Abstract
AIM: To investigate the relationship of changes in expression of marker genes in functional categories or molecular networks comprising one functional category or multiple categories in progression of hepatic fibrosis in hepatitis C (HCV) patients.
METHODS: Marker genes were initially identified using DNA microarray data from a rat liver fibrosis model. The expression level of each fibrosis associated marker gene was analyzed using reverse transcription-polymerase chain reaction (RT-PCR) in clinical biopsy specimens from HCV-positive patients (n = 61). Analysis of changes in expression patterns and interactions of marker genes in functional categories was used to assess the biological mechanism of fibrosis.
RESULTS: The profile data showed several biological changes associated with progression of hepatic fibrosis. Clustered genes in functional categories showed sequential changes in expression. Several sets of clustered genes, including those related to the extracellular matrix (ECM), inflammation, lipid metabolism, steroid metabolism, and some transcription factors important for hepatic biology showed expression changes in the immediate early phase (F1/F2) of fibrosis. Genes associated with aromatic amino acid (AA) metabolism, sulfur-containing AA metabolism and insulin/Wnt signaling showed expression changes in the middle phase (F2/F3), and some genes related to glucose metabolism showed altered expression in the late phase of fibrosis (F3/F4). Therefore, molecular networks showing serial changes in gene expression are present in liver fibrosis progression in hepatitis C patients.
CONCLUSION: Analysis of gene expression profiles from a perspective of functional categories or molecular networks provides an understanding of disease and suggests new diagnostic methods. Selected marker genes have potential utility for biological identification of advanced fibrosis.
Keywords: Hepatitis C, Liver fibrosis, Marker gene, Gene expression, RT-PCR, Molecular network, Metabolism, Transcription factor, Diagnosis
INTRODUCTION
Liver fibrosis is caused by liver disorders such as hepatitis C, hepatitis B, alcoholic hepatitis and non-alcoholic hepatitis. Fibrosis progresses gradually and finally disrupts liver structure and function over several decades, leading to fatal diseases such as cirrhosis and hepatocellular carcinoma (HCC). Classification of fibrosis progression is usually based on histological criteria using the METAVIR scoring system[1], which includes five stages: F0 (no fibrosis), F1, F2, F3, and F4 (cirrhosis). Such a classification is essential in decisions regarding treatment of liver fibrosis. Prominent subjective symptoms do not occur from F1 to F3, but patients begin to be aware of symptoms after F4. However, the F4 stage of cirrhosis is almost incurable and diagnosis of fibrosis at an earlier stage is desirable. The biology after F4 (cirrhosis) has been well studied, due to the interest in diagnosis and therapy for hepatocellular carcinoma (HCC), but progression to HCC may begin in the early stage of fibrosis in hepatitis C[2].
Prevention of HCC and inhibition of fibrosis in the early phase is important, but the detailed hepatic biological changes corresponding to the F stage are unclear. To understand the background of the early stage of fibrosis, we previously identified genes that can be used as markers of biological changes in progression of hepatic fibrosis and diagnosis of fibrotic progression; these data were obtained from DNA microarray data from an experimental DMN (dimethylnitrosamine)-treated rat model of hepatic fibrosis[3]. This work led to marker genes that were arranged in functional categories related to fibrosis, based on genes associated with hepatic cell types such as Kupffer cells, hepatic stellate cells, and hepatocytes.
These marker genes give information on cell-specific and time-dependent behavior of each hepatic cell in fibrogenesis. In the current work, the behavior of marker genes associated with a particular F stage was analyzed using RT-PCR in clinical biopsy specimens from hepatitis C patients. This profile data revealed several biological changes in progression of hepatic fibrosis. Since many functionally clustered genes showed similar changes in expression, we propose serial expression changes in molecular networks associated with liver fibrosis progression in hepatitis C patients. Many functionally clustered genes showed large changes in expression in the early stage of fibrosis, suggest-ing the importance of therapy at this early stage. Alteration of gene expression also suggested qualitative changes in biological status in the transition from F3 to F4, which is a suspected risk factor for development of HCC. We conclude that analysis of gene expression profiles from a perspective of molecular networks provides improved understanding of disease and indicates potential methods of diagnosis.
MATERIALS AND METHODS
Patients in the clinical study
All patients were recruited from the Osaka City University Hospital (Osaka, Japan). Sixty-one patients with seropositive results in diagnosis using the third-generation hepatitis C virus enzyme-linked immunosorbent assay (Lumipulse II Ortho HCV, Ortho-Clinical Diagnostics, Tokyo, Japan) and positive serum HCV-RNA were included in the study. Informed consent was obtained from all patients. Liver biopsies were performed on all patients enrolled in the study, and the histological features of the liver specimens were analyzed and graded using the METAVIR scoring system[1]. The liver fibrotic stage (F stage) and inflammatory activity (A grade) were determined histologically: at least four subjects were found to be in each F stage classification. Determination for chymase 1 exceptionally has been done with three subjects in F4 stage due to the lack of appropriate samples.
Part of the biopsy sample from each patient was imme-diately immersed in RNAlater (QIAGEN, The Netherlands) to inhibit RNAase and then kept at 4°C overnight before being transferred to another tube and frozen at -80°C.
RT-PCR analysis
Total RNA was extracted from liver biopsy samples using an ISOGEN kit (Nippongene) and reverse transcribed using a High Capacity cDNA Archive Kit (ABI, Foster City, CA), in each case according to the manufacturer’s instructions. The total RNA in the final reaction mixture was 10 ng/μL. Real-time PCR was performed on an Applied Biosystems 7500 Real-Time PCR System (ABI) data collection system, and analyses were performed using the accompanying software. RT-PCR was performed using 0.8 μL cDNA in each well, with a final concentration of 1X the probes of the TaqMan® Gene Expression Assay and 1X the Taqman Universal PCR Master Mix (ABI). The final reaction volume was 20 μL. Each sample was analyzed in duplicate. The thermal cycler conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C.
Data were analyzed using the comparative CT method, in which the expression level of a target gene is normalized relative to an endogenous reference. GAPDH was used as the endogenous reference in all experiments. The target CT and endogenous control CT were calculated for each sample, and the target gene expression level was then calculated using the formula 2(34-CT). The average of duplicate measurements was obtained, and the relative expression of each gene in a sample was calculated by setting the expression of GAPDH equal to 1000. PCR fluorogenic probes for all the target genes and the endogenous reference were purchased as TaqMan® Gene Expression Assays (ABI).
Statistical analysis
A Kruskal-Wallis test was applied to select marker genes with statistically significant changes (P < 0.05) in expression level during the fibrosis progression. This calculation was performed using SPSS (SPSS Inc., IL, USA). The gene expression data were subjected to hierarchical clustering analysis using Genowiz™ software (Ocimum Biosolutions).
Pathway analysis of PCR data
The behavior and relationships of marker genes in pathways associated with lipid metabolism were analyzed with bioSpace Explorer, a system for analysis of expression profile data. This system was produced collaboratively by Pharmafrontier Co. Ltd. and World Fusion Co. Ltd. to examine molecular interactions in expression profile data, using both manual and computational text mining.
RESULTS
Expression behavior of marker genes
All marker genes determined in this study are listed in Table 1. For each gene, since the quantitative limitation of biopsy specimens resulted in a difference in sample number for each probe, the type of samples, indicating the number of biopsy specimens, is listed in Table 2. Marker genes were selected as representative members of functional categories or molecular networks based on their expression changes in an experimental hepatic fibrosis model[3]. Marker genes with statistically significant changes in expression are listed in Table 1. Genes that showed statistically insignificant changes in expression are listed with the gene name only in Table 1. Marker genes that showed statistically significant changes by t test during a transition to an adjacent F stage are similar to the selected genes in Table 1. An additional four marker genes selected in the t test analysis were added to the genes in Table 1. Over half of the selected marker genes from the DNA microarray data from the animal model showed statistically significant changes in the human samples, showing the effectiveness of the animal model for selection of genes of significance in humans.
Table 1.
Expression profiles of marker genes with statistically significant changes during fibrosis progression
| Functional category | Description | Gene name | Expression at F1 | Group |
Expression ratio |
Serial | Type | ||
| F2/F1 | F3/F2 | F4/F3 | |||||||
| ECM (or other HSC marker) | Decorin | DCN | 217.6 | 2 | 1.21 | 1.1 | 1.1 | 1 | 1 |
| Matrix metalloproteinase 2 | MMP2 | 11.7 | 2 | 1.51 | 1.2 | 0.9 | 2 | 1 | |
| Hyaluronan-mediated motility receptor | HMMR | 1.2 | 2 | 2.61 | 0.6 | 1.1 | 3 | 1 | |
| Lysyl oxidase | LOX | 1.0 | 2 | 1.41 | 1.2 | 0.9 | 4 | 1 | |
| Lysyl oxidase-like 1 | LOXL1 | 0.8 | 2 | 1.81 | 1.4 | 1.0 | 5 | 1 | |
| Tropomyosin 1 | TPM1 | 35 | 2 | 1.81 | 1.5 | 1.1 | 6 | 1 | |
| Prion | PRNP | 19.4 | 3 | 1.21 | 0.7 | 1.0 | 7 | 1 | |
| Collagen, type I, alpha 1 | COL1A1 | 16.2 | 2 | 1.3 | 2.21 | 0.6 | 8 | 1 | |
| Collagen type III alpha 1 | COL3A1 | 143.9 | 2 | 1.3 | 1.51 | 0.8 | 9 | 1 | |
| Collagen type alpha 1 | COL4A1 | 16.8 | 2 | 1.3 | 1.91 | 0.6 | 10 | 1 | |
| Humican | LUM | 22.8 | 2 | 1.2 | 2.01 | 0.9 | 11 | 1 | |
| Sialoprotein | SPP1 | 13.2 | 2 | 1.2 | 2.21 | 1.4 | 12 | 1 | |
| Glypican 3 | GPC3 | 23.5 | 2 | 1.8 | 2.51 | 1.0 | 13 | 1 | |
| Proline 4-hydroxylase, alpha polypeptide I | P4HA1 | 34 | 1 | 1.1 | 0.42 | 1.2 | 14 | 1 | |
| Insignificant change: MGP, BGN, TAGLN, LGALS1, EDG2, EDG5, TNNT2 | |||||||||
| Inflammation (or apoptosis) | Lysozyme | LYZ | 185.3 | 2 | 2.11 | 0.8 | 1.0 | 15 | 2 |
| TGF beta | TGFB1 | 51.9 | 3 | 1.51 | 0.8 | 0.9 | 16 | 1 | |
| TGF beta 3 | TGFB3 | 3.3 | 3 | 1.41 | 0.9 | 0.9 | 17 | 3 | |
| TNF | TNF | 2.6 | 3 | 1.71 | 0.6 | 1.2 | 18 | 3 | |
| Natural killer cell proteinase 1 | GZMB | 1.7 | 3 | 1.61 | 0.5 | 1.1 | 19 | 1 | |
| IL1 beta | IL1B | 2.6 | 3 | 1.51 | 0.7 | 1.1 | 20 | 3 | |
| Hemopoietic cell kinase | HCK | 22.4 | 3 | 1.31 | 0.6 | 0.9 | 21 | 4 | |
| Interleukin 6 receptor | IL6R | 142.9 | 1 | 0.9 | 0.72 | 1.0 | 22 | 3 | |
| BCL2-related ovarian killer | BOK | 205.9 | 1 | 0.9 | 0.72 | 1.1 | 23 | 5 | |
| Caspase 2 | CASP2 | 1.3 | 1 | 1.2 | 0.72 | 0.8 | 24 | 5 | |
| Chymase 1, mast cell | CMA1 | 0.2 | 2 | 0.7 | 1.8 | 2.21 | 25 | 6 | |
| Insignificant change: LTBP1, LBP, TNFRSF1B, DEFB1, IL1RN, S100A8, BRIC3, CARD12, CASP1, CASP4, CASP8, PAWR, CD19, CD3Z, MS4A1, CD37, TRA@ | |||||||||
| Growth factor | Growth hormone receptor | GHR | 102.3 | 1 | 0.82 | 0.9 | 0.9 | 26 | 4 |
| IGF1 | IGF1 | 56.5 | 1 | 0.9 | 0.72 | 1.1 | 27 | 1 | |
| Insignificant change: PTN, FST, PRLR | |||||||||
| Insulin/ Wnt signal | Cyclin D1 | CCND1 | 190.6 | 2 | 1.71 | 1.0 | 1.0 | 28 | 5 |
| Forkhead box M1 | FOXM1 | 0.6 | 3 | 3.71 | 0.8 | 1.1 | 29 | 4 | |
| Gap junction protein, alpha 1, 43 kDa (connexin 43) | GJA1 | 4 | 3 | 2.41 | 0.5 | 1.0 | 30 | 5 | |
| V-akt murine thymoma viral oncogene homolog 1 | AKT1 | 77.6 | 1 | 0.9 | 0.92 | 1.0 | 31 | 4 | |
| V-akt murine thymoma viral oncogene homolog 2 | AKT2 | 59.6 | 1 | 0.9 | 0.72 | 0.8 | 32 | 7 | |
| Catenin (cadherin-associated protein), beta 1, 88 kDa | CTNNB1 | 159 | 1 | 1.1 | 0.72 | 1.0 | 33 | 5 | |
| Catenin, beta interacting protein 1 | CTNNBIP1 | 24 | 1 | 1.1 | 0.72 | 0.8 | 34 | 5 | |
| Glycogen synthase kinase 3 beta | GSK3B | 44.1 | 1 | 1.1 | 0.82 | 0.8 | 35 | 5 | |
| Dishevelled, dsh homolog 1 (Drosophila) | DVL1 | 12.2 | 1 | 1.1 | 0.72 | 0.9 | 36 | 7 | |
| Membrane-bound transcription factor peptidase, site 1 | MBTPS1 | 72.4 | 1 | 0.9 | 0.72 | 1.0 | 37 | 7 | |
| Membrane-bound transcription factor peptidase, site 2 | MBTPS2 | 13.2 | 1 | 0.9 | 0.72 | 1.0 | 38 | 7 | |
| Tribbles homolog 3 (Drosophila) | TRIB3 | 18.4 | 1 | 1.1 | 0.9 | 0.42 | 39 | 7 | |
| Insignificant change: GSK3A, INSIG1, INSIG2, PRKCB1, PRKCD | |||||||||
| Others signal | Regucalcin (senescence marker protein-30) | RGN | 697.8 | 1 | 0.9 | 0.8 | 0.82 | 56 | 4 |
| Insignificant change: DAB2, PMP22, S100A10, LCN2 | |||||||||
| Transcription factors | CCAAT/enhancer binding protein (C/EBP), alpha | CEBPA | 352.2 | 1 | 0.72 | 1.2 | 0.7 | 40 | 4 |
| Retinoid X receptor, alpha | RXRA | 409.6 | 1 | 0.72 | 1.0 | 0.8 | 41 | 4 | |
| Hepatocyte nuclear factor 4, alpha | HNF4A | 708.7 | 1 | 0.72 | 1.1 | 0.9 | 42 | 4 | |
| Transcription factor 1 (HNF1) | TCF1 | 26.7 | 1 | 0.82 | 0.9 | 0.8 | 43 | 4 | |
| Nuclear receptor subfamily 0, group B, member 2 | NR0B2 | 166.7 | 1 | 0.72 | 0.9 | 0.8 | 44 | 4 | |
| Peroxisome proliferative activated receptor, alpha | PPARA | 62.9 | 1 | 0.72 | 0.8 | 0.9 | 45 | 4 | |
| Inhibitor of DNA binding 1 (splice variation) | ID1 | 463.4 | 3 | 1.61 | 0.5 | 1.4 | 46 | 1 | |
| AE binding protein 1 | AEBP1 | 24.6 | 2 | 1.41 | 1.2 | 1.0 | 47 | 4 | |
| Nuclear receptor subfamily 1, group H, member 2 | NR1H2 (LXRB) | 9 | 1 | 1.2 | 0.72 | 1.0 | 48 | 7 | |
| Nuclear receptor subfamily 1, group H, member 3 | NR1H3 (LXRA) | 27.8 | 1 | 1.0 | 0.72 | 0.8 | 49 | 7 | |
| Nuclear receptor subfamily 1, group H, member 4 | NR1H4 (FXR) | 107 | 1 | 1.0 | 0.72 | 0.9 | 50 | 7 | |
| c/EBPbeta | CEBPB | 274.3 | 1 | 0.9 | 0.72 | 0.9 | 51 | 2 | |
| Upstream transcription factor 2, c-fos interacting | USF2 | 323.7 | 1 | 0.9 | 0.72 | 0.9 | 52 | 4 | |
| Estrogen-related receptor alpha | ESRRA | 79.6 | 1 | 1.2 | 0.62 | 0.8 | 53 | 7 | |
| C-met | MET | 73.5 | 1 | 1.0 | 0.72 | 1.1 | 54 | 3 | |
| Upstream transcription factor 1 | USF1 | 30.8 | 1 | 1.0 | 0.9 | 0.82 | 55 | 4 | |
| Insignificant change: ONECUT1, JUNB, NR3C1, PPARG, PPARGC1B, PPARGC1A, SREBF2, FHL2 | |||||||||
| Transporter | Solute carrier family 6, member 6 | SLC6A6 | 3.7 | 3 | 2.51 | 0.6 | 1.0 | 57 | 5 |
| Solute carrier family 7, member 1 | SLC7A1 | 0.9 | 3 | 2.31 | 0.7 | 1.3 | 58 | 5 | |
| Solute carrier family 38, member 2 Alanine-trasnporter) | SLC38A2 | 68.6 | 3 | 1.31 | 0.6 | 1.0 | 59 | 5 | |
| Solute carrier family 25 member 15 | SLC25A15 | 85.7 | 1 | 1.1 | 0.72 | 0.9 | 60 | 5 | |
| Solute carrier family 7, member 7 | SLC7A7 | 7.1 | 1 | 1.1 | 0.62 | 1.0 | 61 | 5 | |
| Solute carrier family 17 (sodium phosphate), member 1 | SLC17A1 | 23.6 | 1 | 0.9 | 0.72 | 1.0 | 62 | 4 | |
| Insignificant change: SLC38A3, ABCB1, SLC15A4 | |||||||||
| Redox | Catalase | CAT | 1977.7 | 1 | 0.82 | 0.9 | 1.0 | 63 | 4 |
| Paraoxonase 1 | PON1 | 191.1 | 1 | 0.8 | 0.72 | 0.9 | 64 | 4 | |
| Blood coagulation | Coagulation factor X | F10 | 187.5 | 1 | 0.82 | 1.0 | 0.9 | 65 | 4 |
| Angiotensinogen | AGT | 1958.4 | 1 | 0.82 | 1.0 | 1.0 | 66 | 4 | |
| Fibrinogen, A alpha polypeptide | FGA | 8337.5 | 1 | 0.62 | 1.0 | 0.9 | 67 | 4 | |
| Plasminogen | PLG | 6156.1 | 1 | 0.82 | 0.8 | 1.0 | 68 | 4 | |
| Pai I | SERPINE1 | 11.6 | 3 | 0.9 | 2.91 | 0.5 | 69 | 1 | |
| Lipid metabolism | Acyl-Coenzyme A oxidase 2, branched chain | ACOX2 | 122.2 | 1 | 0.82 | 0.9 | 0.9 | 70 | 4 |
| L-3-hydroxyacyl-Coenzyme A dehydrogenase, short chain | HADHSC | 139.8 | 1 | 0.82 | 0.9 | 0.9 | 71 | 4 | |
| Acyl-CoA synthetase long-chain family member 1 | ACSL1 | 2800.8 | 1 | 0.82 | 0.9 | 0.9 | 72 | 4 | |
| Acyl-Coenzyme A oxidase 1, palmitoyl | ACOX1 | 357.7 | 1 | 0.72 | 0.9 | 1.0 | 73 | 4 | |
| Carnitine O-octanoyltransferase | CROT | 10.3 | 1 | 0.72 | 0.8 | 1.1 | 74 | 4 | |
| 2,4-dienoyl CoA reductase 2, peroxisomal | DECR2 | 135.2 | 1 | 0.82 | 0.9 | 0.9 | 75 | 4 | |
| Acetyl-Coenzyme A acyltransferase 2 | ACAA2 | 1167.7 | 1 | 0.82 | 0.8 | 1.1 | 76 | 4 | |
| Acetyl-Coenzyme A acetyltransferase 1 | ACAT1 | 714.2 | 1 | 0.72 | 1.1 | 0.8 | 77 | 4 | |
| Acyl-CoA synthetase long-chain family member 5 | ACSL5 | 57.3 | 1 | 0.82 | 0.9 | 1.0 | 78 | 4 | |
| Dodecenoyl-Coenzyme A delta isomerase | DCI | 327.2 | 1 | 0.92 | 0.8 | 0.9 | 79 | 4 | |
| Enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | ECHS1 | 324.3 | 1 | 0.72 | 1.0 | 0.9 | 80 | 4 | |
| Hydroxyacyl-Coenzyme A dehydrogenase, type II | HADH2 | 422.5 | 1 | 0.72 | 0.9 | 0.9 | 81 | 4 | |
| Hydroxyacyl-Coenzyme A dehydrogenase, beta subunit | HADHB | 398.8 | 1 | 0.82 | 0.9 | 1.1 | 82 | 4 | |
| Lipase, hepatic | LIPC | 561.5 | 1 | 0.52 | 1.5 | 0.9 | 83 | 4 | |
| Hydroxyacyl-Coenzyme A dehydrogenase, alpha subunit | HADHA | 103.2 | 1 | 0.9 | 0.92 | 1.0 | 84 | 4 | |
| Palmitoyl-protein thioesterase 1 | PPT1 | 82.1 | 1 | 1.0 | 0.82 | 0.8 | 85 | 4 | |
| Fatty acid synthase | FASN | 105.3 | 3 | 0.8 | 1.81 | 0.6 | 86 | 4 | |
| Peroxisomal D3, D2-enoyl-CoA isomerase | PECI | 292.8 | 1 | 0.8 | 0.8 | 0.82 | 87 | 4 | |
| Acyl-CoA synthetase long-chain family member 4 | ACSL4 | 5.8 | 2 | 1.1 | 1.4 | 2.31 | 88 | 4 | |
| Insignificant change: BHHADH, ACAA1, CPT1A, ACADM, ACACA, CPT2 | |||||||||
| Steroid (or drug) metabolism | Aldo-keto reductase family 1, member D1 | AKR1D1 | 248.3 | 1 | 0.62 | 0.8 | 0.6 | 89 | 4 |
| HMT1 hnRNP methyltransferase-like 2 | HRMT1L2 | 6.6 | 1 | 0.82 | 1.0 | 0.8 | 90 | 4 | |
| Hydroxysteroid (11-beta) dehydrogenase 1 | HSD11B1 | 1064.6 | 1 | 0.72 | 0.8 | 0.8 | 91 | 4 | |
| Hydroxysteroid (17-beta) dehydrogenase 4 | HSD17B4 | 66.7 | 1 | 0.82 | 0.9 | 0.9 | 92 | 4 | |
| Steroid-5-alpha-reductase, alpha polypeptide 1 | SRD5A1 | 74.3 | 1 | 0.72 | 0.8 | 0.9 | 93 | 4 | |
| UDP glycosyltransferase 2 family, polypeptide B7 | UGT2B7 | 530.3 | 1 | 0.82 | 0.8 | 0.9 | 94 | 4 | |
| Sulfotransferase family 1E, estrogen-preferring, member 1 | SULT1E1 | 52.8 | 1 | 0.52 | 0.5 | 1.4 | 95 | 7 | |
| Aldo-keto reductase family 1, member C4 | AKR1C4 | 137.1 | 1 | 0.9 | 0.62 | 1.2 | 96 | 4 | |
| Hydroxysteroid (17-beta) dehydrogenase 2 | HSD17B2 | 496.9 | 1 | 1.0 | 0.82 | 1.1 | 97 | 4 | |
| Sulfotransferase family, cytosolic, 2A, member 1 | SULT2A1 | 689 | 1 | 0.9 | 0.82 | 0.9 | 98 | 4 | |
| Hydroxysteroid (17-beta) dehydrogenase 8 | HSD17B8 | 72.2 | 1 | 0.7 | 0.72 | 1.0 | 99 | 7 | |
| Steroid sulfatase (microsomal), arylsulfatase C, isozyme S | STS | 10.9 | 1 | 1.0 | 1.0 | 0.62 | 100 | 4 | |
| Emopamil binding protein (sterol isomerase) | EBP | 197 | 1 | 1.0 | 0.7 | 0.72 | 101 | 5 | |
| Farnesyl-diphosphate farnesyltransferase 1 | FDFT1 | 252.6 | 1 | 0.7 | 0.9 | 0.72 | 102 | 5 | |
| Insignificant change: HSD17B2, HSD3B1, LCMT1, SULT2A1, HMGCR, DHCR7, CES2 | |||||||||
| Bile acid metabolism | Sterol O-acyltransferase 1 | SOAT1 | 9.1 | 1 | 1.0 | 0.82 | 0.9 | 103 | 5 |
| Alcohol dehydrogenase 1C (class I), gamma polypeptide | ADH1C | 312.7 | 1 | 0.7 | 0.62 | 1.1 | 104 | 5 | |
| Alcohol dehydrogenase, iron containing, 1 | ADHFE1 | 112.7 | 1 | 0.7 | 0.72 | 0.8 | 105 | 5 | |
| Cytochrome P450, family 7, subfamily A, polypeptide 1 | CYP7A1 | 104.1 | 1 | 0.7 | 0.62 | 2.9 | 106 | 5 | |
| Prostanoid | Arachidonate 5-lipoxygenase-activating protein | ALOX5AP | 7.9 | 2 | 1.71 | 0.7 | 1.0 | 107 | 3 |
| Leukotriene B4 receptor 2 | LTB4R2 | 2.5 | 1 | 1.1 | 0.72 | 1.0 | 108 | 3 | |
| Insignificant change: LTA4H, CYSLTR1, CYSLTR2, LTC4S, PPT1 | |||||||||
| Aromatic amino acid metabolism | Dopa decarboxylase | DDC | 51.4 | 1 | 1.0 | 0.82 | 0.8 | 109 | 4 |
| Monoamine oxidase B | MAOB | 426.6 | 1 | 0.8 | 0.82 | 1.0 | 110 | 4 | |
| Kynurenine 3-monooxygenase | KMO | 51.7 | 1 | 1.0 | 0.72 | 1.0 | 111 | 4 | |
| Kynureninase | KYNU | 63.3 | 1 | 0.9 | 0.72 | 0.8 | 112 | 4 | |
| Tyrosine aminotransferase | TAT | 658.2 | 1 | 0.9 | 0.52 | 1.3 | 113 | 4 | |
| GTP cyclohydrolase 1 | GCH1 | 49.6 | 1 | 0.9 | 0.72 | 0.9 | 114 | 4 | |
| Insignificant change: HPD | |||||||||
| Sulfur-containing amino acid metabolism | MAT2 | MAT2B | 111.7 | 1 | 1.1 | 0.72 | 1.1 | 115 | 3 |
| Cystathionase (cystathionine gamma-lyase) | CTH | 112 | 1 | 1.0 | 0.72 | 1.1 | 116 | 3 | |
| Cystathionine-beta-synthase | CBS | 350.4 | 1 | 1.1 | 0.72 | 0.9 | 117 | 3 | |
| Betaine-homocysteine methyltransferase | BHMT | 1032.1 | 1 | 1.0 | 0.72 | 1.0 | 118 | 3 | |
| Methionine adenosyltransferase I, alpha | MAT1A | 547.6 | 1 | 1.0 | 0.52 | 1.1 | 119 | 2 | |
| Cysteine dioxygenase, type I | CDO1 | 84.4 | 1 | 0.9 | 0.72 | 1 | 120 | 1 | |
| Glutamate-cysteine ligase, catalytic subunit | GCLC | 175.9 | 1 | 1.1 | 0.62 | 1.1 | 121 | 1 | |
| Glutathione S-transferase A1 | GSTA1 | 2812.3 | 1 | 1.0 | 0.72 | 1.0 | 122 | 1 | |
| Alanyl (membrane) aminopeptidase | ANPEP | 501.9 | 1 | 0.9 | 0.82 | 0.9 | 124 | 5 | |
| Bile acid Coenzyme A: amino acid N-acyltransferase | BAAT | 284.4 | 1 | 1.0 | 0.62 | 1.0 | 125 | 5 | |
| Glutathione synthetase | GSS | 70.1 | 1 | 1.1 | 0.82 | 0.8 | 126 | 5 | |
| Lactate dehydrogenase A | LDHA | 716.3 | 1 | 0.8 | 0.72 | 1.0 | 127 | 5 | |
| Mercaptopyruvate sulfurtransferase | MPST | 684.3 | 1 | 0.9 | 0.72 | 0.9 | 128 | 5 | |
| Serine dehydratase | SDS | 440.4 | 1 | 0.9 | 0.42 | 1.0 | 129 | 5 | |
| Methionine adenosyltransferase II, alpha | MAT2A | 122.9 | 1 | 1.0 | 0.9 | 0.82 | 130 | 5 | |
| Insignificant change: GGT1, GSR, MTR, DNMMT1, CSAD, GCLM, LDHB | |||||||||
| Energy source amino acid metabolism | Phosphoenolpyruvate carboxykinase 2 (mitochondrial) | PCK2 | 791.8 | 1 | 0.82 | 0.9 | 1.0 | 131 | 4 |
| Alanine-glyoxylate aminotransferase | AGXT | 3352.3 | 1 | 0.72 | 0.9 | 1.0 | 132 | 4 | |
| Alanine-glyoxylate aminotransferase 2 | AGXT2 | 122.8 | 1 | 0.62 | 0.8 | 1.0 | 133 | 4 | |
| Aldehyde dehydrogenase 2 family (mitochondrial) | ALDH2 | 2027.7 | 1 | 0.72 | 0.9 | 0.9 | 134 | 4 | |
| Aldehyde dehydrogenase 9 family, member A1 | ALDH9A1 | 160.9 | 1 | 0.82 | 0.8 | 0.9 | 135 | 4 | |
| Pyruvate kinase, liver and RBC | PKLR | 211.3 | 1 | 0.62 | 1.1 | 0.7 | 136 | 4 | |
| Aldehyde dehydrogenase 3 family, member A2 | ALDH3A2 | 237.4 | 1 | 0.9 | 0.82 | 1.1 | 137 | 4 | |
| Phosphoenolpyruvate carboxykinase 1 (soluble) | PCK1 | 3166.8 | 1 | 1.0 | 0.52 | 1.2 | 138 | 4 | |
| Dihydrolipoamide dehydrogenase | DLD | 72.3 | 1 | 0.9 | 0.82 | 0.9 | 139 | 4 | |
| Glutaminase 2 (liver, mitochondrial) | GLS2 | 133.8 | 1 | 0.8 | 0.72 | 1.0 | 140 | 4 | |
| Glutamate-ammonia ligase | GLUL | 6.2 | 1 | 1.0 | 0.62 | 1.2 | 141 | 4 | |
| Glutamic-oxaloacetic transaminase 1, soluble | GOT1 | 388.2 | 1 | 0.9 | 0.72 | 0.8 | 142 | 4 | |
| Glutamic-pyruvate transaminase | GPT | 298.8 | 1 | 0.8 | 0.62 | 1.1 | 143 | 4 | |
| Pyruvate carboxylase | PC | 27.8 | 1 | 0.7 | 0.72 | 0.9 | 144 | 4 | |
| Phosphoglucomutase 1 | PGM1 | 218.8 | 1 | 0.8 | 0.82 | 1.0 | 145 | 4 | |
| Pyruvate dehydrogenase kinase, isoenzyme 2 | PDK2 | 24 | 1 | 0.9 | 0.82 | 0.9 | 146 | 7 | |
| Pyruvate dehydrogenase kinase, isoenzyme 4 | PDK4 | 85.6 | 1 | 0.7 | 0.52 | 1.4 | 147 | 7 | |
| Fumarylacetoacetate hydrolase | FAH | 85.8 | 1 | 0.9 | 0.9 | 0.82 | 148 | 4 | |
| Malic enzyme 1, NADP (+) -dependent, cytosolic | ME1 | 4.9 | 2 | 1.0 | 1.0 | 1.31 | 149 | 4 | |
| Insignificant change: ALDOA, ASNS, GOT2, MGC33309, PDHB, PDK1 | |||||||||
| Glucose metabolism | Phosphorylase, glycogen; liver | PYGL | 37.2 | 1 | 0.82 | 1.0 | 1.0 | 150 | 4 |
| Aldolase B, fructose-bisphosphate | ALDOB | 13141.8 | 1 | 0.7 | 0.82 | 0.9 | 151 | 4 | |
| Hexokinase 3 (white cell) | HK3 | 3.3 | 1 | 1.2 | 0.62 | 0.9 | 152 | 4 | |
| Glycogen synthase 2 (liver) | GYS2 | 276.6 | 1 | 0.9 | 0.62 | 0.9 | 153 | 7 | |
| Sterol regulatory element binding transcription factor 1 | SREBF1 | 10.6 | 2 | 1.1 | 1.21 | 0.9 | 154 | 4 | |
| Glycerol-3-phosphate dehydrogenase 1 (soluble) | GPD1 | 130.8 | 1 | 0.8 | 1.0 | 0.82 | 155 | 4 | |
| Ketohexokinase (fructokinase) | KHK | 411.8 | 1 | 0.7 | 1.0 | 0.82 | 156 | 4 | |
| Glucokinase (hexokinase 4) regulator | GCKR | 204.8 | 1 | 0.7 | 1.0 | 0.82 | 157 | 4 | |
| Aldolase C, fructose-bisphosphate | ALDOC | 36.8 | 1 | 0.7 | 0.9 | 0.62 | 158 | 4 | |
| Insignificant change: GCK, PFKFB1, G6PC, HK2 | |||||||||
| Urea cycle | Carbamoyl-phosphate synthetase 1, mitochondrial | CPS1 | 1230.8 | 1 | 0.72 | 0.9 | 0.9 | 159 | 7 |
| Ornithine aminotransferase (gyrate atrophy) | OAT | 94.8 | 1 | 0.52 | 0.6 | 1.6 | 160 | 7 | |
| Insignificant change: OTC | |||||||||
Liver biopsy samples were analyzed by RT-PCR using a TaqMan® Gene Expression Assay probe, as described in the Materials and Methods. The genes were selected from DNA microarray data from a rat fibrosis model presented in a previous paper 3. Genes in the ECM category (or other HSC markers) were selected as marker genes for HSC. Genes in the inflammation category were selected as markers of inflammatory cells in liver. Other genes were selected as markers of hepatocytes. Expression profiles of marker genes that showed statistically significant changes during fibrosis by a Kruskal-Wallis test are listed. Genes that showed statistically insignificant changes in expression are listed with the gene name only in Table 1. Columns from left to right indicate functional category, gene description, gene name, expression intensity at F1 stage, group classification (expression of marker genes in group 1 decreased almost linearly along with the F stages, expression of marker genes in group 2 increased almost linearly along with the F stages, and expression of marker genes in group 3 had a peak in the middle of the F stages), expression ratio between neighbouring F stages (1an increased ratio, 2a decreased ratio), serial gene number and types of samples, which indicates the number of biopsy specimens, as shown in the Table 2. Actual alteration behavior of some characteristic genes are shown in Figure 1.
Table 2.
Types of samples indicating the number of biopsy specimens
| Type of sample |
Number of samples |
|||
| F1 | F2 | F3 | F4 | |
| 1 | 25 | 13 | 14 | 9 |
| 2 | 23 | 13 | 14 | 8 |
| 3 | 22 | 12 | 14 | 8 |
| 4 | 12 | 6 | 9 | 4 |
| 5 | 7 | 4 | 5 | 4 |
| 6 | 6 | 4 | 4 | 3 |
| 7 | 5 | 4 | 5 | 5 |
Quantitative limitation of biopsy specimens resulted in a difference in sample number for each probe, according to the interest in the particular gene.
Genes showing changes in expression were roughly divided into three groups: genes in group 1 showed an almost linear decrease in expression along with an increase in F stage; genes in group 2 showed an almost linear increase with an increase in F stage; and genes in group 3 showed a peak in the middle of the F stage scale. Almost all genes in one category belonged to one or two groups, suggesting that genes in one category showed similar changes in expression during progression of fibrosis. The expression ratio between F stages is also shown in Table 1, with the peak ratio shown in bold. The peak for almost all genes in a given category occurred at the transition to the same F stage, again suggesting that genes in one category underwent changes in expression under similar mechanistic control.
Expression changes in gene clusters in the early phase (F1 to F2) of fibrosis
The functional categories showing a peak change in express-ion ratio in the early phase of fibrosis were inflammation, ECM, blood coagulation, lipid metabolism, half of the genes in steroid metabolism, half of those in energy source amino AA metabolism, and half of transcription factors. Expression of marker genes in the inflammation and ECM categories started to increase in the early phase. Expression of marker genes related to inflammation, such as LYZ, GZMB, IL1B, TNF and TGFB1, occurred in clusters and reached a peak at fibrotic stage F2, as shown in Figure 1A, suggesting that inflammatory events are particularly active at the F2 stage. However, histological classification of inflammatory activity shows a tendency for an increase in inflammation that is correlated with an increase in F stage; therefore, the conclusion regarding inflammatory events based on expression of marker genes appears to differ from that based on histological classification. Gene expression in the ECM category also increased until F3 or F4, as shown in Figure 1B; expression of such genes might indicate an inflammatory response for wound healing. Increased expression of some clustered genes related to the cell cycle, CCND1, FOXM1 and GJA1 (Connexin 43), was also found at an early stage, as shown in Figure 1C, and might reflect a response to hepatic cell injury.
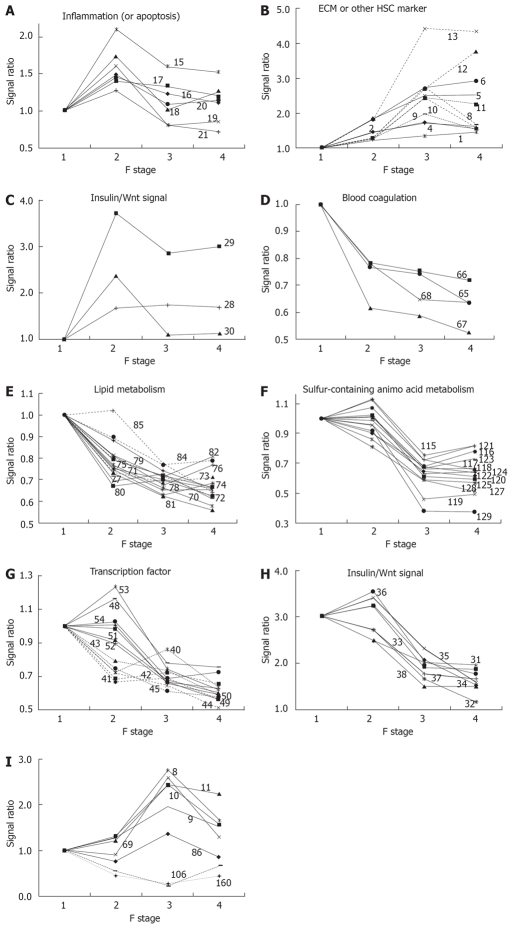
Figure 1.
Clusters of genes in functional categories. The characteristic behavior of gene clusters for each functional category is shown separately in Figure 1A-I. The numbers in each figure refer to the serial number of genes in Table 1. The expression ratio relative to the F1 stage is plotted in each graph. A: Inflammation gene cluster with a peak at F2; B: ECM gene cluster showing increased expression along with fibrosis progression; C: Insulin/Wnt signaling gene cluster with increased expression in the early phase of fibrosis; D: Blood coagulation factor gene cluster showing decreased expression along with fibrosis progression; E: Lipid metabolism gene cluster showing decreased expression along with fibrosis progression; F: Sulfurcontaining amino acid metabolism gene cluster showing a synchronous decrease in expression from F2 to F3; G: Two transcription factor gene clusters showing sequential decreases in expression; H: Insulin/Wnt signaling gene cluster showing decreased expression from F2 to F3; I: A gene cluster showing a peak or bottom at F3.
Almost all other genes were linearly down-regulated. Several genes related to blood coagulation, i.e. F10, AGT, FGA and PLG, were down-regulated as a cluster in the early phase, as shown in Figure 1D; this down-regulation may be linked to prolongation of the blood coagulation time in cirrhosis. An early response of many clustered genes associated with lipid metabolism was also found, as shown in Figure 1E. Expression of these genes decreased consecutively during fibrosis and the early response of genes affecting lipid metabolism is of interest.
Expression changes in gene clusters in the middle phase (F2 to F3) of fibrosis
The peak change in the expression ratio of marker genes in metabolism of sulfur-containing AA and aromatic AA was delayed, compared to genes associated with other kinds of metabolism. Marker genes for sulfur-containing AA metabolism decreased remarkably as a cluster in the phase from F2 to F3, as shown in Figure 1F. Decreased expression of marker genes for steroid metabolism, energy source AA, and transcription factors were separable into two groups: early-response and middle-response genes.
All the down-regulated transcription factors, including TCF1 (HNF-1), HNF4A (HNF-4), CEBPA (C/EBP alpha), CEBPB (C/EBP beta), PPARA (PPAR alpha), RXRA (RXR alpha), NR1H3 (LXRA), NR1H2 (LXRB), NR1H4 (FXR), USF-1, USF-2, and NR0B2 are important in hepatic metabolism and other regulatory mechanisms. Alteration of expression of these genes might be related to abnormal expression of metabolic enzymes. Two clusters of transcription factors were clearly distinguishable based on their expression pattern, as shown in Figure 1G: the first cluster, including HNF-4, C/EBPA, RXR, TCF1 (HNF1), PPARA, and NR0B2, which showed altered expression in the early phase, might influence expression of the second cluster, including CEBPB, NR1H2(LXRB), NR1H3(LXRA), NR1H4(FXR), ESRRA and USF2, which showed altered expression in the middle phase of fibrosis.
A cluster of genes associated with insulin/Wnt signaling were down-regulated, as shown in Figure 1H, suggesting a common regulatory mechanism. These expression changes are likely to be related to changes in expression of transcription factors and genes in metabolic networks. The down-regulated genes included GSK3B and CTNNB1 (catenin beta 1), which participates in Wg/Wnt signaling for regulation of cell proliferation and differentiation[4]; GJA1 (connexin 43), which forms gap junctions and is regulated by Wg/Wnt signaling[5]; and FOXM1, which is associated with cell proliferation[6] and liver regeneration[7]. All of these genes had peak expression at F2 in a clustered manner, as shown in Figure 1C and G. Enhancement of cell proliferation for wound healing might be linked with a peak of inflammation at F2, and expression of genes such as CCND1, GJA1 and FOXM1 in the downstream part of the insulin/Wnt pathway were altered ahead of genes involved in insulin/Wnt signaling. The relationship between these genes requires further study.
Expression changes in gene clusters in the late phase (F3 to F4) of fibrosis
Few genes showed altered expression in the late phase of fibrosis, but a cluster of genes in the glucose metabolism category showed decreased expression. It was also of interest that expression of several genes reversed direction or abruptly altered in the late phase, as shown in Figure 1I. These results suggest different biological changes from the start of the late stage in the transition from F3 to F4.
Serial expression changes for the functional categories are summarized in Table 3 and these data indicate associations between clustered genes in one category and inter-category relationships.
Table 3.
Classification of gene clusters based on serial alteration of gene expression along with the fibrosis progression
| F1-F2 | F2-F3 | F3-F4 |
| Inflammation | ||
| Wound-healing (ECM) | ||
| Blood coagulation | ||
| Transcription factors (cluster 1) | Transcription factors (cluster 2) | |
| Insulin/Wnt signal | ||
| Lipid metabolism | ||
| Steroid metabolism (cluster 1) | Steroid metabolism (cluster 1) | |
| Bile acid metabolism | ||
| Energy source AA metabolism (cluster 1) | Energy source AA metabolism (cluster 1) | |
| Aromatic AA metabolism | ||
| Sulfur-containing AA metabolism | ||
| Glucose metabolism (cluster 1) | Glucose metabolism (cluster 2) |
Gene clusters are indicated under the name of gene category. The pattern of serial alteration for gene cluster was determined based on the maximum changing point of the expression ratio shown in Table 1.
Down-regulated individual marker genes (group 1)
Regeneration of hepatic cells is suppressed during fibrosis and such suppression is thought to then cause further fibrosis. IGF1, GHR and IL6R (inflammation) support the regeneration of hepatocytes and down-regulation of the expression of these genes may be linked directly to formation of fibrosis. Down-regulation of PON1, which associates with HDL (high-density lipoprotein) and regulates the cellular redox state, and PPT, which is known as a lysosomal hydrolase of long chain fatty acyl CoA and has a role in maintenance of synaptic function, may be related to mitochondrial damage, as we and others have suggested[3,8,9]. RGN, a calcium-binding protein that plays a pivotal role in maintaining cell homeostasis and function, was also down-regulated. Down-regulation of these genes may impair liver function. TRIB3 inhibits Akt/PKB activation by insulin[10], and this gene was suddenly down-regulated from the F3 to F4 phase, suggesting new conditions in the insulin signaling pathway in the transition from F3 to F4.
Up-regulated individual marker genes (group 2)
Most inflammatory marker genes showed peak expression in the middle phase of fibrosis, as shown in Figure 1A, but CMA1 (chymase 1), which is produced by mast cells, underwent a linear increase in expression with progression of fibrosis. This is of note, since chymase has been reported to be involved in chronic hepatic fibrosis in autoimmune hepatitis and primary biliary cirrhosis[11], and mast cells may have a special role in fibrogenesis. The role of mast cells in chronic inflammation, however, deserves further study, because of the shortage of determined sample and low expression of CMA1.
Individual marker genes with peak expression in the middle phase of fibrosis (group 3)
The only hepatic stellate cell (HSC)-specific marker gene to show peak expression at the F2 stage was PRNP, which is reported to be a marker for the early phase of HSC activation. The amino acid transporters SLC38A2 (ATA2), SLC6A6 (TAUT) and SLC7A1 (CAT-1) showed peak expression at F2, which may also suggest enhancement of cell proliferation at this stage of fibrosis. Increased expression of SLC38A2, which preferentially transports alanine, has been reported in regeneration of hepatocytes[12], since hepatocytes require alanine as an energy source[13]. Based on our results, down-regulation of SLC38A2 in the late phase of fibrosis suggests that utilization of alanine as an energy source decreases at this stage of fibrosis. Up-regulation of SLC6A6, a taurine transporter, in the early phase of fibrosis can be understood as protective behavior against injury of hepatocytes[14]. FASN, a marker gene for fatty acid metabolism, showed peak expression at the F3 stage of fibrosis. We have observed suppression of biosynthesis and degradation of fatty acids in the liver in a CCl4-induced cirrhotic rat model, and clinical results show temporal enhancement of fatty acid biosynthesis before subsequent suppression in an advanced phase of fibrosis; this may be a compensative action related to suppression of other forms of energy metabolism in fibrosis, as we have previously suggested[3].
Candidate marker genes for diagnosis of fibrosis and discrimination of fibrotic stages
Diagnosis in the early stage of fibrosis may be important for monitoring progression of fibrosis and hepatic biological changes. Candidate marker genes at each step of fibrosis were selected based on an expression change ratio > 1.5 and an intensity > 10. Up-regulation of LYZ and down-regulation of FGA, OAT and AGXT2 were noteworthy in the transition from F1 to F2, and up-regulation of genes in the ECM category and down-regulation of genes in metabolism of energy source AA, aromatic AA, steroids and sulfur-containing AA occurred in the transition from F2 to F3. Diagnosis of the late stage of fibrosis (the transition from F3 to F4) is important because of the risk of tumorigenesis. Some genes showed a reversal of expression in the F3 to F4 transition, suggesting that biological changes in the stage from F3 to F4 are qualitatively different from those at earlier stages. Remarkable down-regulation of TRIB3, which inhibits insulin signaling and NFκB signaling, was noteworthy in the F3 to F4 transition. The reversal in expression of FASN in this phase may indicate changes in lipid metabolism and that of OAT indicates changes in ornithine metabolism or the urea cycle. The early increase in collagen expression began to decline in the F3 to F4 stage and the similar decline of SERPINE1 (Pai1) expression may reflect increased fibrolysis. Consecutive analysis of marker molecules in plasma will be important for monitoring progression of fibrosis at each step, and the data in this paper provides useful information for the selection of serum markers and interpretation of changes in the levels of these markers.
Coordinated regulation of functional categories
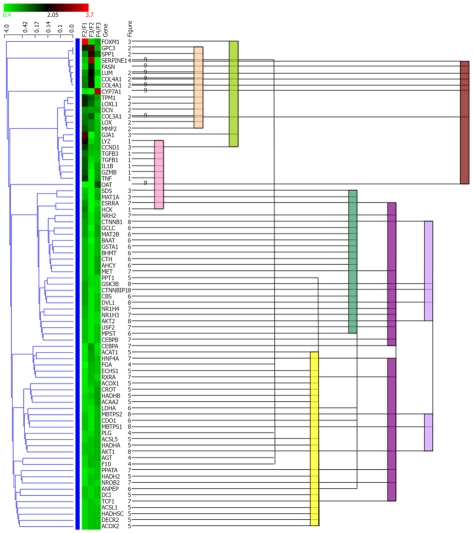
All genes in Figure 1 were subjected to hierarchical cluster-ing analysis. Statistical clustering of the expression ratio between neighboring F stages for the genes in Table 1 was combined with functional categories, as shown in Figure 2. Functional categories clearly corresponded to the statistical clusters, suggesting coordinated regulation of genes in one functional category. Overlap of functional categories in statistical clusters suggested that these categories might be regulated by correlated mechanisms. In contrast, separation of members of a category into multiple positions of a statistical cluster suggests that the functional category may be divided into subgroups with respect to regulation.
Figure 2.
Comparison between statistical clustering analysis and functional cluster. All genes in Figure 1 were subjected to hierarchical clustering analysis using Genowiz™. Statistical clustering of the expression ratio between neighboring F stages for the genes in Table 1 was combined with functional categories. The color coding in the center column shows the expression ratio between neighboring F stages for the genes in Table 1: Ratio increases from green to red. The left dendrogram shows the result of hierarchical clustering by Genowiz™ software. The numbers on the right of gene names indicate the subgraph numer in Figure 1, which represents functional categories, and the same numbers are tied with color bars.
DISCUSSION
Biological interpretation of changes in gene expression
Changes in expression of hepatic cell-specific marker genes reflects biological changes in the progression of hepatic fibrosis. Shimizu et al[15] reported co-localization of chymase with fibrotic tissue, and we have reported increased expression of marker genes for mast cells, including chymase, in progression of fibrosis in a DNM-induced rat fibrosis model. The results reported here also show correlation of the expression of chymase with the stage of fibrosis. Therefore, these results suggest that a certain cell population producing chymase has an important role in the pathogenesis of fibrosis.
Many marker genes related to inflammation showed peak expression at F2. Inflammation has been reported to induce activation of HSCs[16], and we have shown a peak in activated inflammatory cells in a DMN-induced fibrosis model. These results suggest that a temporal strong inflammatory reaction is required for initiation of auto-stimulatory activation of HSCs. In the DMN-induced fibrosis model, inflammation was suppressed after it reached a peak, but weaker inflammation might still be sufficient to maintain activation of HSCs.
Expression of HSC-specific marker genes such as COL1A1, LUM, TAGLN, MGP, SERPINE1 (PAI-1) and LOX did not increase or decrease continuously from F3 to F4, as shown in Figure 1B and I, whereas expression of DCN, BGN and GPC3 increased comparatively from F3 to F4, as shown in Figure 1B. This suggests some changes in the behavior of HSCs between F3 and F4, and it has been hypothesized that the perpetuation phase[17] may change to a new phase in fibrosis of stage F4. Therefore, profiles of HSC-specific marker genes may be useful for indication of the phase of fibrosis.
A cluster of down-regulated genes associated with sulfur-containing AA metabolism is shown in Figure 1F. Changes in expression of the clustered genes occurred simultaneously between the F2 and F3 stages; this transition may reflect a biological change causing fibrotic progression, and the change in expression of the clustered genes seems to suggest an increased demand for glutathione[18]. Abnormalities in metabolism of sulfur-containing AA in thioacetamide-induced cirrhosis in rat liver have been found in a proteomics analysis[19], and similar abnormalities were also reported based on hepatic gene expression changes in patients with alcoholic hepatitis[20].
There is only limited information on altered expression of transcription factors in fibrotic liver, but down-regulation of HNF-4 in human cirrhosis[21] and of PPARs in hepatitis C virus genotype 3[22] have been reported. C/EBP alpha and C/EBP beta regulate proliferation of hepatocytes[23] and glucose and lipid homeostasis[24–27], and HNF-1 and HNF-4 broadly regulate hepatic functions such as carbohydrate metabolism[28], lipid metabolism[29], bile acid metabolism and HDL-cholesterol metabolism[30]; expression of HNF-1 is also regulated by HNF-4[31]. USF1 and USF2 have been reported as glucose signals[32,33], and the USF1 and USF2 homodimers and the USF1-USF2 heterodimer regulate expression of liver-specific genes such as apolipoprotein A2 and pyruvate kinase. HNF-4 and USF2a bind to the enhancer sequence cooperatively[34]. HNF-4 also regulates PPAR alpha[35], which in turn regulates glucose[36], lipid[37] and cholesterol metabolism[38]. RXR alpha regulates lipid, bile acid and cholesterol homeostasis[39], and LXR alpha, LXR beta and FXR are associated with lipid[40,41], bile acid and cholesterol homeostasis[40,42]. RXR and FXR form heterodimers with other transcription factors, including other members of the same family or with PPAR alpha[40,43,44]. NR0B2 (SHP) regulates cholesterol metabolism[45], glucose metabolism[45] and bile acid synthesis[46], and interacts with LXR[47]. AKT1 and AKT2 are important kinases in the pathway of insulin regulation of glucose homeostasis[48] and in fatty acid synthesis[49]. Hence, the altered expression of these transcription factors may relate to altered expression of metabolic enzymes in glucose and lipid metabolism in the fibrotic liver of hepatitis C patients.
The continuous increase in expression of cyclin D1 (CCND1) correlated with F stage, as shown in Figure 1C, and appears to be important for hepatic tumorigenesis. The association of Wnt signaling[50,51] and cyclin D[52] with tumorigenesis is well known. Catenin beta 1 (CTNNB1) regulates cyclin D expression[53] and is itself regulated through phosphorylation by GSK3B[54] or other kinases[55]. Down-regulation of expression of CTNNB1 occurred in advanced fibrotic stages, as shown in Figure 1H, and beta interacting protein 1 (CTNNBIP1), which is a negative regulator of CTNNB1, was also down-regulated, as also shown in Figure 1H. From this perspective, it was interesting that alterations in expression of genes associated with insulin signaling, including GSK3B, CTNNB1, CTNNBIP1 and downstream genes such as CCND1 and GJA1 (connexin 43), were clustered, as shown in Figures 1C and H. Suppression of insulin signaling has been reported in cirrhosis[56]; however, the response of downstream genes in this pathway was inconsistent with suppression of the insulin signal. Therefore, further studies will be necessary to clarify whether this inconsistency arises from differences between expression levels and the activity of the protein products of these genes, or if another signal[57] is involved in the Wnt and insulin signaling pathways.
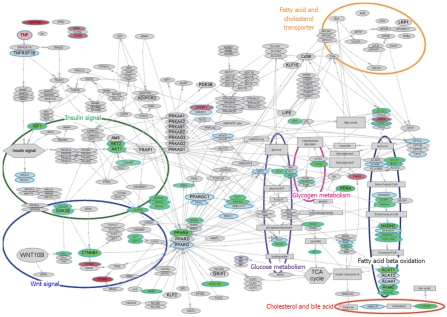
The bioSpace Explorer is a system for analysis of DNA microarray data that may allow an understanding of the molecular relationships underlying the above results. The network of genes from previous reports revealed integral relationships among insulin/Wnt signal, CEBPA, PPARA, RXRA, glucose metabolism, lipid synthesis and lipid metabolism. An initial version of bioSpace Explorer was constructed based on molecular networks related to lipid metabolism, with entries showing relationships between molecules via a line between the molecules. This bird’s eye view of the pathway including lipid metabolism with the input PCR data is illustrated in Figure 3; molecules related to inflammation were up-regulated and many genes related to lipid metabolism were down-regulated.
Figure 3.
Molecular network associated with lipid metabolism. Gene expression changes in pathways related to lipid metabolism, illustrated with bioSpace Explorer (a system for analysis of DNA microarray data for lipid metabolism; Pharma Frontier Co. Ltd / World Fusion Co. Ltd.; see texts for details). The bird’s-eye view of the lipid metabolism is displayed. The up-regulated and down-regulated gene expression ratios at F3 vs F1 in Table 1 are displayed in red and green, respectively, with the color gradation proportional to the ratio. Genes in Table 1 that did not show a statistically significant change in expression are indicated with a blue circle with gray background. An entry with a gray background only indicates no input data. Most of the entity names in Figure 2 are the same as the gene name in Table 1, but the names “C20orf97”, “SCEH”, “Acaa2”, and “ACS5” in Figure 2 refer to “TRIB3”, “ECHS1”, “ACAA2”, and “ACSL5”, respectively, in Table 1. These differences are due to the software used in bioSpace Explorer.
Hypothetical causes of biological changes in progression of fibrosis
Biological changes in fibrosis can be summarized as follows. Initially, Kupffer cells or other inflammatory cells are activated in the transition from F1 to F2. This event immediately influences production of ECM and cell cycle genes for wound-healing[58]. Blood coagulation is quickly suppressed in moving from F1 to F2, as shown by down-regulation of coagulation factor genes and up-regulation of the inhibitor, PAI-1. Several transcription factor genes are also immediately influenced, probably due to inflammation, as shown by the down-regulation of CEBPA, HNF4A, TCF1 and NR0B2. Expression of many genes associated with lipid metabolism also changed quickly in the transition from F1 to F2. Down-regulation of these genes may be controlled by down-regulation of the transcription factors, especially RXRA, PPARA, LXRs and FXR. Some genes related to steroid metabolism also responded quickly for control of inflammation.
Expression of genes associated with sulfur-containing AA metabolism and aromatic AA metabolism changed simultaneously in the transition from F2 to F3. The first type of metabolism relates to the redox state[59] and the second is associated with production of active metabolites such as catecholamines and serotonin. Such important biological states are controlled to maintain homeostasis through several mechanisms[60] and this may explain the delayed change in expression of these genes. The expression of many genes related to intracellular signaling, including insulin/Wnt signaling, also changed simultaneously in the transition from F2 to F3. This delayed change may also reflect compensative action for hepatic cellular defects on metabolism for energy supply and/or hepatic cellular proliferation. The main molecules in fibrosis, such as collagens, increase in expression from F2 to F3 and cause development of fibrosis through activation of HSCs through a stimulatory cycle involving inflammatory cells, HSCs and hepatocytes, as described previously[3].
Some quantitative biological changes started in the transition from F3 to F4. Down-regulation of genes associated with sugar metabolism and fatty acid synthesis at this stage might induce persistent defects in energy storage and supply to the liver. The liver transits into an inescapable negative cycle between this defect in the hepatic energy state and mitochondrial damage in cirrhosis. This negative cycle will be discussed in a future paper describing DNA microarray analysis of an animal model of cirrhosis.
Coordinated regulation of functional categories
Statistical cluster analysis showed coordinated regulation of functional categories in liver fibrosis. These regulatory mechanisms can be examined prospectively using bioSpace Explorer, and these results will be discussed in a future paper. In the current work, the clinical gene expression profiles assembled using RT-PCR, using genes originally selected based on DNA microarray data from an experimental animal model, provided an improved understanding of disease and suggested new methods of diagnosis. Therefore, statistical analysis and functional clustering or network analysis of the transcriptome, alone or in combination, can provide an overview of a dynamic biological system.
COMMENTS
Background
Information from clinical specimens is very important, but there is a limitation in sample preparation. With DNA microarray, it is difficult to determine gene expression profile from a small amount of sample such as a clinical needle biopsy sample. RT-PCR with TaqMan probe can make it possible with high quality. We have to get maximum information with a minimum number of TaqMan probes because the amount of samples is limited and probes are expensive. Selection of probes is a key factor. We extensively determined gene expression profiles from animal models of liver fibrosis with DNA microarray (WJG 2006; 12: 6473). The gene marker sets were arranged to show the change of gene expression of a molecular network or functionally clustered gene sets. In this paper, the selected gene marker sets effectively showed the dynamic behavior of global gene network change during liver fibrosis.
Research frontiers
Dynamic behavior of genome-wide genes expression is now measured with DNA microarray. This technology must be applied to clinical samples. Such information can greatly advance the study of disease pathogenesis, diagnosis and therapy. One problem is the interpretation of huge expression profile data from DNA microarray. Advanced technology of computational text-mining has recently shown the genome-wide molecular networks or functional molecular clusters. This genome-wide network is going to be applied to the analysis of DNA microarray data. When expression profile data are arranged as a change in their networks of functional clusters, these huge data are expected to suggest effectively the biological meaning in terms of broad aspects of research interest. Therefore, we are developing an analysis system genome-wide molecular network which was made possible by a combination system based on both computational and manual text-mining. This system was partially applied on the interpretation of gene expression profile in this paper. Prevention therapy for individual patients at an early stage is required because genomic polymorphism is going to reveal the personal risk of diseases. The biological background of progression to disease onset must be understood for development of prevention therapy. Prediction of liver cancer risk is going to be tried in the early stage of liver fibrosis before cirrhosis. The accumulation process of hepatic stress which leads to onset of liver cancer has to be elucidated. For example, there is a question why BCAA (branched chain amino acids), which improves hepatic metabolism, reduces onset of liver cancer.
Innovations and breakthroughs
We have already prepared the gene marker sets which can show the change of molecular networks or functionally clustered genes by DNA microarray experiment on liver fibrosis of animal models. The gene marker sets were linked to biological events in each hepatic cell such as hepatocytes, immune cells and stellate cells. Application of gene marker sets and RT-PCR with TaqMan probe technology on clinical specimens successfully showed the serial change of gene expression in molecular network or functionally clustered gene sets in the progression of liver fibrosis. It was proved that network analysis is a powerful tool for biological research. Our sequential approach (animal model/DNA microarray → selection of appropriate gene marker sets in molecular networks → clinical samples/RT-PCR → analysis functionally clustered gene markers in molecular networks → analysis of the relation between molecular networks) can effectively advance clinical research. Serial change of clustered gene expression during liver fibrosis progression, which was made clear in this paper, will reveal the molecular mechanism of many symptoms before and after the onset of cirrhosis and liver cancer.
Application
Gene marker sets and RT-PCR on clinical specimens as well as analytical methods with genome-wide gene networks can be applied to get the information of dynamic biological progression on various diseases. Serial change of clustered gene expression during liver fibrosis progression can provide a method of diagnosis and therapy in liver fibrosis. For example, the question why BCAA, which improves hepatic metabolism, reduces onset of liver cancer, will be solved.
Terminology
Text-mining: all published information about molecular interaction and their function are collected and arranged to make new valuable information such as gene-wide molecular networks; TaqMan-PCR probe: Applied Biosystems offers more than 700 000 TaqMan® Gene Expression Assays, the most comprehensive set of pre-designed Real-Time PCR assays available. All TaqMan® Gene Expression Assays have been designed using validated bioinformatics pipeline of Applied Biosystems, and run with the same PCR protocol, eliminating the need for primer design or PCR optimization.
Peer review
This paper revealed that metabolic deficiency occurs before the onset of cirrhosis. It had already been found in animal models with hepatic toxic substances in the preceding paper. Metabolic deficiency in hepatitis which was caused by a virus was found to be the same as animal models in this paper. Gene marker sets, selected from analysis of animal models, and analysis methods using molecular networks can lead to success in finding the serial change of functionally clustered genes expression during liver fibrosis progression. A set of appropriate gene markers in each network was a key to analysis. The sequential approach (animal model/DNA microarray → appropriate gene marker sets in molecular networks → clinical samples/RT-PCR → analysis functionally clustered gene markers in molecular network → analysis of the relation between molecular networks) is useful to elucidate the molecular mechanism of disease.
Acknowledgments
We thank Ms. Tomoko Kobayashi of Pharmafrontier Co. Ltd. and Mr. Ken Osaki of World Fusion Co. Ltd for statistical analysis and operation of the bioSpace Explorer for analysis of DNA microarray data for lipid metabolism.
Peer reviewer: Richard A Rippe, Dr, Department of Medicine, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7038, United States
S- Editor Liu Y L- Editor Roberts SE E- Editor Liu Y
References
- 1.Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. The French METAVIR Cooperative Study Group. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- 2.Gobel T, Vorderwulbecke S, Hauck K, Fey H, Haussinger D, Erhardt A. New multi protein patterns differentiate liver fibrosis stages and hepatocellular carcinoma in chronic hepatitis C serum samples. World J Gastroenterol. 2006;12:7604–7612. doi: 10.3748/wjg.v12.i47.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahara Y, Takahashi M, Wagatsuma H, Yokoya F, Zhang QW, Yamaguchi M, Aburatani H, Kawada N. Gene expression profiles of hepatic cell-type specific marker genes in progression of liver fibrosis. World J Gastroenterol. 2006;12:6473–6499. doi: 10.3748/wjg.v12.i40.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dierick H, Bejsovec A. Cellular mechanisms of wingless/Wnt signal transduction. Curr Top Dev Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- 5.Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, Costa RH. Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 2003;11:149–162. doi: 10.3727/000000003108749044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferre N, Marsillach J, Camps J, Mackness B, Mackness M, Riu F, Coll B, Tous M, Joven J. Paraoxonase-1 is associated with oxidative stress, fibrosis and FAS expression in chronic liver diseases. J Hepatol. 2006;45:51–59. doi: 10.1016/j.jhep.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Kim SJ, Zhang Z, Lee YC, Mukherjee AB. Palmitoyl-protein thioesterase-1 deficiency leads to the activation of caspase-9 and contributes to rapid neurodegeneration in INCL. Hum Mol Genet. 2006;15:1580–1586. doi: 10.1093/hmg/ddl078. [DOI] [PubMed] [Google Scholar]
- 10.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 11.Satomura K, Yin M, Shimizu S, Kato Y, Nagano T, Komeichi H, Ohsuga M, Katsuta Y, Aramaki T, Omoto Y. Increased chymase in livers with autoimmune disease: colocalization with fibrosis. J Nippon Med Sch. 2003;70:490–495. doi: 10.1272/jnms.70.490. [DOI] [PubMed] [Google Scholar]
- 12.Fowler FC, Banks RK, Mailliard ME. Characterization of sodium-dependent amino acid transport activity during liver regeneration. Hepatology. 1992;16:1187–1194. [PubMed] [Google Scholar]
- 13.Freeman TL, Ngo HQ, Mailliard ME. Inhibition of system A amino acid transport and hepatocyte proliferation following partial hepatectomy in the rat. Hepatology. 1999;30:437–444. doi: 10.1002/hep.510300212. [DOI] [PubMed] [Google Scholar]
- 14.Warskulat U, Borsch E, Reinehr R, Heller-Stilb B, Monnighoff I, Buchczyk D, Donner M, Flogel U, Kappert G, Soboll S, et al. Chronic liver disease is triggered by taurine transporter knockout in the mouse. FASEB J. 2006;20:574–576. doi: 10.1096/fj.05-5016fje. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu S, Satomura K, Aramaki T, Katsuta Y, Takano T, Omoto Y. Hepatic chymase level in chronic hepatitis: co-localization of chymase with fibrosis. Hepatol Res. 2003;27:62–66. doi: 10.1016/s1386-6346(03)00194-3. [DOI] [PubMed] [Google Scholar]
- 16.Baroni GS, Pastorelli A, Manzin A, Benedetti A, Marucci L, Solforosi L, Di Sario A, Brunelli E, Orlandi F, Clementi M, et al. Hepatic stellate cell activation and liver fibrosis are associated with necroinflammatory injury and Th1-like response in chronic hepatitis C. Liver. 1999;19:212–219. doi: 10.1111/j.1478-3231.1999.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 17.Gressner AM. Transdifferentiation of hepatic stellate cells (Ito cells) to myofibroblasts: a key event in hepatic fibrogenesis. Kidney Int Suppl. 1996;54:S39–S45. [PubMed] [Google Scholar]
- 18.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 19.Low TY, Leow CK, Salto-Tellez M, Chung MC. A proteomic analysis of thioacetamide-induced hepatotoxicity and cirrhosis in rat livers. Proteomics. 2004;4:3960–3974. doi: 10.1002/pmic.200400852. [DOI] [PubMed] [Google Scholar]
- 20.Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2004;28:173–181. doi: 10.1097/01.ALC.0000108654.77178.03. [DOI] [PubMed] [Google Scholar]
- 21.Berasain C, Herrero JI, Garcia-Trevijano ER, Avila MA, Esteban JI, Mato JM, Prieto J. Expression of Wilms' tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology. 2003;38:148–157. doi: 10.1053/jhep.2003.50269. [DOI] [PubMed] [Google Scholar]
- 22.de Gottardi A, Pazienza V, Pugnale P, Bruttin F, Rubbia-Brandt L, Juge-Aubry CE, Meier CA, Hadengue A, Negro F. Peroxisome proliferator-activated receptor-alpha and -gamma mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment Pharmacol Ther. 2006;23:107–114. doi: 10.1111/j.1365-2036.2006.02729.x. [DOI] [PubMed] [Google Scholar]
- 23.Greenbaum LE, Cressman DE, Haber BA, Taub R. Coexistence of C/EBP alpha, beta, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. Implications for maintenance of the differentiated state during liver growth. J Clin Invest. 1995;96:1351–1365. doi: 10.1172/JCI118170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arizmendi C, Liu S, Croniger C, Poli V, Friedman JE. The transcription factor CCAAT/enhancer-binding protein beta regulates gluconeogenesis and phosphoenolpyruvate carboxykinase (GTP) gene transcription during diabetes. J Biol Chem. 1999;274:13033–13040. doi: 10.1074/jbc.274.19.13033. [DOI] [PubMed] [Google Scholar]
- 25.Gautier-Stein A, Mithieux G, Rajas F. A distal region involving hepatocyte nuclear factor 4alpha and CAAT/enhancer binding protein markedly potentiates the protein kinase A stimulation of the glucose-6-phosphatase promoter. Mol Endocrinol. 2005;19:163–174. doi: 10.1210/me.2004-0105. [DOI] [PubMed] [Google Scholar]
- 26.Qiao L, MacLean PS, You H, Schaack J, Shao J. knocking down liver ccaat/enhancer-binding protein alpha by adenovirus-transduced silent interfering ribonucleic acid improves hepatic gluconeogenesis and lipid homeostasis in db/db mice. Endocrinology. 2006;147:3060–3069. doi: 10.1210/en.2005-1507. [DOI] [PubMed] [Google Scholar]
- 27.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 28.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J Am Soc Nephrol. 2000;11 Suppl 16:S140–S143. [PubMed] [Google Scholar]
- 29.Louet JF, Hayhurst G, Gonzalez FJ, Girard J, Decaux JF. The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 alpha and cAMP-response element-binding protein (CREB) J Biol Chem. 2002;277:37991–38000. doi: 10.1074/jbc.M205087200. [DOI] [PubMed] [Google Scholar]
- 30.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 31.Miura N, Tanaka K. Analysis of the rat hepatocyte nuclear factor (HNF) 1 gene promoter: synergistic activation by HNF4 and HNF1 proteins. Nucleic Acids Res. 1993;21:3731–3736. doi: 10.1093/nar/21.16.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 33.Kahn A. From the glycogenic function of the liver to gene regulation by glucose. C R Seances Soc Biol Fil. 1998;192:813–827. [PubMed] [Google Scholar]
- 34.Moriizumi S, Gourdon L, Lefrancois-Martinez AM, Kahn A, Raymondjean M. Effect of different basic helix-loop-helix leucine zipper factors on the glucose response unit of the L-type pyruvate kinase gene. Gene Expr. 1998;7:103–113. [PMC free article] [PubMed] [Google Scholar]
- 35.Pineda Torra I, Jamshidi Y, Flavell DM, Fruchart JC, Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol Endocrinol. 2002;16:1013–1028. doi: 10.1210/mend.16.5.0833. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Chang V, Joseph SB, Trujillo C, Bassilian S, Saad MF, Lee WN, Kurland IJ. Peroxisomal proliferator-activated receptor alpha deficiency diminishes insulin-responsiveness of gluconeogenic/glycolytic/pentose gene expression and substrate cycle flux. Endocrinology. 2004;145:1087–1095. doi: 10.1210/en.2003-1173. [DOI] [PubMed] [Google Scholar]
- 37.Lee SS, Chan WY, Lo CK, Wan DC, Tsang DS, Cheung WT. Requirement of PPARalpha in maintaining phospholipid and triacylglycerol homeostasis during energy deprivation. J Lipid Res. 2004;45:2025–2037. doi: 10.1194/jlr.M400078-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. "New" hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Wan YJ, Cai Y, Lungo W, Fu P, Locker J, French S, Sucov HM. Peroxisome proliferator-activated receptor alpha-mediated pathways are altered in hepatocyte-specific retinoid X receptor alpha-deficient mice. J Biol Chem. 2000;275:28285–28290. doi: 10.1074/jbc.M000934200. [DOI] [PubMed] [Google Scholar]
- 40.Edwards PA, Kennedy MA, Mak PA. LXRs; oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vascul Pharmacol. 2002;38:249–256. doi: 10.1016/s1537-1891(02)00175-1. [DOI] [PubMed] [Google Scholar]
- 41.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 42.Lambert G, Amar MJ, Guo G, Brewer HB Jr, Gonzalez FJ, Sinal CJ. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J Biol Chem. 2003;278:2563–2570. doi: 10.1074/jbc.M209525200. [DOI] [PubMed] [Google Scholar]
- 43.Vidal H. PPAR receptors: recent data. Ann Endocrinol (Paris) 2005;66:1S5–1S9. [PubMed] [Google Scholar]
- 44.Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HJ, Kim JY, Kim JY, Park SK, Seo JH, Kim JB, Lee IK, Kim KS, Choi HS. Differential regulation of human and mouse orphan nuclear receptor small heterodimer partner promoter by sterol regulatory element binding protein-1. J Biol Chem. 2004;279:28122–28131. doi: 10.1074/jbc.M313302200. [DOI] [PubMed] [Google Scholar]
- 46.Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol. 2002;16:2065–2076. doi: 10.1210/me.2001-0194. [DOI] [PubMed] [Google Scholar]
- 48.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 49.Ono H, Shimano H, Katagiri H, Yahagi N, Sakoda H, Onishi Y, Anai M, Ogihara T, Fujishiro M, Viana AY, et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52:2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- 50.Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, Giovannini M, Perret C. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci USA. 2004;101:17216–17221. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murata M, Miyoshi Y, Ohsawa M, Shibata K, Ohta T, Imai Y, Nishikawa M, Iwao K, Tateishi H, Shimano T, et al. Accumulation of beta-catenin in the cytoplasm and the nuclei during the early hepatic tumorigenesis. Hepatol Res. 2001;21:126–135. doi: 10.1016/s1386-6346(01)00116-4. [DOI] [PubMed] [Google Scholar]
- 52.Yamaoka H, Ohtsu K, Sueda T, Yokoyama T, Hiyama E. Diagnostic and prognostic impact of beta-catenin alterations in pediatric liver tumors. Oncol Rep. 2006;15:551–556. [PubMed] [Google Scholar]
- 53.Kolligs FT, Bommer G, Goke B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002;66:131–144. doi: 10.1159/000066755. [DOI] [PubMed] [Google Scholar]
- 54.Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971–9976. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- 56.Picardi A, D'Avola D, Gentilucci UV, Galati G, Fiori E, Spataro S, Afeltra A. Diabetes in chronic liver disease: from old concepts to new evidence. Diabetes Metab Res Rev. 2006;22:274–283. doi: 10.1002/dmrr.636. [DOI] [PubMed] [Google Scholar]
- 57.Gotoh J, Obata M, Yoshie M, Kasai S, Ogawa K. Cyclin D1 over-expression correlates with beta-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3 beta and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis. 2003;24:435–442. doi: 10.1093/carcin/24.3.435. [DOI] [PubMed] [Google Scholar]
- 58.Kershenobich Stalnikowitz D, Weissbrod AB. Liver fibrosis and inflammation. A review. Ann Hepatol. 2003;2:159–163. [PubMed] [Google Scholar]
- 59.Nagahara N, Katayama A. Post-translational regulation of mercaptopyruvate sulfurtransferase via a low redox potential cysteine-sulfenate in the maintenance of redox homeostasis. J Biol Chem. 2005;280:34569–34576. doi: 10.1074/jbc.M505643200. [DOI] [PubMed] [Google Scholar]
- 60.Fitzpatrick PF. The aromatic amino acid hydroxylases. Adv Enzymol Relat Areas Mol Biol. 2000;74:235–294. doi: 10.1002/9780470123201.ch6. [DOI] [PubMed] [Google Scholar]