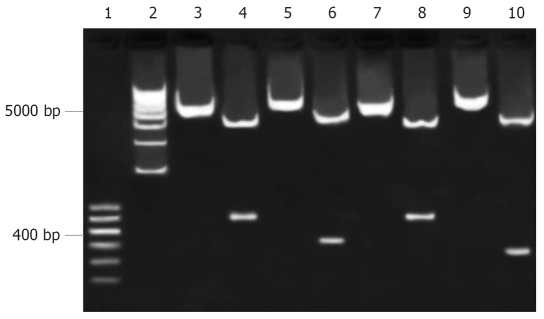
After ligation of promoter fragments into pGEM-T vector, PCR products (Figure 2) were sequenced by Bioasia (Shanghai, China). Using BLAST searches of Entrez (NCBI), the sequencing results were proved to be consistent with the template (p3.6II). The four recombinants pEGFP-Sp, pEGFP-preSp, pEGFP-Cp and pEGFP-Xp were identified by restriction enzymes (Figure 3).
Figure 2.
The HBV promoters obtained from PCR were separated by agarose gel electrophoresis. Lane 1: Marker; Lane 2: preSp; Lane 3: Sp; Lane 4: Cp; Lane 5: Xp.
Figure 3.
Restriction analysis of recombinant vectors. Lanes 1, 2: Marker; Lanes 3, 4: pEGFP-Sp before and after being digested by Acc65Iand AgeI; Lanes 5, 6: pEGFP-Cp before and after being digested by Acc65Iand AgeI; Lanes 7, 8: pEGFP-Xp before and after being digested by Acc65Iand AgeI; Lanes 9, 10: pEGFP-preSp before and after being digested by Acc65Iand AgeI.
Detection of EGFP-positive cells
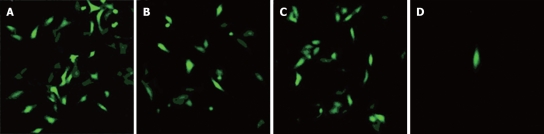
EGFP-positive cells could be seen by fluorescence microscopy among HepG2 cells transfected with pEGFP-Sp, pEGFP-Cp and pEGFP-Xp, but few could be seen among HepG2 cells transfected with pEGFP-preSp (Figure 4). These findings suggested HBV promoters could control the expression of EGFP.
Figure 4.
Forty-eight hours after transfection, EGFP positive cells were detected by fluorescence microscopy (× 100). A: HepG2 cells transfected with pEGFP-Sp; B: HepG2 cells transfected with pEGFP-Cp; C: HepG2 cells transfected with pEGFP-Xp; D: HepG2 cells transfected with pEGFP-preSp.
FACS analysis
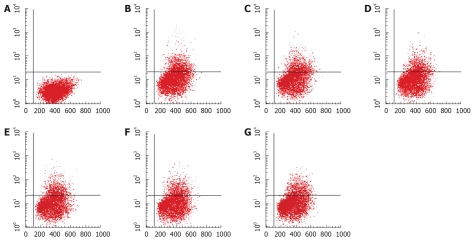
The percentage of EGFP-positive cells in each well was obtained by FACS. β-L-D4A inhibited the expressions of EGFP under the control of Sp (Figure 5, one representative picture was chosen from each group) and Xp in a dose-dependent manner, but had no effect on the expression of EGFP under the control of preSp and Cp; by contrast, β-LPA inhibited the expression of EGFP under the control of Cp and Xp in a dose-dependent manner, but had no effect on the expression of EGFP under the control of preSp and Sp; lamivudine could not inhibit the expression of EGFP under the control of any HBV promoter. The results are summarized in Tables 2 and 3.
Figure 5.
β-L-D4A inhibited the expression of EGFP in HepG2 cells transfected with pEGFP-Sp, as determined by FACS analysis. A: HepG2 cells not transfected with pEGFP-Sp, not treated with drug, the percentage of EGFP-positive cells was 0; B: HepG2 cells transfected with pEGFP-Sp, not treated with drug, the percentage was 21.42%; C: HepG2 cells transfected with pEGFP-Sp, treated with Lamivudine at 1 μmol/L, the percentage was 21.14%; D-G: HepG2 cells transfected with pEGFP-Sp, treated with β-L-D4A at various concentrations (0.08, 0.4, 2, 10 μmol/L), the percentages were 18.76%, 17.31%, 15.53%, 13.65% respectively.
Table 2.
Effect-dosage relationship of the inhibition of the expression of EGFP under the control of the Sp and Xp promoters by β-L-D4A
| Dosage (μmol/L) | n |
EGFP-positive cells (%) (mean ± SD) |
Inhibition rate (%) |
||
| Sp | Xp | Sp | Xp | ||
| Control | 3 | 21.26 ± 0.25 | 18.52 ± 1.25 | 0.0 | 0.0 |
| (Lamivudine) | 4 | 21.00 ± 0.43 | 18.37 ± 1.01 | 1.2 | 0.8 |
| 0.08 | 4 | 18.76 ± 0.41b | 16.47 ± 0.49b | 11.8 | 11.1 |
| 0.4 | 4 | 17.31 ± 0.41b | 15.68 ± 0.36b | 18.6 | 15.3 |
| 2 | 4 | 15.54 ± 0.48b | 13.54 ± 0.59b | 26.9 | 26.9 |
| 10 | 4 | 11.33 ± 0.32b | 10.84 ± 0.81b | 46.7 | 41.5 |
P < 0.01 vs Blank control. EGFP: Enhanced green fluorescent protein.
Table 3.
Effect-dosage relationship of the inhibition of the expression of EGFP under the control of the Cp and Xp promoters by β-LPA
| Dosage (μmol/L) | n |
EGFP-positive cells (%) (mean ± SD) |
Inhibition rate (%) |
||
| Cp | Xp | Cp | Xp | ||
| Control | 3 | 13.99 ± 0.29 | 18.58 ± 0.39 | 0.0 | 0.0 |
| (Lamivudine) | 4 | 13.80 ± 0.63 | 18.46 ± 0.49 | 1.3 | 0.6 |
| 0.002 | 4 | 12.98 ± 0.16b | 17.54 ± 0.31b | 7.2 | 5.6 |
| 0.01 | 4 | 11.96 ± 0.75b | 16.18 ± 0.21b | 14.5 | 12.9 |
| 0.05 | 4 | 10.88 ± 0.43b | 14.63 ± 0.37b | 22.2 | 21.3 |
| 1 | 4 | 8.79 ± 0.56b | 11.94 ± 1.37b | 37.7 | 35.7 |
P < 0.01 vs Blank control. EGFP: Enhanced green fluorescent protein.
DISCUSSION
HBV infection remains a major public health problem worldwide. Antiviral treatment of chronic Hepatitis B currently relies on immune modulators such as interferon alpha and its pegylated form, and viral polymerase inhibitors which belong to the nucleoside and nucleotide analog family[24]. Unfortunately, interferon alpha therapy is associated with several side effects, and the response rate for those receiving treatment has been unsatisfactory[25,26]. Because of the slow kinetics of viral clearance and spontaneous viral genome variability, viral mutants resistant to nucleoside analogs may be selected[27,28]. Thus, drugs targeting other unique viral targets are needed.
Our results indicate β-L-D4A and β-LPA can inhibit the activities of HBV promoters in a dose-dependent manner. On the other hand, expression of EGFP was not inhibited by lamivudine. This shows lamivudine has no effect on HBV promoters and suggests the anti-HBV effects of the two novel nucleoside analogues are mediated by mechanisms different from those used by lamivudine. HBV, a causative agent of hepatitis and hepatocellular carcinoma, contains a 3.2-kb partially double-stranded DNA genome. Upon infection of a host, the viral genome is transcribed to generate a 3.5-kb pregenomic RNA used as a template for viral replication. The pregenomic/core promoter is responsible for the synthesis of this 3.5-kb pregenomic RNA; therefore, regulation of this promoter is important in the viral life cycle[29]. The 3.5-kb RNA also serves as a template for the synthesis of polymerase and nucleocapsid core protein. In addition to the 3.5-kb RNA, three more transcripts are generated from the HBV genome. The large surface antigen is synthesized from a 2.4-kb RNA, and the major and middle antigens are synthesized from 2.1-kb transcripts. The X-gene product (HBx) is synthesized from the smallest 0.8-kb RNA[29]. The transcriptions of these RNAs are governed by the pre-S, surface, and X promoters, respectively[30,31]. Thus, HBV promoters are crucial for HBV transcription and play an important role in the HBV replicative cycle. Our previous study shows that β-L-D4A and β-LPA possess potent inhibitory effects on the replication of HBV in vitro with little cytotoxicity or mitochondrial toxicity, and can inhibit the expression of HBV antigens at high concentrations[10,13]; these findings can be explained by a model in which the two compounds inhibit the activities of HBV promoters, as shown in the present study. Thus, HBV promoters may be molecular targets of these two compounds. To confirm this, DNaseIfootprinting assays should be performed in the future. Two main points are worthy of mention here. First, although HBV promoters are crucial to the HBV life cycle, no research on anti-HBV drugs using promoters as molecular targets has been reported to date. Therefore, our effort to explore the effects of these two novel nucleoside analogues on HBV promoters is valuable and necessary. Second, four EGFP expression vectors containing different HBV promoters were successfully constructed by us. These vectors offer us the ability to monitor the activities of HBV promoters and provide an effective way to detect the effects of novel anti-HBV drugs on HBV promoters. Compared with the chloramphenicol acetyltransferase (CAT) reporter gene, the EGFP reporter gene has more advantages. Analysis of EGFP expression easier and there is no pollution from radiation. In summary, we have shown that β-L-D4A can inhibit the activities of Sp and Xp promoters, and that β-LPA can inhibit the activities of Cp and Xp promoters, in dose-dependent manners. These findings may help us to explain the mechanisms of action of these two novel compounds.
COMMENTS
Background
Hepatitis B virus (HBV) infection remains a global health problem. Currently, antiviral treatment of chronic Hepatitis B relies on interferon alpha and nucleoside analogs that inhibit viral polymerase. However, interferon alpha therapy has many side effects, while the use of nucleoside analogs can lead to the emergence of resistant viral mutants. Thus, development of novel antiviral agents against HBV is an extremely important undertaking.
Research frontiers
All of the approved chemotherapeutic drugs for the treatment of HBV hepatitis are nucleoside analogs targeting HBV DNA polymerase. Drugs targeting other unique viral targets are needed.
Innovations and breakthroughs
Although HBV promoters are crucial to HBV’s life cycle, research on anti-HBV drugs targeting HBV promoters has not yet been reported. Therefore, our efforts to explore the effects of two novel nucleoside analogues on HBV promoters are valuable and necessary.
Applications
This work may help to explain the mechanisms underlying the anti-HBV actions of β-L-D4A and β-LPA, which possess potent inhibitory effects on the replication of HBV, with little cytotoxicity or mitochondrial toxicity. Therefore, they are expected to be developed as new clinical anti-HBV drugs.
Terminology
Green fluorescent protein (GFP) was firstly used as a marker of gene expression by Chalfie (Science 1994; 263: 802-805), and later developed as an EGFP reporter gene, which uses GFP to monitor gene expression and protein localization in living organisms.
Peer review
The authors explored nucleoside analogues β-L-D4A and β-LPA’s effects on HBV promoters.
Supported by The National Natural Science Foundation of China, No. 30330680
Peer reviewer: Valeria Ghisetti, Dr, Laboratory of Microbiology, Molinette Hospital, Corso Bramante 88/90, 10126 Torino, Italy
S- Editor Zhu LH L- Editor Kerr C E- Editor Liu Y
References
- 1.O’Connor JA. Acute and chronic viral hepatitis. Adolesc Med. 2000;11:279–292. [PubMed] [Google Scholar]
- 2.Sun Z, Ming L, Zhu X, Lu J. Prevention and control of hepatitis B in China. J Med Virol. 2002;67:447–450. doi: 10.1002/jmv.10094. [DOI] [PubMed] [Google Scholar]
- 3.Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395–403. doi: 10.1016/s1473-3099(02)00315-8. [DOI] [PubMed] [Google Scholar]
- 4.Robertson SE, Mayans MV, El-Husseiny A, Clemens JD, Ivanoff B. The WHO Vaccine Trial Registry. Vaccine. 2001;20:31–41. doi: 10.1016/s0264-410x(01)00261-4. [DOI] [PubMed] [Google Scholar]
- 5.Ocama P, Opio CK, Lee WM. Hepatitis B virus infection: current status. Am J Med. 2005;118:1413. doi: 10.1016/j.amjmed.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 7.Liaw YF. Therapy of chronic hepatitis B: current challenges and opportunities. J Viral Hepat. 2002;9:393–399. doi: 10.1046/j.1365-2893.2002.00388.x. [DOI] [PubMed] [Google Scholar]
- 8.Hantz O, Kraus JL, Zoulim F. Design and evaluation of hepatitis B virus inhibitors. Curr Pharm Des. 2000;6:503–523. doi: 10.2174/1381612003400740. [DOI] [PubMed] [Google Scholar]
- 9.Cheng YC, Ying CX, Leung CH, Li Y. New targets and inhibitors of HBV replication to combat drug resistance. J Clin Virol. 2005;34 Suppl 1:S147–S150. doi: 10.1016/s1386-6532(05)80026-5. [DOI] [PubMed] [Google Scholar]
- 10.Wu JM, Lin JS, Xie N, Liang KH. Inhibition of hepatitis B virus by a novel L-nucleoside, beta-L-D4A and related analogues. World J Gastroenterol. 2003;9:1840–1843. doi: 10.3748/wjg.v9.i8.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JM, Lin JS, Xie N, Jiang FC, Liang KH. Effect and mechanism of beta-L-D4A (a novel nucleoside analog) against hepatitis B virus. Zhonghua Ganzangbing Zazhi. 2003;11:268–270. [PubMed] [Google Scholar]
- 12.Wu JM, Lin JS, Xie N, Qiu GF, Hu XM. Synthesis of a novel L-nucleoside, beta-L-D4A and its inhibition on the replication of hepatitis B virus in vitro. Yao Xue Xue Bao. 2005;40(9):825–829. [PubMed] [Google Scholar]
- 13.Qiu YL, Ptak RG, Breitenbach JM, Lin JS, Cheng YC, Drach JC, Kern ER, Zemlicka J. Synthesis and antiviral activity of phosphoralaninate derivatives of methylenecyclopropane analogues of nucleosides. Antiviral Res. 1999;43:37–53. doi: 10.1016/s0166-3542(99)00029-7. [DOI] [PubMed] [Google Scholar]
- 14.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 15.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 16.Carroll JA, Stewart PE, Rosa P, Elias AF, Garon CF. An enhanced GFP reporter system to monitor gene expression in Borrelia burgdorferi. Microbiology. 2003;149:1819–1828. doi: 10.1099/mic.0.26165-0. [DOI] [PubMed] [Google Scholar]
- 17.Lu SY, Sui YF, Li ZS, Pan CE, Ye J, Wang WY. Construction of a regulable gene therapy vector targeting for hepatocellular carcinoma. World J Gastroenterol. 2003;9:688–691. doi: 10.3748/wjg.v9.i4.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitzweg C, Zhang S, Bergert ER, Castro MR, McIver B, Heufelder AE, Tindall DJ, Young CY, Morris JC. Prostate-specific antigen (PSA) promoter-driven androgen-inducible expression of sodium iodide symporter in prostate cancer cell lines. Cancer Res. 1999;59:2136–2141. [PubMed] [Google Scholar]
- 19.Ha-Lee YM, Lee J, Pyun H, Kim Y, Sohn J, Cho YJ, Kim Y. Sequence variations of hepatitis B virus promoter regions in persistently infected patients. Arch Virol. 2001;146:279–292. doi: 10.1007/s007050170175. [DOI] [PubMed] [Google Scholar]
- 20.Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 21.Kramvis A, Kew MC. The core promoter of hepatitis B virus. J Viral Hepat. 1999;6:415–427. doi: 10.1046/j.1365-2893.1999.00189.x. [DOI] [PubMed] [Google Scholar]
- 22.Bock CT, Kubicka S, Manns MP, Trautwein C. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology. 1999;29:1236–1247. doi: 10.1002/hep.510290426. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Raney AK, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 24.Zoulim F. Antiviral therapy of chronic hepatitis B. Antiviral Res. 2006;71:206–215. doi: 10.1016/j.antiviral.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Wong DK, Cheung AM, O'Rourke K, Naylor CD, Detsky AS, Heathcote J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–323. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 26.Manns MP. Current state of interferon therapy in the treatment of chronic hepatitis B. Semin Liver Dis. 2002;22 Suppl 1:7–13. doi: 10.1055/s-2002-35695. [DOI] [PubMed] [Google Scholar]
- 27.Zoulim F, Poynard T, Degos F, Slama A, El Hasnaoui A, Blin P, Mercier F, Deny P, Landais P, Parvaz P, et al. A prospective study of the evolution of lamivudine resistance mutations in patients with chronic hepatitis B treated with lamivudine. J Viral Hepat. 2006;13:278–288. doi: 10.1111/j.1365-2893.2005.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P, et al. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–3507. doi: 10.1128/AAC.48.9.3498-3507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 30.Shaul Y, Rutter WJ, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jameel S, Siddiqui A. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol Cell Biol. 1986;6:710–715. doi: 10.1128/mcb.6.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]