Abstract
Phenanthroindolizidine-based tylophora alkaloids have been reported to have potential antitumor, anti-immuno and anti-inflammatory activity. The structure-activity relationships of a series of tylophora alkaloids were studied to guide future drug design. Our results indicate that although these compounds are structural analogs, their potency of cytotoxicity, selectivity against NF-κB signaling pathway, and their inhibitory effects against protein and nucleic acid synthesis are different. Because they do not have an identical spectrum of targets, the studied compounds are structural, but may not be functional analogs.
Keywords: Tylophora alkaloids, Structure, activity relationship, Protein, DNA and RNA synthesis
Tylophora alkaloids originate from various plants of the Asclepiadaceae family, such as Tylophora, that are native of India and Southeast Asia.1 They have antitumor,2–6 anti-inflammatory,7 anti-arthritis,8 and anti-lupus activity in vivo.9 Due to their diverse and potent pharmacological actions, they continue to be targets for synthesis, modification, structure-activity relationship (SAR) studies since their first isolation in 1935.10 Their molecular mechanisms of action of antitumor and anti-inflammatory activity include: inhibitory effect on protein synthesis2 and nucleic acid synthesis,2,11 inhibitory effect on RNA transcription which are controlled by cyclic AMP response elements (CREs), activator protein-1 (AP-1) sites, and NF-κB binding sites,6 and ability to suppress the expression of a subset of proteins, such as cyclin D1, cyclin B1 and CDK4.12 Tylocrebrine, a positional isomer of tylophorine, was found in clinical trials to have intolerable central nervous system (CNS) side effects. To offset such CNS side effects, a series of phenanthrene–based tylophorine derivatives (PBTs) with increased polarity was synthesized to limit the crossing of the blood-brain barrier.13 It was assumed that these analogs would have the same mechanism of action and behave as functional analogs of the tylophora alkaloids.
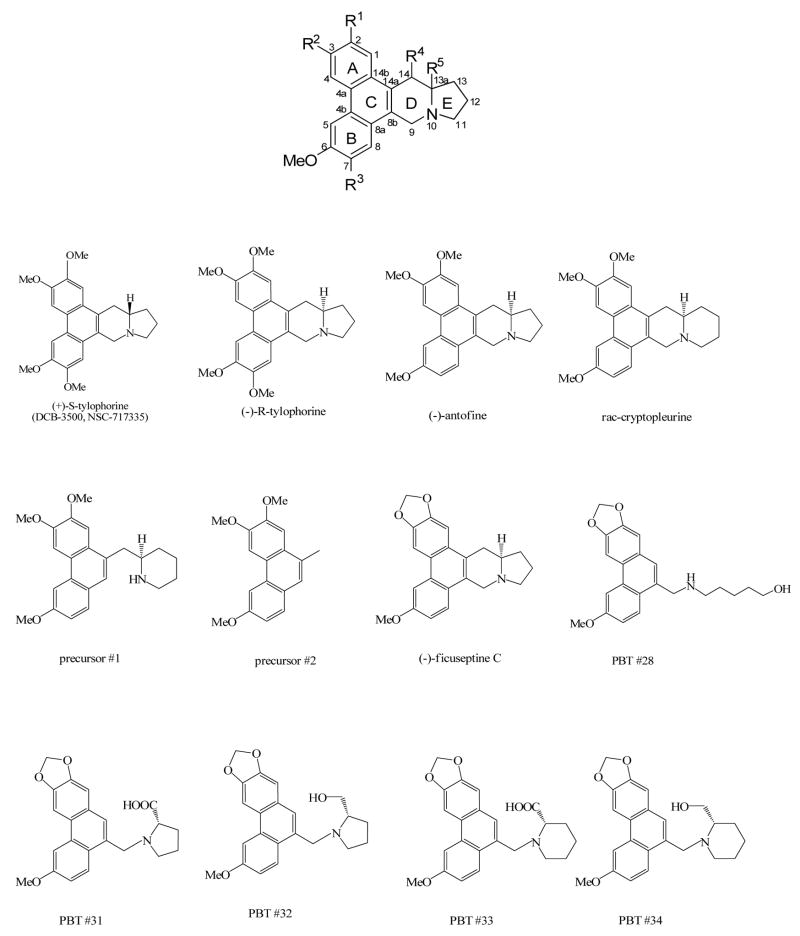
Previous structure–activity relationships of phenanthroindolizidine alkaloids have concluded that the rigid phenanthrene structure (Figure 1) on the phenanthrene ring is required to maintain potent cytotoxicity, and that the lack of an indolizidine ring or the presence of OMe ether at position 2 leads to the loss of cytotoxicity.14–16 To continue the structure–activity relationship studies using the tylophora alkaloids rac-cryptopleurine, (−)-antofine, (−)-tylophorine, and (−)-ficuseptine C recently obtained by a concise and modular total synthesis approach,17 and to verify whether the synthetic PBT compounds are functional analogs of the tylophora alkaloids, we compared their cytotoxicity, inhibitory effects on NF-κB, AP-1, CRE and glucocorticoid response element (GRE), and some of their impact on protein, DNA and RNA synthesis. We hypothesized that opening of the D-ring would have affect the potency; if the PBT compounds are functional tylophora alkaloid analogs, they might have comparable effects on the different intracellular pathways. Phenanthrene-based derivatives PBTs #28 and #31–34 were synthesized by Dr. K. H. Lee’s group.13 The tested tylophora alkaloids, rac-cryptopleurine, (−)-antofine, (−)-tylophorine, (−)-ficuseptine C, precursors #1 and precursors #2 were provided by Dr. Alois Fürstner.17 (+)-S-Tylophorine (DCB-3500) was synthesized by Dr. David C. Baker’s laboratory. 16
Figure 1.
The chemical structures of tylophora alkaloids, and phenanthrene-based tylophorine derivatives.
To analyze the structure-activity relationships of tylophora alkaloids, we first compared their cytotoxicity in HepG2, PANC-1, and CEM cells, followed by the procedures described previously.6,16 (+)-(S)-tylophorine and (−)-(R)-tylophorine differ only in the absolute configuration of the chiral center at the 13a position, the (R)-configured compound led to an approximately 3~4-fold decrease in cytotoxicity (Table 1). The difference in the structures of (−)-antofine and (−)-(R)-tylophorine is at the R3 position (Figure 1). The absence of an OMe substituent at R3 led to a 6-fold increase in cytotoxicity in HepG2 cells, and more than 10-fold increase in cytotoxicity in PANC-1, and CEM cells (Table 1). The structural difference between (−)-antofine and rac-cryptopleurine lies in the size of the E-ring. The presence of the six-membered E-ring in the phenanthroquinolizidine skeleton of rac-cryptopleurine led to a 2~4-fold increase in cytotoxicity in comparison to (−)-antofine which has a five-membered Ering (1.5±0.2 nM vs. 4.9±0.4 nM in HepG2; 0.5±0.1 nM vs. 2.2±0.3 nM in PANC-1; and 2±0.5 nM vs. 5.2±0.5 nM in CEM as shown in Table 1). In contrast, precursor #1 and #2 were the least potent compounds among the tylophora-related alkaloids listed in Table 1. The structural difference between rac-cryptopleurine and precursor #1 is that the D-ring is formally opened in precursor #1, which leads to a dramatic decrease of cytotoxicity. For instance, in HepG2 cells, the GI50 value of rac-cryptopleurine is 1.5±0.2 nM, whereas the GI50 value of precursor #1 is 1723±417 nM. This represents a more than 1000-fold decrease of cytotoxicity. Precursor #2 represents the bare phenanthrene backbone of the tylophora alkaloids lacking the fused aliphatic heterocyclic domain. This structural change is not tolerated and results in very poor cytotoxicity, with a GI50 value of over 10 μM in HepG2 and PANC-1 cells (Table 1). The difference in the structures of (−)-ficuseptine C and (−)-antofine consists in the replacement of the OMe groups at R1 and R2 in (−)-antofine by a cyclic methylenedioxy unit in (−)-ficuseptine C. This structural change resulted in a more than 60-fold decrease of cytotoxicity in HepG2, PANC-1 and CEM cells (Table 1, compare 371±27 nM vs. 4.9±0.4 nM in HepG2; 156±26 nM vs. 2.2±0.3 nM in PANC-1; and 323±13 nM vs. 5.2±0.5 nM in CEM). Other phenanthrene-based tylophorine derivatives were designed to increase the polarity as mentioned previously which was achieved by formal opening of the indolizidine.13 However, this structural modification dramatically impaired the cytotoxicity. As shown in Table 1 (lower panel), the GI50s of all PBTs tested is between 1.6 to 8.6 μM in HepG2, PANC-1, and CEM cells.
Table 1.
The GI50 of tylophora alkaloids, and PBTs on the growth inhibition of HepG2, PANC-1 and CEM cells.
| HepG2 GI50a (nM) |
PANC-1 GI50a (nm) |
CEM GI50a (nm) |
|
|---|---|---|---|
| (+)-(S)-tylophorine | 11 ± 4b | 12 ± 2 | 15 ± 3 |
| (−)-(R)-tylophorine | 33 ± 2 | 29 ± 6 | 67 ± 6 |
| (−)-antofine | 4.9 ± 0.4 | 2.2 ± 0.3 | 5.2 ± 0.5 |
| Rac-cryptopleurine | 1.5 ± 0.2 | 0.5 ± 0.1 | 2 ± 0.5 |
| precursor #1 | 1723 ± 417 | 904 ± 297 | 567 ± 58 |
| precursor #2 | >10000 | >10000 | >2000 |
| (−)-ficuseptine C | 371 ± 27 | 156 ± 26 | 323 ± 13 |
|
| |||
| HepG2 GI50a (μM) |
PANC-1 GI50a (μM) |
CEM GI50a (μM) |
|
|
| |||
| PBT #28 | 2.3 ± 0.3 | 2.2 ± 0.3 | 2.0 ± 0.3 |
| PBT #31 | 8.6 ± 3.6 | 7.4 ± 0.3 | 6.2 ± 0.2 |
| PBT #32 | 5.4 ± 1.4 | 6.2 ± 0.7 | 2.7 ± 0.1 |
| PBT #33 | 3 ± 0.7 | 2.3 ± 0.2 | 1.7 ± 0.3 |
| PBT #34 | 3.1 ± 1.0 | 2.3 ± 0.2 | 1.6 ± 0.5 |
Values are means ± SD of three experiments, with each data point done in triplicate.
Published.6
Previously, we had shown that DCB-3503 and its analogs could inhibit NF-κB, CRE and AP-1 mediated transcription, more selectively against NF-κB signaling pathway.6 NF-κB is a family of transcription factors that play pivotal roles in chronic and acute inflammatory diseases, autoimmune diseases and different types of cancer.18 Recently, NF-κB and the signaling pathways that regulate its activity have become a focus of intense drug discovery and development efforts.18 The potent inhibitory effects of DCB-3503 on NF-κB-mediated transcription may be one of the mechanisms of its antitumor activity. Here we compared the effects of the tylophora alkaloids (Figure 1) against NF-κB, CRE, AP-1, and GRE mediated transcription. HepG2-NF-κB-luc, HepG2-CRE-luc and HepG2-AP-1-luc stable cell lines were generated as previously described.16 GRE mediated transcription was analyzed by transient transfection of HepG2 cells with pGRE-luc (Clontech). The IC50 of the inhibitory effect of the tylophora alkaloids against stimulator induced NF-κB, CRE, AP-1, GRE activity is listed in Table 2. The IC50 of the inhibitory effect of tylophora alkaloids against endogenous NF-κB, CRE, AP-1, and GRE mediated transcription is shown in “Supplementary data Table S1”. Among the four signaling pathways, NF-κB is the most sensitive signaling pathway inhibited by all the tylophora alkaloids (listed in Table 2, upper panel) except precursor #2. For instance, the IC50 of (−)-antofine for NF-κB, CRE, AP-1, and GRE are 7.3±1.9 nM, 167±42 nM, 135±16 nM, and 24±0.5 nM, respectively. NF-κB is about 20-fold more sensitive in terms of inhibition than CRE and AP-1 pathways. In addition, the rank order of their potency in terms of NF-κB inhibition was rac-cryptopleurine > (−)-antofine > (+)-(S)-tylophorine > (−)-(R)-tylophorine > (−)-ficuseptine C > precursor #1 > precursor #2. Similar rank order was observed in regards to the potency of cytotoxicity (Table 1). The rank order of potency and selectivity against endogenous NF-κB mediated transcription also had the same trend (Supplementary data Table 1). In terms of relative activity against different signaling pathways, the tylohora alkaloids have demonstrated some degree of selectivity. However, the activity of all the PBTs tested against all the four signaling pathways is very poor, with IC50 of more than 6 μM (Table 2, lower panel). If we calculate the relative ratio of IC50 (Table 2) and GI50 (Table 1, Column HepG2) for (−)-antofine, the value would be 1.5, 34, 28, and 4.9 for NF-κB, CRE, AP-1 and GRE respectively. If we apply the same calculation for precursor #1, the value would be 1.3, 7, 5.7, and 2.6 for NF-κB, CRE, AP-1 and GRE respectively. This indicated that (−)-antofine selectively inhibited NF-κB, whereas precursor #1 showed less selectivity against the four signaling pathways. These data suggested that the different analogs might act on multiple target sites, and affect different pathway through different mechanisms.
Table 2.
The IC50 of the inhibitory effect of tylophora alkaloids, and PBTs against stimulated NF-κB, CRE, AP-1 and GRE pathways in HepG2 cells.
| NF-κB IC50a (nm) |
CRE IC50a (nm) |
AP-1 IC50a (nm) |
GRE IC50a (nm) |
|
|---|---|---|---|---|
| (+)-S-tylophorine | 41 ± 20b | >300 b | >300 b | 229 ± 33 |
| (−)-R-tylophorine | 317 ± 51 | 7910 ± 344 | 2257 ± 123 | 534 ± 113 |
| (−)-antofine | 7.3 ± 1.9 | 167± 42 | 135 ±16 | 24 ± 0.5 |
| Rac-cryptopleurine | 1.4 ± 0.6 | 25 ± 0.5 | 18.6 ± 0.8 | 10 ± 0.2 |
| precursor #1 | 2171 ± 351 | 12025 ± 830 | 9776 ± 625 | 4500 ± 312 |
| precursor #2 | >10000 | >15000 | >15000 | >15000 |
| (−)-ficuseptine C | 1244 ± 93 | 2566 ± 352 | 3383 ± 158 | 2440 ± 651 |
|
| ||||
| NF-κB IC50a (μM) |
CRE IC50a (μM) |
AP-1 IC50a (μM) |
GRE IC50a (μM) |
|
|
| ||||
| PBT #28 | 7.9 ± 1.1 | 8.6 ± 0.6 | 8.7 ± 2.1 | 9.2 ± 1 |
| PBT #31 | 23.5 ± 2.2 | 25.0 ± 6 | 27 ± 4.4 | >30 |
| PBT #32 | 21 ± 0.5 | 24 ± 0.8 | >30 | 19 ± 4.2 |
| PBT #33 | 7 ± 0.2 | 12.1 ± 0.4 | 6.2 ± 0.3 | 7.2 ± 1.9 |
| PBT #34 | 10.2 ± 1.1 | 18.5 ± 2.1 | 10 ± 1.8 | 20.7 ± 2.8 |
Values are means ± SD of three experiments, with each data point done in triplicate.
Published.16
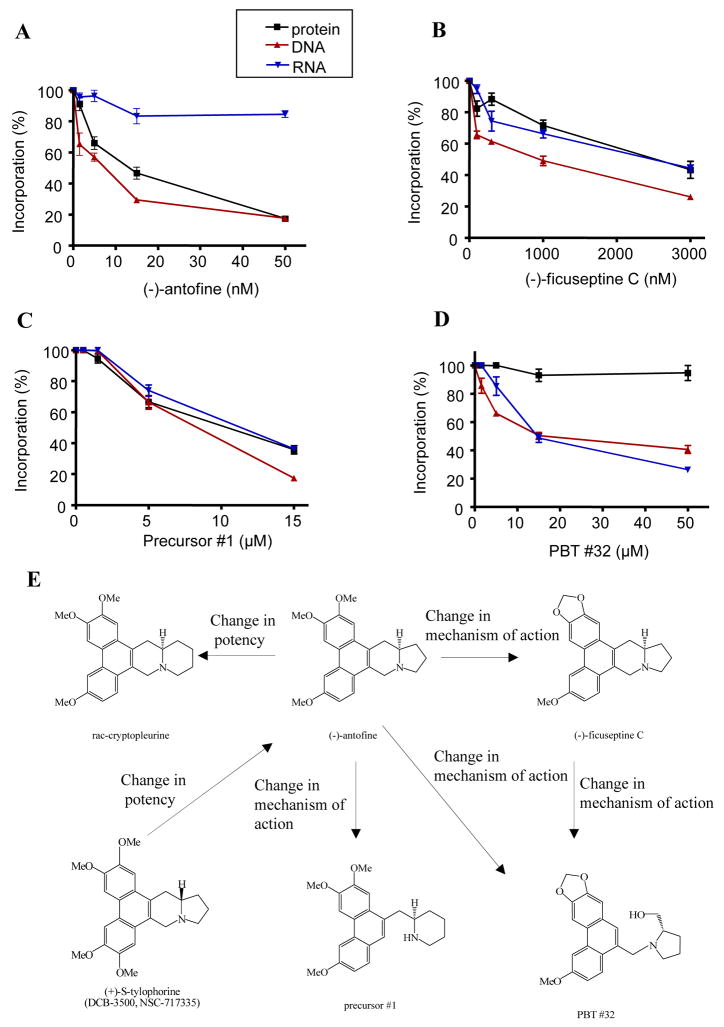
Early studies in the 1970’s demonstrated that the phenanthrene alkaloid tylocrebrine inhibit protein and nucleic acid synthesis.11 During our studies of the mechanism of action of the tylophorine analog DCB-3503, we also observed that this compound could inhibit amino acid and thymidine, but not uridine incorporation in a time-dependent and dose-dependent manner (Gao et al, unpublished data). We observed that (−)-ficuseptine C, precursor #1, and PBTs differ significantly in terms of their cytotoxicities and their potency and selectivity against the NF-κB signaling pathway (Table 1 and 2); we decided to investigate their effects against protein, DNA and RNA synthesis, by comparing their effects against L-[35S]-methionine incorporation into protein, [14C]-thymidine incorporation into DNA, and [14C]-uridine incorporation into RNA. The experimental procedures are available in “Supplementary data”. The concentrations tested were based on each analog’s specific cytotoxicity (1/3, 1, 3, and 10 EC50).
As shown in Figure 2A and 2B, both (−)-antofine and (−)-ficuseptine C inhibited DNA and protein synthesis in a dose-dependent manner. In contrast, (−)-antofine inhibited RNA synthesis only slightly, whereas (−)-ficuseptine C also inhibited RNA synthesis in a dose-dependent manner. There are two major structural differences between PBTs and the rest of the tylophora alkaloids listed in Figure 1: 1) the OMe groups at R1 and R2 are replaced by a cyclic methylenedioxy moiety; 2) the indolizidine ring of the natural products has been formally opened. By comparing the effects of (−)-antofine and (−)-ficuseptine C, the role of the replacement of the OMe groups at R1 and R2 by a methylenedioxy bridge was clarified. Therefore our results suggest that the replacement of the OMe ethers at R1 and R2 by a methylenedioxy moiety significantly changes the compound’s mechanism of action. The major difference between (−)-antofine and precursor #1 is the opening of D-ring. (−)-ficuseptine C shares the methylenedioxy motif with the PBTs and has a similar pyrrolidine ring as compound PBTs #32. Therefore, by comparing the effects of (−)-antofine and precursor #1, and the effects of (−)-ficuseptine C with those of PBT #32, the effect of opening the indolizidine ring was clarified. As shown in Figure 2C and 2D, both precursor #1 and PBT #32 inhibited DNA and RNA synthesis in a dose-dependent manner. In contrast, PBT #32 only sightly inhibited protein synthesis, whereas precursor #1 also inhibited protein synthesis in a dose-dependent manner. These results indicate that the formal opening of the D-ring or the indolizidine ring significantly changes the mechanism of action of these compounds (compare the profile changes between Figure 2A and 2C, Figure 2B and 2D). In addition, the distinct effects between (−)-antofine and PBT #32 [(−)-antofine more selectively inhibited protein synthesis, but not RNA synthesis (Figure 2A), whereas PBT #32 more selectively inhibited RNA synthesis, but not protein synthesis (Figure 2D)] demonstrate that replacement of the OMe groups at R1 and R2 by a methylenedioxy unit and the opening of the indolizidine ring totally changes their mechanism of action. Due to the significantly divergent effects of (−)-antofine, PBT #32, (−)-ficuseptine C and precursor #1 against protein and nucleic acid synthesis, we conclude that these compounds, although being structurally fairly close, may not constitute the same class of compounds in functional terms. The concept of structural analogs with different mechanisms of action has been reported previously.19,20 Kimball et al. recently described a series of tylophora analogs derived from Tyloindicine I, operated through an unknown mechanism of action different from the parental compound.19 Another example is the comparison of podophyllotoxin and etoposide (VP-16), podophyllotoxin is a potent inhibitor of microtubule assembly, whereas its derivative etoposide, currently used in clinical treatment of many cancers such as small cell lung carcinoma and testicular cancer, does not inhibit tubulin polymerization, but rather acts as an inhibitor of DNA topo-isomerase II, which causes double strand breaks in DNA.20
Figure 2. The effects of tylophora alkaloids, and phenanthrene-based tylophorine derivatives on L-[35S]-methionine, [14C]-thymidine and [14C]-uridine incorporation.
HepG2 cells were pretreated with serial dilutions of drugs for 5 min and labeled with L-[35S]-methionine, [14C]-thymidine or [14C]-uridine, respectively. Their incorporation percentage (compared with control) was shown. A. HepG2 cells were pretreated with (−)-antofine 1.5 nM, 5 nM, 15 nM and 50 nM, respectively. B. HepG2 cells were pretreated with (−)-ficuseptine C 100 nM, 300 nM, 1 μM and 3 μM, respectively. C. HepG2 cells were pretreated with precursor #1 0.5 μM 1.5 μM, 5 μM and 15 μM, respectively. D. HepG2 cells were pretreated with PBT #32 1.5 μM, 5 μM, 15 μM, and 50 μM, respectively. E. Schematic description of the SAR of tylophora alkaloids and PBTs.
In summary, phenanthrene based compounds exhibit pronounced structure-activity relationships; both the replacement of the OMe ethers at R1 and R2 by a methylenedioxy bridge (Figure 1) and the opening of the indolizidine ring results in dramatically reduced cytotoxic potency, whereas the loss of the OMe group at R3 and the replacement of the five-membered E-ring by a six-membered E-ring significantly increased the observed cytotoxicity. Although these compounds are close structural analogs, their cytotoxic potency, selectivity against NF-κB signaling pathway, and their effects against protein, DNA, and RNA synthesis are so different that they should not be considered as the same class of compounds. That is, various tylophora alkaloids may be structural but not functional analogs.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr. David C. Baker for providing the compound (+)-(S)-tylophorine. We thank Dr. Elijah Paintsil for the critical reading of the manuscript. Annie Pei-Chun Chen was supported by Taiwan Merit Scholarship (TMS-094-2-B-014). A. Fürstner acknowledges financial support by the Chemical Genomics Center (CGC) initiative of the Max Planck Gesellschaft and the Fonds der Chemischen Industrie. This investigation was supported in part by grant CA 17625 from the National Cancer Institute, NIH awarded to K. H. Lee, and a fellowship from National Foundation for Cancer Research to Y.C. Cheng.
Footnotes
Supplementary data
Supplementary data associated with this article (Synthesis of phenanthrene-based tylophorine derivatives described in Scheme 1, synthesis of tylophora alkaloids exemplified in Scheme 2, IC50s of the inhibitory effect of tylophora alkaloids and PBTs against endogenous NF-κB, CRE, AP-1 and GRE mediated transcription in HepG2 cells listed in Table S1) can be found, in the online version, at doi: xxxxxxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Li Z, Zhong J, Huang R. Synthesis. 2001;16:2365. [Google Scholar]
- 2.Donaldson GR, Atkinson MR, Murray AW. Biochem Biophys Res Commun. 1968;31:104. doi: 10.1016/0006-291x(68)90037-5. [DOI] [PubMed] [Google Scholar]
- 3.Rao KN, Venkatachalam SR. Toxicol In Vitro. 2000;14:53. doi: 10.1016/s0887-2333(99)00092-2. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu H, Watanabe M, Ohyama M, Enya T, Koyama K, Kanazawa T, Kawahara N, Sugimura T, Wakabayashi K. J Med Chem. 2001;44:1833. doi: 10.1021/jm0004042. [DOI] [PubMed] [Google Scholar]
- 5.Staerk D, Lykkeberg AK, Christensen J, Budnik BA, Abe F, Jaroszewski JW. J Nat Prod. 2002;65:1299. doi: 10.1021/np0106384. [DOI] [PubMed] [Google Scholar]
- 6.Gao W, Lam W, Zhong S, Kaczmarek C, Baker DC, Cheng YC. Cancer Res. 2004;64:678. doi: 10.1158/0008-5472.can-03-1904. [DOI] [PubMed] [Google Scholar]
- 7.Yang CW, Chen WL, Wu PL, Tseng HY, Lee S. J Mol Pharmacol. 2006;69:749. doi: 10.1124/mol.105.017764. [DOI] [PubMed] [Google Scholar]
- 8.You X, Pan M, Gao W, Shiah HS, Tao J, Zhang D, Koumpouras F, Wang S, Zhao H, Madri JA, Baker D, Cheng YC, Yin Z. Arthritis Rheum. 2006;54:877. doi: 10.1002/art.21640. [DOI] [PubMed] [Google Scholar]
- 9.Choi JY, Gao W, Odegard J, Shiah HS, Kashgarian M, McNiff JM, Baker DC, Cheng YC, Craft J. Arthritis Rheum. 2006;54:3277. doi: 10.1002/art.22119. [DOI] [PubMed] [Google Scholar]
- 10.Chopra RNL-C, De NN, Chakerburty M. Ind J Med Res. 1935;23:263. [Google Scholar]
- 11.Huang MT, Grollman AP. Mol Pharmacol. 1972;8:538. [PubMed] [Google Scholar]
- 12.Shiah HS, Gao W, Baker DC, Cheng YC. Mol Cancer Ther. 2006;5:2484. doi: 10.1158/1535-7163.MCT-06-0146. [DOI] [PubMed] [Google Scholar]
- 13.Wei L, Brossi A, Kendall R, Bastow KF, Morris-Natschke SL, Shi Q, Lee KH. Bioorg Med Chem. 2006;14:6560. doi: 10.1016/j.bmc.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Fu Y, Lee SK, Min HY, Lee T, Lee J, Cheng M, Kim S. Bioorg Med Chem Lett. 2007;17:97. doi: 10.1016/j.bmcl.2006.09.080. [DOI] [PubMed] [Google Scholar]
- 15.Chuang TH, Lee SJ, Yang CW, Wu PL. Org Biomol Chem. 2006;4:860. doi: 10.1039/b516152e. [DOI] [PubMed] [Google Scholar]
- 16.Gao W, Bussom S, Grill SP, Gullen EA, Hu YC, Huang X, Zhong S, Kaczmarek C, Gutierrez J, Francis S, Baker DC, Yu S, Cheng YC. Bioorg Med Chem Lett. 2007;17:4338. doi: 10.1016/j.bmcl.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Fürstner A, Kennedy JW. Chem Eur J. 2006;12:7398. doi: 10.1002/chem.200600592. [DOI] [PubMed] [Google Scholar]
- 18.Karin M, Yamamoto Y, Wang QM. Nat Rev Drug Disc. 2004;3:17. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 19.Kimball FS, Tunoori AR, Victory SF, Dutta D, White JM, Himes RH, Georg GI. Bioorg Med Chem Lett. 2007;17:4703. doi: 10.1016/j.bmcl.2007.05.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damayanthi Y, Lown JW. Podophyllotoxins: current status and recent developments. Curr Med Chem. 1998;5:205. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.