Abstract
The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) are potent pulmonary carcinogens in rats. NNK and NNAL require metabolic activation to express their carcinogenicity. Cytochrome P450-catalyzed α-hydroxylation at the methyl position of NNK or NNAL generates reactive intermediates, which alkylate DNA to form pyridyloxobutyl (POB)-DNA adducts or pyridylhydroxybutyl (PHB)-DNA adducts. NNK is metabolized to NNAL in a reversible and stereoselective manner, and the tissue-specific retention of (S)-NNAL is believed to be important to the carcinogenicity of NNK. In the present study, we investigated the formation of POB- and PHB-DNA adducts in extra-hepatic tissues of F344 rats treated chronically with NNK, (R)- and (S)-NNAL (10 ppm in the drinking water, 1–20 weeks). POB- and PHB-DNA adducts were quantified in nasal olfactory mucosa, nasal respiratory mucosa, oral mucosa, and pancreas of treated rats. Adduct formation in the nasal respiratory mucosa exceeded that in the other tissues. O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dThd) or O2-[4-(3-pyridyl)-4-hydroxybut-1-yl]thymidine (O2-PHB-dThd) was the major adduct, followed by 7-[4-(3-pyridyl)-4-oxobut-1-yl]guanine (7-POB-Gua) or 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]guanine (7-PHB-Gua). There was a remarkable similarity in adduct formation between the NNK and (S)-NNAL groups, both of which were distinctively different from that in the (R)-NNAL group. For example, in the nasal olfactory mucosa, POB-DNA adduct levels in the NNK and (S)-NNAL groups were not significantly different from each other, while (R)-NNAL treatment generated 6 – 33 times lower of POB-DNA adducts than did NNK treatment. In contrast, (R)-NNAL treatment produced significantly higher levels of PHB-DNA adducts than did NNK or (S)-NNAL treatment. Similar trends were observed in the nasal respiratory mucosa, oral mucosa and pancreas. These results suggest extensive retention of (S)-NNAL in various tissues of NNK-treated rats, and support a mechanism in which the preferential metabolism of NNK to (S)-NNAL, followed by sequestration of (S)-NNAL in the target tissues and reoxidation to NNK, is important to NNK tumorigenesis.
Keywords: Pyridyloxobutyl DNA adducts, Pyridylhydroxybutyl DNA adducts, 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone, 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol
Introduction
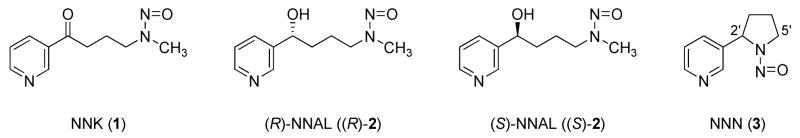
Tobacco-specific nitrosamines, a group of carcinogens found in tobacco products, are believed to play an important role in tobacco carcinogenesis (1, 2). Among these compounds, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK, 1, Chart 1), its primary metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL, 2), and N′-nitrosonornicotine (NNN, 3) are the most carcinogenic. NNK is a potent lung carcinogen in rodents, inducing tumors of this site independent of the route of administration (3). It also causes tumors of the pancreas, nasal mucosa, and liver (3). When given by oral swabbing to rats, a combination of NNK and NNN induces oral tumors as well (4). NNK and NNN have been evaluated as human carcinogens by the International Agency for Research on Cancer (2).
Chart 1.
Structures of NNK, (R)- and (S)-NNAL, and NNN.
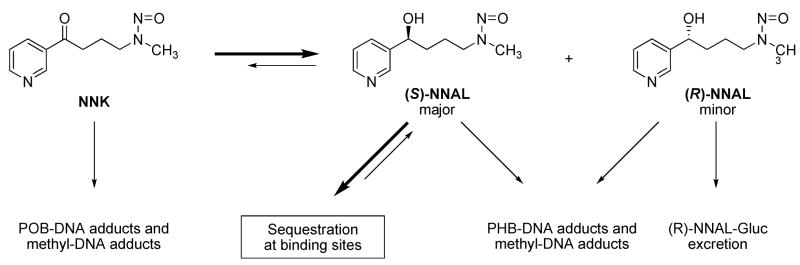
NNK is metabolically converted, in a reversible reaction, to NNAL. NNAL has a chiral center, and there are remarkable differences between (R)- and (S)-NNAL in terms of tumorigenicity, metabolism, and pharmacokinetics. In A/J mice, (S)-NNAL was as potent as NNK as a lung tumorigen, while (R)-NNAL was much less active (5). (S)-NNAL is the predominantly formed enantiomer from metabolic reduction of NNK in rat liver cytosol and microsomes, and rat lung cytosol, accounting for 63–97% of the total NNAL formed, while divergent results have been obtained with rat lung microsomes (6, 7). Multiple enzymes seem to be involved in NNK reduction in human and rat tissues, including cytosolic aldo-keto reductases (AKR) and carbonyl reductase, and microsomal 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1). All of them favor the formation of (S)-NNAL from NNK (7). (S)-NNAL is reoxidized much faster than (R)-NNAL in rat liver and lung, but little stereoselectivity was observed in human liver (6). Various cytochrome P450s expressed in mice, rats, and human lung catalyze the oxidation of (S)-NNAL more efficiently than (R)-NNAL (8, 9). NNAL can be subsequently glucuronidated and excreted in the urine. While (R)-NNAL is extensively conjugated in this way, (S)-NNAL tends to be retained in the lung and reconverted back to NNK (10, 11). In smokers and smokeless tobacco users, the ratios of (S)/(R)-NNAL and their glucuronides in the urine were significantly higher 7 days after cessation than at baseline, indicating a stereoselective retention of (S)-NNAL (12). Although NNAL is not present in substantial amounts in tobacco products, these observations imply that (S)-NNAL may play a significant role in the mechanisms of tumor induction by NNK.
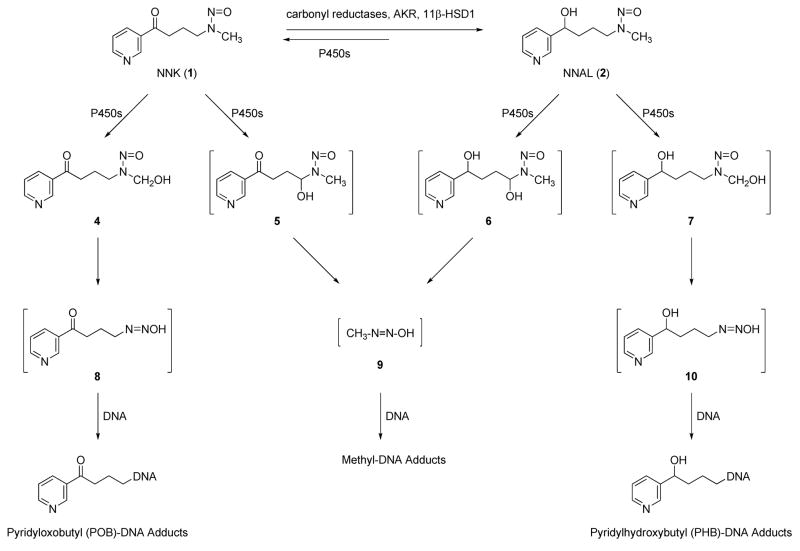
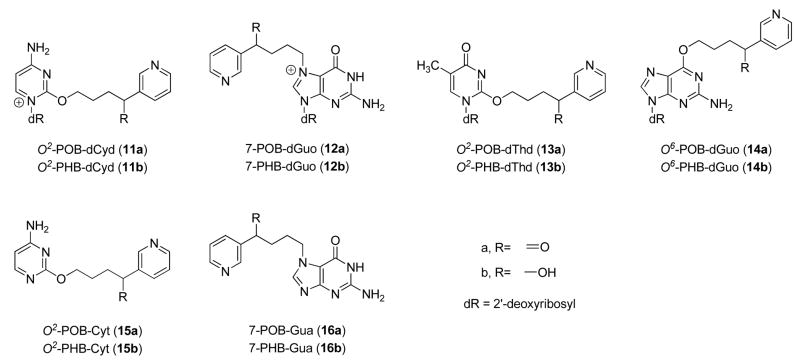
Both NNK and NNAL are metabolically activated through cytochrome P450-catalyzed α-hydroxylation, and DNA adduct formation is critical to their carcinogenicity (13). As shown in Scheme 1, α-methyl hydroxylation of NNK results in a reactive intermediate 4, which spontaneously decomposes to 4-(3-pyridyl)-4-oxobutanediazohydroxide (8). This intermediate, or its corresponding diazonium ion, can alkylate DNA to form pyridyloxobutyl (POB)-DNA adducts. The formation of POB-DNA adducts is believed to play an important role in NNK tumorigenesis in rodents (14–18). Similarly, α-methyl hydroxylation of NNAL yields intermediate 10, resulting in pyridylhydroxybutyl (PHB)-DNA adducts. α-Hydroxylation of NNK or NNAL at the methylene group gives methanediazohydroxide (9), which produces methyl adducts in DNA. Four specific POB-DNA adducts, O2-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxycytidine (O2-POB-dCyd, 11a, Chart 2), 7-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (7-POB-dGuo, 12a), O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dThd, 13a), and O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (O6-POB-dGuo, 14a), have been characterized previously (19–21). The corresponding PHB-DNA adducts (11b–14b) have also been identified (21, 22). Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) methods have been developed for the analysis of these POB- and PHB-DNA adducts in DNA (23, 24). The unstable adducts 11a, 12a, and 12b were analyzed as the bases O2-[4-(3-pyridyl)-4-oxobut-1-yl]cytosine (O2-POB-Cyt, 15a), 7-[4-(3-pyridyl)-4-oxobut-1-yl]guanine (7-POB-Gua, 16a), and 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]guanine (7-PHB-Gua, 16b) after loss of deoxyribose upon neutral thermal hydrolysis. Adduct 11b was not included in the analysis, because of the low level of its corresponding POB-DNA adduct 11a observed in NNK-treated rats (25).
Scheme 1.
Overview of NNK and NNAL metabolism and DNA adduct formation.
Chart 2.
Structures of POB- and PHB-DNA adducts.
Previously, four POB-DNA adducts and three PHB-DNA adducts were quantitated in liver and lung DNA of F344 rats treated chronically with NNK, (R)-NNAL, and (S)-NNAL in the drinking water (24, 25). Adduct levels were similar in NNK- and (S)-NNAL-treated rats, and significantly different from those in (R)-NNAL-treated rats. NNK and (S)-NNAL treatment generated higher levels of POB-DNA adducts than (R)-NNAL treatment, while the reverse was observed with PHB-DNA adducts. These results supported a hypothesis that preferential metabolism of NNK to (S)-NNAL, followed by retention of (S)-NNAL in the lung and reoxidation to NNK, is important for NNK tumorigenesis in the lung. Although lung is the major target tissue of NNK in the rat, NNK also causes tumors in other tissues. Therefore, in the present study, we extended our research to other target tissues–pancreas, nasal olfactory and respiratory mucosa, and oral mucosa. DNA was isolated from these tissues of NNK-, (R)-NNAL- and (S)-NNAL-treated rats, and POB- and PHB-DNA adducts were quantified in a simultaneous analysis using LC-ESI-MS/MS.
Materials and Methods
CAUTION
NNK and NNAL are carcinogenic. They should be handled in a well-ventilated hood with extreme care and appropriate protective equipment.
Chemicals
NNK was purchased from Toronto Research Chemicals (Toronto, Ontario, Canada). Purity was greater than 98%. (R)- and (S)-NNAL, POB- and PHB-DNA adducts and internal standards were synthesized (23, 24, 26, 27). Puregene DNA purification solutions were obtained from Qiagen (Valencia, CA). Calf thymus DNA, micrococcal nuclease (from Staphylococcus aureus), and phosphodiesterase II (from bovine spleen) were procured from Worthington Biochemical Co. (Lakewood, NJ). Alkaline phosphatase (from calf intestine) was purchased from Roche Diagnostics Corporation (Indianapolis, IN). All other chemicals were obtained from Sigma-Aldrich.
Animal Experiment
This has been described (25). Briefly, 216 male F344 rats were randomly divided into four groups of 54 rats: (1) control; (2) NNK; (3) (R)-NNAL; and (4) (S)-NNAL. The rats in the treatment groups received 10 ppm of the appropriate carcinogen in the drinking water, and the control rats were given tap water. Nine rats per group were sacrificed by CO2 overdose at 1, 2, 5, 10, 16, and 20 weeks. Tissues were harvested and stored at −80 °C until DNA isolation.
Isolation of nasal olfactory and respiratory mucosa
This was performed as previously described (14). After the rats were sacrificed, the mandibula of the head was removed. The head was placed on the necropsy board with the palate facing up. The head was split using a bone mallet and a scalpel, cutting longitudinally on the median line, through the hard palate. Respiratory mucosa was retrieved from the naso- and maxilloturbinates, the lateral walls of the nasal passages, and the median septum anterior to the olfactory area. Olfactory mucosa was obtained from the ethmoturbinates and the lateral wall and septum of the olfactory area. The tissues were stored in PBS buffer at −80 °C.
Quantitation of POB- and PHB-DNA adducts by LC-ESI-MS/MS
Our method was adapted from previous studies (23, 24), which enabled us to simultaneously analyze both POB- and PHB-DNA adducts from rat DNA. DNA isolation was performed following the Puregene DNA isolation protocol (Qiagen) with some modifications (28). Pancreatic DNA was isolated from three rats at each time point, while nasal olfactory, nasal respiratory, and oral mucosa from nine rats in each group were divided into three pools of three for DNA isolation. Depending on the starting amount of tissue, the reagents were scaled accordingly. The DNA (0.1–1 mg), plus four deuterated internal standards of POB-DNA adducts and three deuterated internal standards of PHB-DNA adducts, was subjected to neutral thermal hydrolysis (100 °C, 30 min), followed by enzymatic hydrolysis with micrococcal nuclease, phosphodiesterase II, and alkaline phosphatase. A 10 μL aliquot was removed for dGuo quantitation, and the remaining hydrolysate was purified on a solid phase extraction (SPE) cartridge [Strata-X, 33 μm, 30 mg/1 mL (Phenomenex, Torrance, CA)]. After the sample was applied, the cartridge was washed with 2 mL H2O and 1 mL 10% CH3OH/H2O, and the analytes were eluted with 2 mL CH3OH. The eluants were evaporated to dryness, and dissolved in 20 μL of 2% NH4OAc for LC-ESI-MS/MS analysis. An 8 μl aliquot was analyzed by LC-ESI-MS/MS. A buffer control which lacked DNA and a calf thymus DNA sample were prepared each time and processed in the same way as negative controls.
LC-ESI-MS/MS analysis was carried out with an Agilent 1100 capillary flow HPLC (Agilent Technologies, Palo Alto, CA) equipped with a Luna 250 mm × 0.5 mm 5 μm C18 column (Phenomenex) and coupled to a Discovery Max (ThermoElectron, San Jose, CA) triple quadrupole mass spectrometer. The solvent elution program was a gradient from 5 to 65% CH3OH in 15 mM NH4OAc buffer in 30 min at a flow rate of 10 μL/min at 30 °C. The ESI source was operated in the positive ion mode. The adducts were analyzed by MS/MS using selected reaction monitoring (SRM). Ion transitions and their collision energies are listed in Table 1. Other MS parameters were optimized to achieve maximum signal intensity. Calibration curves were constructed before each analysis using standard solutions containing varying amounts of each adduct with a constant amount of the corresponding deuterated internal standard in 2% NH4OAc. The amount of DNA was calculated from the dGuo content as determined by HPLC (23), considering that 1 mg of DNA contains 3 μmol of nucleotides (29), and dGuo accounts for 22% of the total nucleotides in rat DNA, as previously determined in our lab. Adduct levels were expressed as fmol adduct/mg DNA.
Table 1.
SRM transitions of POB- and PHB-DNA adducts and corresponding internal standards
| Adducts | Parent ion([M + 1]+, m/z) | Daughter ion (m/z) | Collision energy (eV) |
|---|---|---|---|
| O2-POB-Cyt (15a) | 259.1 | 148.1 [POB]+ | 17 |
| [pyridine-D4]O2-POB-Cyt | 263.1 | 152.1 ([pyridine-D4]POB])+ | 17 |
| 7-POB-Gua (16a) | 299.1 | 148.1 [POB]+ | 17 |
| [pyridine-D4]7-POB-Gua | 303.1 | 152.1 ([pyridine-D4]POB])+ and [Gua+H]+ | 17 |
| O2-POB-dThd (13a) | 390.1 | 148.1 [POB]+ | 20 |
| [pyridine-D4]O2-POB-dThd | 394.1 | 152.1 ([pyridine-D4]POB])+ | 20 |
| O6-POB-dGuo (14a) | 415.1 | 148.1 [POB]+ | 20 |
| [pyridine-D4]O6-POB-dGuo | 419.1 | 152.1 ([pyridine-D4]POB])+ and [Gua+H]+ | 20 |
| 7-PHB-Gua (16b) | 301.1 | 150.1 [PHB]+ | 17 |
| [pyridine-D4]7-PHB-Gua | 305.1 | 154.1 ([pyridine-D4]PHB])+ | 17 |
| O2-PHB-dThd (13b) | 392.1 | 150.1 [PHB]+ | 20 |
| [pyridine-D4]O2-PHB-dThd | 396.1 | 154.1 ([pyridine-D4]PHB])+ | 20 |
| O6-PHB-dGuo (14b) | 417.1 | 150.1 [PHB]+ | 20 |
| [pyridine-D4]O2-PHB-dGuo | 421.1 | 154.1 ([pyridine-D4]PHB])+ | 20 |
Statistical Analyses
Because of a highly skewed distribution, the amounts of POB- and PHB-DNA adducts were transformed to the natural log scale. The following three statistical analyses were performed for POB- and PHB-DNA adducts. (1) Total POB- and PHB-DNA adduct levels were compared between different tissue types. The adducts were measured in nasal olfactory, nasal respiratory, and oral mucosa from the same 9 rats (3 pools of 3 rats), whereas they were quantified in pancreas from only three rat. Therefore two different statistical tests were employed. First, a repeated measures analysis of variance (ANOVA) was used to compare nasal olfactory, nasal respiratory, and oral mucosa. Tissue type was the repeated factor and time was the fixed effect. The model assumes a linear trend over time. If this global test across all six time points was significant for tissue type, then the least-squares means for the three tissues, derived from the ANOVA model, were compared two at a time for each time point separately. The Tukey procedure was utilized to control for multiple comparisons among the tissues. Second, an analysis of covariance (ANCOVA) was applied to compare pancreas to each of the other tissues. The grouping factor was tissue type and the covariate was time. If this overall test was significant, then the least-squares means for the pancreas and each of the other tissues, considered one at a time, were compared at each time point separately. (2) An ANCOVA was used to compare total POB- and PHB-DNA adduct levels among different treatments (NNK, (R)-NNAL, and (S)-NNAL). The grouping factor was treatment and the covariate was time. If this overall test was significant for treatment, then the least-squares means for the treatments (based on the ANCOVA model) were compared two at a time for each time point separately. The Tukey procedure was applied to control for multiple comparisons among treatments. (3) A repeated measures ANOVA was employed to compare individual POB- and PHB-DNA adducts for each tissue and treatment. Time points were included in the model as a fixed effect. The analysis was carried out separately for each combination of tissue and treatment. If an adduct was missing or not detected for all or most of the data for a particular tissue and treatment, it was excluded for that analysis. If the global test including adducts and time points was significant, then additional tests were performed to compare the least-squares means of individual adducts at each time point. The Tukey procedure was applied to control for multiple comparisons between the adducts evaluated two at a time. Statistical significance was set at P < 0.05.
Results
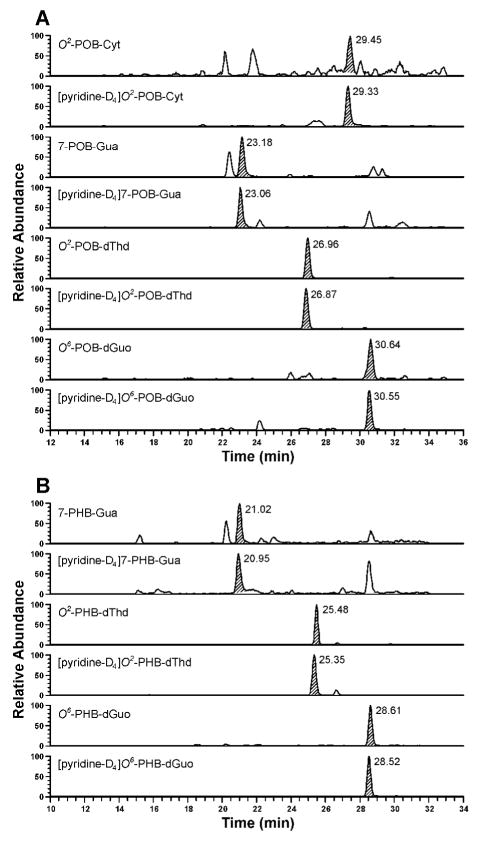
No POB- or PHB-DNA adducts were found in the pancreas, nasal and oral mucosa from control rats, but they were readily detected in DNA from NNK-, (R)-NNAL- and (S)-NNAL-treated rats. Representative chromatograms from the analyses of POB- and PHB-DNA adducts from the same sample of nasal respiratory mucosa DNA are illustrated in Figure 1. Peaks corresponding to the retention times of all adducts were observed, and each coeluted with the appropriate internal standard. The accuracy, precision, limits of detection, and recoveries of this method were established previously (23, 24).
Figure 1.
LC-ESI-MS/MS chromatograms obtained upon the analyses of (A) POB-DNA adducts; and (B) PHB-DNA adducts in nasal respiratory mucosa DNA of rats treated with NNK for 16 weeks. Individual POB- and PHB-DNA adducts and their internal standards were monitored as indicated on each channel. SRM transitions are summarized in Table 1.
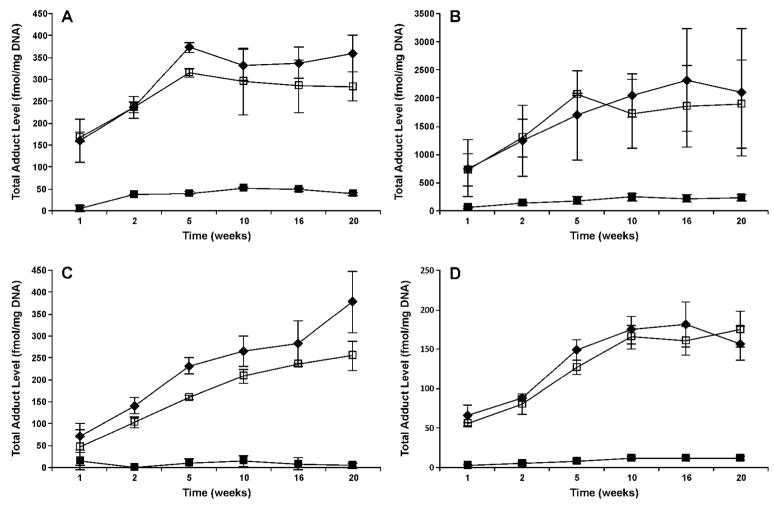
Levels of total POB-DNA adducts in the nasal olfactory mucosa, nasal respiratory mucosa, oral mucosa, and pancreas of rats treated with NNK, (R)- and (S)-NNAL are summarized in Figure 2A–D, and individual POB-DNA adduct levels from the NNK group are illustrated in Figure 3A–D (also see Table 1S in the Supporting Information). Overall, the formation and accumulation of POB-DNA adducts in the different tissues were similar, with remarkably higher levels observed in the NNK and (S)-NNAL groups than in the (R)-NNAL group. For example, in the nasal olfactory mucosa (Figure 2A), total POB-DNA adducts in the NNK- and (S)-NNAL-treated rats increased rapidly during the initial period of treatment, and reached a plateau after 5 weeks at about 300–400 fmol/mg DNA. Levels of individual adducts in the nasal olfactory mucosa of NNK-treated rats are shown in Figure 3A. O2-POB-dThd exceeded 7-POB-Gua at all time points, and the differences were significant after 10 weeks (P < 0.05). O6-POB-dGuo was detected at 1, 2, 5, and 10 weeks, and O2-POB-Cyt was only observed at 10 weeks. Their levels were both significantly lower than those of O2-POB-dThd and 7-POB-Gua (P < 0.05). As the major adduct in the olfactory mucosa, O2-POB-dThd accounted for 58–82% of the total adducts, and its level increased quickly during the initial NNK treatment. 7-POB-Gua accounted for 18–40 % of total adducts, and its level was relatively stable throughout the treatment. Similar results were observed in (S)-NNAL-treated rats, with the maximum amount (315 fmol/mg DNA) slightly lower than that in NNK-treated rats (Figure 2A). Individual POB-DNA adduct levels in (S)-NNAL-treated rats were also similar to those in NNK-treated rats (Supporting Information Table 1S). However, (R)-NNAL treatment generated a much lower level of POB-DNA adducts. The total amount was about 6–33 times lower than in the NNK-treated rats, and the differences were significant at all time points (P < 0.05). Only O2-POB-dThd and 7-POB-Gua were observed in the (R)-NNAL group.
Figure 2.
Total POB-DNA adduct levels (fmol/mg DNA) ± SD vs time (weeks) in the (A) nasal olfactory mucosa; (B) nasal respiratory mucosa; (C) oral mucosa; and (D) pancreas of NNK-, (R)-NNAL-, and (S)-NNAL-treated rats. Symbol designations are: ◆, NNK; ■, (R)-NNAL; and □, (S)-NNAL.
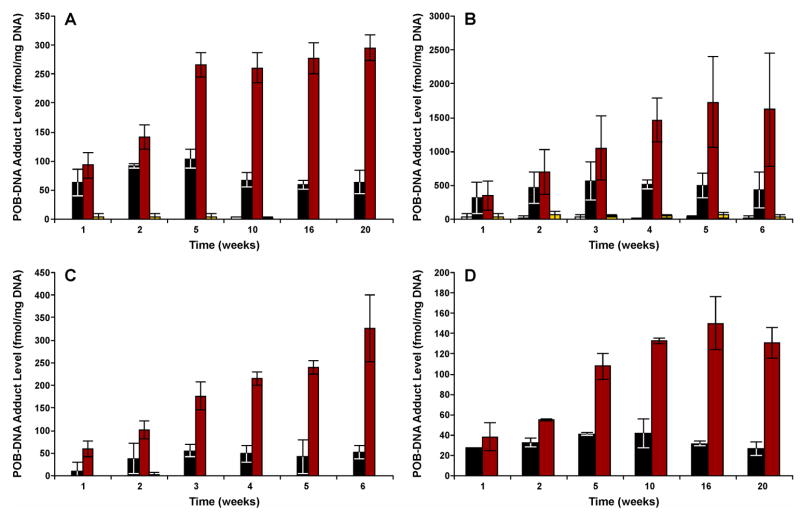
Figure 3.
Levels of each POB-DNA adduct in the (A) nasal olfactory mucosa; (B) nasal respiratory mucosa; (C) oral mucosa; and (D) pancreas of NNK treated rats. Adduct levels are shown in sequence at each time point, as follows: □, O2-POB-Cyt; ■, 7-POB-Gua;  , O2-POB-dThd;
, O2-POB-dThd;  , O6-POB-dGuo.
, O6-POB-dGuo.
POB-DNA adduct patterns seen in the pancreas, nasal respiratory mucosa and oral mucosa were very similar to that in the olfactory mucosa (Figure 2B–D and Figure 3B–D). Total adduct levels in the NNK and (S)-NNAL groups were not significantly different from each other (P > 0.05). Among these tissues, nasal respiratory mucosa had significantly higher levels of POB-DNA adducts than any other tissue in NNK- and (S)-NNAL-treated rats (P < 0.05). The maximum amount was observed at 16 weeks (2316 fmol/mg DNA) in the NNK treatment group (Figure 2B). O2-POB-dThd was the major adduct in all the tissue types, followed by 7-POB-Gua. O2-POB-Cyt and O6-POB-dGuo were mainly detected in the nasal respiratory mucosa of NNK- and (S)-NNAL-treated rats, with levels significantly lower than the other two adducts (P < 0.05).
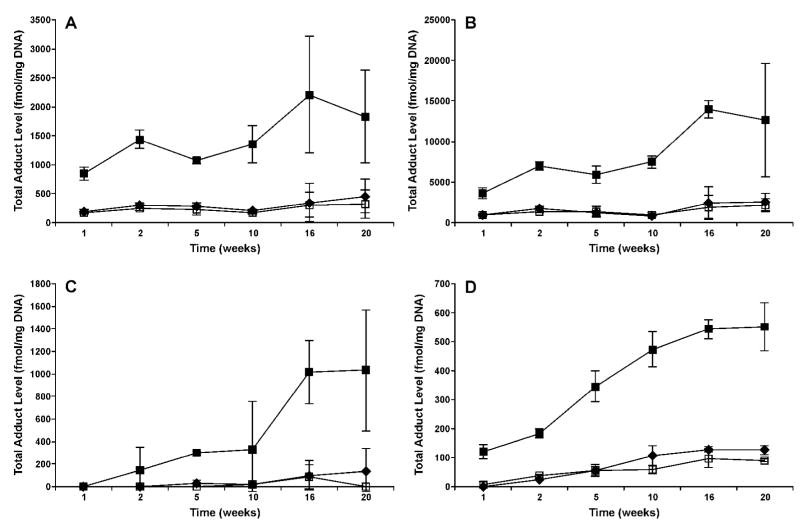
In contrast to POB-DNA adducts, PHB-DNA adduct levels were much higher in (R)-NNAL-treated rats than in NNK- and (S)-NNAL-treated rats. Levels of total and individual PHB-DNA adducts are summarized in Figure 4A–D and Figure 5A–D (also see Table 2S in the Supporting Information). In the pancreas, for example, (R)-NNAL treatment generated 4–7 times higher levels of PHB-DNA adducts than NNK treatment (Figure 4D). The differences were significant at all time points except 20 weeks (P < 0.05). The highest adduct level was 552 fmol/mg DNA, observed at 20 weeks. Levels of the three individual PHB-DNA adducts were in the following order: O2-PHB-dThd > 7-PHB-Gua > O6-PHB-dGuo (Figure 5D). This order was similar to the corresponding POB-DNA adducts, expect that O6-POB-dGuo was not detected in the pancreas. Levels of O2-PHB-dThd generally increased throughout the treatment period, while 7-PHB-Gua levels remained relatively stable. O2-PHB-dThd accounted for 56–86 % of total adducts, and 7-PHB-Gua accounted for 13–40 %. These differences between O2-PHB-dThd and 7-PHB-Gua were significant after 5 weeks (P < 0.05). Levels of O6-PHB-dGuo were less than 4%, and were significantly lower than those of O2-PHB-dThd and 7-PHB-Gua (P < 0.05). (S)-NNAL treatment generated similar adduct levels as NNK treatment.
Figure 4.
Total PHB-DNA adduct levels (fmol/mg DNA) ± SD vs time (weeks) in the (A) nasal olfactory mucosa; (B) nasal respiratory mucosa; (C) oral mucosa; and (D) pancreas of NNK-, (R)-NNAL-, and (S)-NNAL-treated rats. Symbol designations are: ◆, NNK; ■, (R)-NNAL; and □, (S)-NNAL.
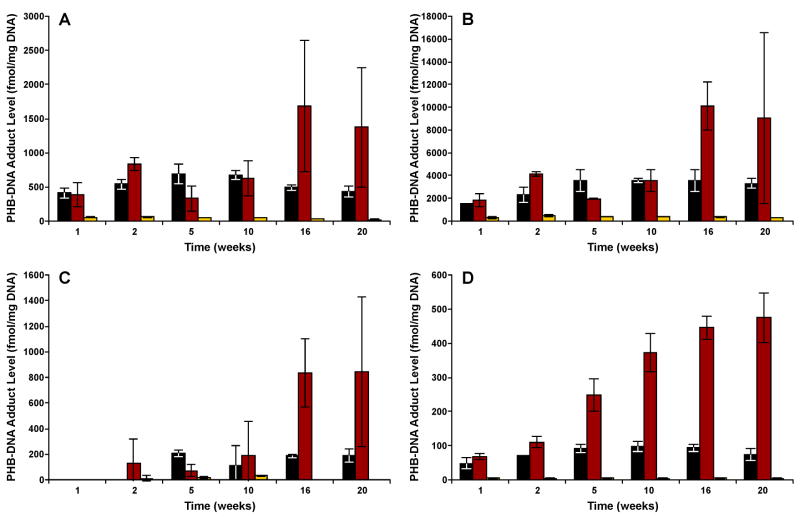
Figure 5.
Levels of each PHB-DNA adduct in the (A) nasal olfactory mucosa; (B) nasal respiratory mucosa; (C) oral mucosa; and (D) pancreas of (R)-NNAL treated rats. Adduct levels are shown in sequence at each time point, as follows: ■, 7-POB-Gua;  , O2-POB-dThd;
, O2-POB-dThd;  , O6-POB-dGuo.
, O6-POB-dGuo.
Overall, patterns of PHB-DNA adduct formation in nasal olfactory and respiratory mucosa and oral mucosa were similar to pancreas, with a remarkable kinship between the NNK and (S)-NNAL groups, both of which were lower than (R)-NNAL group (Figure 4A–C). The highest PHB-DNA adduct levels were clearly observed in nasal respiratory mucosa. In nasal olfactory and respiratory mucosa of (R)-NNAL-treated rats, a slight decrease in total adduct levels was seen at 5 weeks, followed by an increase afterwards, and the maximum amounts were both observed at 16 weeks, with 14010 fmol/mg DNA in respiratory and 2204 fmol/mg DNA in olfactory mucosa. In terms of individual PHB-DNA adducts, O2-PHB-dThd decreased in both nasal mucosae at 5 weeks, and then accumulated at high levels after 16 weeks (Figure 5A–B). Levels of O2-PHB-dThd were significantly greater than 7-PHB-Gua in the nasal olfactory mucosa after 16 weeks, and in the nasal respiratory mucosa after 10 weeks (P < 0.05). O6-PHB-dGuo was significantly lower than either O2-PHB-dThd or 7-PHB-Gua at all time points (P < 0.05). In the oral mucosa of (R)-NNAL-treated rats, PHB-DNA adducts were not detected after 1 week of treatment, and the total levels increased after 2 weeks, reaching a maximum of 1033 fmol/mg DNA at 20 weeks. However, the global test indicated no significant differences among individual PHB-DNA adducts (P = 0.35).
It is possible that POB- and PHB-DNA adducts, or their corresponding precursors are interchangeable in vivo. To test this possibility, DNA samples from nasal and oral mucosa of (R)- and (S)-NNN-treated rats were analyzed for PHB-DNA adducts. NNN, through metabolic activation by 2′-hydroxylation, only directly forms POB-DNA adducts (3). If the reduction pathway occurs, PHB-DNA adducts would be detected in these NNN-treated rats. We did not detect any PHB-DNA adducts in these tissues, excluding the possibility of in vivo reduction of POB-DNA adducts.
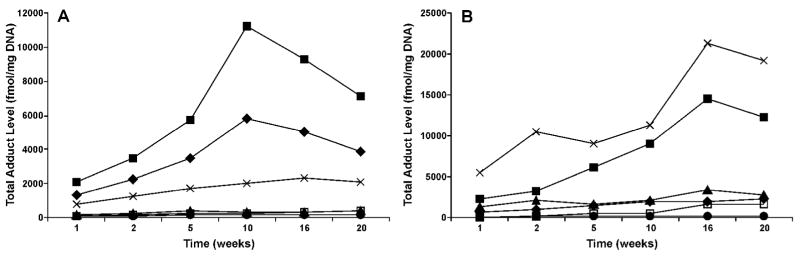
Comparative levels of POB-DNA adducts in NNK-treated rats and PHB-DNA adducts in (R)-NNAL-treated rats are illustrated in Figure 6. The levels in the lung and liver were measured previously (24, 25). Highest levels of POB-DNA adducts were observed in the lung, followed by the liver and nasal respiratory mucosa. Levels in the nasal olfactory mucosa, oral mucosa, and pancreas were lower. The order of PHB-DNA adduct formation was: nasal respiratory mucosa > lung > nasal olfactory mucosa > liver > oral mucosa > pancreas.
Figure 6.
Comparison of adduct levels in the liver, lung, nasal olfactory mucosa, nasal respiratory mucosa, oral mucosa, and pancreas of treated rats: (A) total POB-DNA adduct levels (fmol/mg DNA) vs time (weeks) in NNK-treated rats; (B) total PHB-DNA adduct levels (fmol/mg DNA) vs time (weeks) in (R)-NNAL-treated rats. Levels of POB- and PHB-DNA adducts in the liver and lung were measured previously (24, 25). Symbol designations are: ◆, liver; ■, lung; ▲, nasal olfactory mucosa; X, nasal respiratory mucosa; □, oral mucosa; and ● pancreas.
Discussion
We used a LC-ESI-MS/MS method to analyze POB- and PHB-DNA adducts in DNA from tissues of rats treated with NNK and enantiomers of NNAL. Both types of adducts were detected in all the extra-hepatic tissues investigated here: pancreas, nasal olfactory mucosa, nasal respiratory mucosa, and oral mucosa. Our results demonstrated a remarkable similarity in adduct formation from NNK and (S)-NNAL treatment, both of which were dramatically different from (R)-NNAL treatment. Total POB-DNA adducts in these tissues were significantly higher in NNK- and (S)-NNAL-treated rats than in (R)-NNAL-treated rats, while the opposite was observed with PHB-DNA adducts. These observations are consistent with previous results obtained in the liver and lung of rats receiving the same treatment (24, 25).
NNK and NNAL interconvert in vivo. Previous studies indicated that NNK was preferentially converted to (S)-NNAL in rats (6, 7). (S)-NNAL had a large volume of distribution, and was selectively retained in the lung, while (R)-NNAL was extensively glucuronidated and excreted in the urine (10, 11). Both NNK and NNAL can be metabolically activated by cytochrome P450-catalyzed α-hydroxylation, and POB- and PHB-DNA adducts are formed exclusively from their corresponding precursors, as shown in Scheme 1. Therefore, analyzing POB- and PHB-DNA adducts within the same tissue of rats treated with NNK and enantiomers of NNAL provides valuable information about the interconversion of these compounds at a given time point. Our previous studies investigated POB- and PHB-DNA adduct formation in liver and lung, and we have proposed a mechanism for the interconversion of NNK and NNAL in vivo in rats, shown in Scheme 2. In this hypothesis, NNK is reduced to NNAL with a preference for (S)-NNAL, which is sequestered at receptor sites in rat lung and perhaps other tissues as well. The sequestration of (S)-NNAL protects it from α-methyl hydroxylation and PHB-DNA adduct formation. It is then released from the protected site and oxidized to NNK, which forms POB-DNA adducts. (R)-NNAL, on the other hand, undergoes glucuronidation as well as α-hydroxylation to form PHB-DNA and methyl-DNA adducts. Our results in this study fully supported this hypothesis, and indicate that the sequestration of (S)-NNAL not only occurs in the lung, but also in other target tissues of NNK. In fact, Wu et al. (10) showed the presence of (S)-NNAL in liver and kidney 24 h after NNK or NNAL administration, although higher concentrations were found in the lung. Our results suggest extensive distribution and retention of (S)-NNAL, possibly at receptor sites, in various extra-hepatic tissues including pancreas, nasal and oral mucosa. NNK binds to nicotinic acetylcholine and β-adrenergic receptors (30–32). It is likely that NNAL could also bind to these receptors stereoselectively. In fact, (R)-epinephrine, the natural substrate for the β2-adrenergic receptor, is not structurally dissimilar to (S)-NNAL (12). Experiments are in progress to test the binding affinity of (R)- and (S)-NNAL to these receptors. After binding, (S)-NNAL is sequestered and protected from α-hydroxylation. This tissue-specific sequestration of (S)-NNAL, followed by its reoxidation to NNK and local activation of NNK, is likely to be important in NNK tumorigenesis in rats.
Scheme 2.
Proposed mechanisms for the interconversion of NNK and NNAL, and their DNA adduct formation in rats.
There was extensive formation of PHB-DNA adducts from (R)-NNAL-treated rats, which was also observed previously in the rat liver and lung (24). In A/J mice, (S)-NNAL, with potency similar to that of NNK, induced more lung tumors than (R)-NNAL (5). Therefore, we might have expected low PHB-DNA adduct formation in (R)-NNAL treated rats. However, our results demonstrate equal or higher levels of PHB-DNA adducts in the (R)-NNAL group compared with the POB-DNA adduct counterparts in the NNK and (S)-NNAL groups. Although less potent than NNK or (S)-NNAL, (R)-NNAL is still a fairly strong carcinogen in A/J mice (5). Studies also suggested that DNA methylation was critically important in NNK tumorigenesis in A/J mice (33). The difference between the carcinogenicity of (R)- and (S)-NNAL in A/J mice might be in part due to differences in methyl-DNA adduct formation. The mutation properties of PHB-DNA adducts have not been evaluated. O6-PHB-dGuo may be mutagenic as its structural analogue O6-POB-dGuo, which induces G to A transitions and G to T transversions in E. coli. and human cells (34). However, this adduct only constitutes a small percentage of the total PHB-DNA adducts. Moreover, the carcinogenicity of (R)- and (S)-NNAL in rats is not known either, and it might be different from that in A/J mice. Therefore, the importance of the PHB-DNA adducts in (R)-NNAL-treated rats is not clear. Studies to investigate the carcinogenicity of NNK and enantiomers of NNAL in rats would help to answer this question.
The highest levels of POB- and PHB-DNA adduct formation were clearly observed in the nasal respiratory mucosa. Nasal cavity is the second most common target tissue when NNK is administered by s.c. injection (3). A total dose of 0.33 mmol/kg NNK by s.c. injection induced nasal tumors in 22 % of F344 rats (35). However, NNK given in the drinking water is less potent as a nasal carcinogen. Fewer nasal tumors were induced when NNK was given by this route at a total dose of 0.68 mmol/kg (36). This difference is believed to be related to the liver clearance of orally administered NNK. DNA pyridyloxobutylation was shown to be important in rat nasal carcinogenesis by NNK in a study using deuterated NNK analogues (14). NNK and NNN, both of which can pyridyloxobutylate DNA, have similar carcinogenic activities toward the rat nose, but N-nitrosodimethylamine (NDMA), which can only methylate DNA, has little effect (3). POB-DNA adducts, as measured by released 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB), were detected in nasal olfactory and respiratory mucosa of NNK-treated rats, in similar amounts (14). In our study, POB-DNA adducts were significantly higher in the respiratory than in the olfactory mucosa. This could be due to the different doses and routes of administration used in the two studies.
The efficient formation of POB- and PHB-DNA adducts in rat nasal respiratory mucosa requires a highly active P450 enzyme to catalyze the α-hydroxylation of NNK and NNAL. Rat nasal mucosa contains significant levels of P450s (37). Rat P450 2A3 appears to be the most efficient P450 for NNK metabolic activation(38). It also catalyzes α-hydroxylation of NNAL, but the catalytic efficiency is about 2 orders of magnitude lower (8). P450 2A3 is expressed in the rat lung and olfactory mucosa, but not in the respiratory mucosa (39). Considering these properties of P450 2A3, there must be other P450s present in the nasal respiratory mucosa that are responsible for the α-hydroxylation of NNK and NNAL. Or, it is possible that intermediates formed in the olfactory mucosa are transported to the respiratory mucosa. Some other rat nasal P450s, such as P450s 1A1, 2B1, are expressed in both olfactory and respiratory mucosa, and may be involved in NNK activation (38, 40). The differences between the adduct levels in olfactory and respiratory mucosa could also be attributed to differences in DNA repair.
Peterson et al. showed that rat pancreatic microsomes were not able to catalyze α-hydroxylation of NNK or NNAL (41). However in our experiment, POB- and PHB-DNA adducts, which result from α-hydroxylation of NNK and NNAL, were both detected in the pancreas of treated rats. There are some possible explanations to this apparent contradiction. First, in vivo metabolism of NNK and NNAL in the rat pancreas might be different from that observed in in vitro experiments using microsomes. Second, activated forms of NNK and NNAL could be transported to the pancreas, then alkylate DNA to produce adducts. One previous study demonstrated the presence of the O-glucoronide of α-hydroxymethylNNK (4, Scheme 1) in the urine of NNK-treated rats (42). Glucuronidation stabilized this intermediate. This glucuronide may be transported to the pancreas, and deglucuronidated, thus producing DNA reactive species to form adducts. NNK is a pancreatic carcinogen in rats when administered in the drinking water (3). Although the origin of these POB- and PHB-DNA adducts in rat pancreas needs to be further investigated, our results indicate they might play a role in the rat pancreatic carcinogenesis by NNK.
Numerous epidemiology studies have associated cigarette smoking with pancreatic cancer in human (43). NNK was detected in human pancreatic juice (44). However in a subsequent study, DNA pyridyloxobutylation, analyzed as HPB-releasing adducts, was not observed in smokers’ pancreas (45). This study might have been limited by lack of sensitivity to detect low levels of adducts in the pancreas. Furthermore, human exposure to NNK from smoking is far less than the dose used in our animal experiment. Our highly sensitive LC-ESI-MS/MS method will be useful to investigate the POB- and PHB-DNA adducts in human pancreas.
A combination of NNK and NNN, given through oral swabbing, induced tumors in the oral cavity of rats (4), but NNK alone did not cause oral cavity tumors (46). Our recent studies indicated the potential importance of NNN in the induction of oral tumors (data not shown), but NNK might also contribute to this carcinogenic effect. Our results clearly demonstrated the presence of POB- and PHB-DNA adducts in oral mucosa of NNK-, (R)-NNAL- and (S)-NNAL-treated rats, with levels and formation patterns similar to those in other tissues. NNN, through metabolic activation, forms the same POB-DNA adducts as does NNK. Therefore, co-administration of NNK might enhance the carcinogenicity of NNN by producing more POB-DNA adducts, or by enhancing the mutagenicity of NNN-specific adducts.
We have so far analyzed six tissues in NNK- and NNAL-treated rats, and POB- and PHB-DNA adducts were detected in all of them. The highest levels of POB-DNA adducts were observed in the lung of NNK-treated rats (Figure 6A), consistent with the potent carcinogenicity of NNK in the rat lung. Liver had the second most abundant POB-DNA adducts, followed by the nasal respiratory mucosa. Both of them are well-established target tissues of NNK. POB-DNA adduct levels in the nasal olfactory mucosa, oral mucosa, and pancreas were much lower. However, we haven’t found any tissue in treated rats that are free of POB- or PHB-DNA adducts. Although the oral mucosa is generally considered as non-target tissue for NNK tumorigenesis, it may be subjected to NNK tumorigenesis to some extent, as discussed above. Experiments will be carried out to investigate other non-target tissues such as the kidney. Whether these adducts exist in non-target tissues will help to establish the importance of POB- and PHB-DNA adducts in NNK-induced tumorigenesis in rats.
To summarize, we have used LC-ESI-MS/MS to quantitatively analyze POB- and PHB-DNA adducts in pancreas, nasal and oral mucosa of rats treated chronically with NNK, (R)-NNAL and (S)-NNAL. Levels of adducts from NNK and (S)-NNAL treatment exhibited remarkable similarity, and were distinctively different from those formed from (R)-NNAL treatment. These results provide further evidence for our previously proposed mechanism in which (S)-NNAL, formed preferentially in the metabolism of NNK, is sequestered in target tissues, and slowly released and reoxidized to NNK. These events are potentially important in NNK tumorigenesis, and might partially explain its tissue selectivity.
Supplementary Material
Acknowledgments
Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource, and statistical analyses were performed in the Biostatistics and Informatics Shared Resource of the Masonic Cancer Center. These shared resources are supported in part by Grant CA-77598 from the National Cancer Institute. We thank Drs. Fekadu Kassie and Roland Gunther for help with the isolation of rat nasal mucosa, Shannon Nord for help with DNA isolation, and Bob Carlson for manuscript preparation. Stephen S. Hecht is an American Cancer Society Research Professor, supported in part by ACS grant RP-00-138. Siyi Zhang was partially supported by a Graduate School Doctoral Dissertation Fellowship from the University of Minnesota. This research was supported by Grant CA-81301 from the National Cancer Institute.
References
- 1.Hecht SS, Hoffmann D. Tobacco-specific nitrosamines, an important group of carcinogens in tobacco and tobacco smoke. Carcinogenesis. 1988;9:875–884. doi: 10.1093/carcin/9.6.875. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 89. IARC; Lyon, France: 2007. Smokeless tobacco and tobacco-specific nitrosamines. [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS, Rivenson A, Braley J, DiBello J, Adams JD, Hoffmann D. Induction of oral cavity tumors in F344 rats by tobacco-specific nitrosamines and snuff. Cancer Res. 1986;46:4162–4166. [PubMed] [Google Scholar]
- 5.Upadhyaya P, Kenney PM, Hochalter JB, Wang M, Hecht SS. Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers and metabolites in the A/J mouse. Carcinogenesis. 1999;20:1577–1582. doi: 10.1093/carcin/20.8.1577. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyaya P, Carmella SG, Guengerich FP, Hecht SS. Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis. 2000;21:1233–1238. [PubMed] [Google Scholar]
- 7.Breyer-Pfaff U, Martin HJ, Ernst M, Maser E. Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab Dispos. 2004;32:915–922. [PubMed] [Google Scholar]
- 8.Jalas JR, Ding X, Murphy SE. Comparative metabolism of the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol by rat cytochrome P450 2A3 and human cytochrome P450 2A13. Drug Metab Dispos. 2003;31:1199–1202. doi: 10.1124/dmd.31.10.1199. [DOI] [PubMed] [Google Scholar]
- 9.Jalas JR, Hecht SS. Synthesis of stereospecifically deuterated 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) diastereomers and metabolism by A/J mouse lung microsomes and cytochrome P450 2A5. Chem Res Toxicol. 2003;16:782–793. doi: 10.1021/tx034021t. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Upadhyaya P, Carmella SG, Hecht SS, Zimmerman CL. Disposition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in bile duct-cannulated rats: stereoselective metabolism and tissue distribution. Carcinogenesis. 2002;23:171–179. doi: 10.1093/carcin/23.1.171. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman CL, Wu Z, Upadhyaya P, Hecht SS. Stereoselective metabolism and tissue retention in rats of the individual enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), metabolites of the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Carcinogenesis. 2004;25:1237–1242. doi: 10.1093/carcin/bgh120. [DOI] [PubMed] [Google Scholar]
- 12.Hecht SS, Carmella SG, Ye M, Le KA, Jensen JA, Zimmerman CL, Hatsukami DK. Quantitation of metabolites of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone after cessation of smokeless tobacco use. Cancer Res. 2002;62:129–134. [PubMed] [Google Scholar]
- 13.Preussmann R, Stewart BW. N-Nitroso carcinogens. In: Searle CE, editor. Chemical Carcinogens. Washington, DC: American Chemical Society; 1984. pp. 643–828. [Google Scholar]
- 14.Trushin N, Rivenson A, Hecht SS. Evidence supporting the role of DNA pyridyloxobutylation in rat nasal carcinogenesis by tobacco-specific nitrosamines. Cancer Res. 1994;54:1205–1211. [PubMed] [Google Scholar]
- 15.Morse MA, Wang CX, Stoner GD, Mandal S, Conran PB, Amin SG, Hecht SS, Chung FL. Inhibition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced DNA adduct formation and tumorigenicity in lung of F344 rats by dietary phenethyl isothiocyanate. Cancer Res. 1989;49:549–553. [PubMed] [Google Scholar]
- 16.Staretz ME, Foiles PG, Miglietta LM, Hecht SS. Evidence for and important role of DNA pyridyloxobutylation in rat lung carcinogenesis by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: effects of dose and phenethyl isothiocyanate. Cancer Res. 1997;57:259–266. [PubMed] [Google Scholar]
- 17.Sticha KRK, Kenney PMJ, Boysen G, Liang H, Su X, Wang M, Upadhyaya P, Hecht SS. Effects of benzyl isothiocyanate and phenethyl isothiocyanate on DNA adduct formation by a mixture of benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in A/J mouse lung. Carcinogenesis. 2002;23:1433–1439. doi: 10.1093/carcin/23.9.1433. [DOI] [PubMed] [Google Scholar]
- 18.Boysen G, Kenney PMJ, Upadhyaya P, Wang M, Hecht SS. Effects of benzyl isothiocyanate and 2-phenethyl isothiocyanate on benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone metabolism in F-344 rats. Carcinogenesis. 2003;24:517–525. doi: 10.1093/carcin/24.3.517. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco specific carcinogens. Chem Res Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 21.Hecht SS, Villalta PW, Sturla SJ, Cheng G, Yu N, Upadhyaya P, Wang M. Identification of O2-substituted pyrimidine adducts formed in reactions of 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanol with DNA. Chem Res Toxicol. 2004;17:588–597. doi: 10.1021/tx034263t. [DOI] [PubMed] [Google Scholar]
- 22.Upadhyaya P, Sturla SJ, Tretyakova N, Ziegel R, Villalta PW, Wang M, Hecht SS. Identification of adducts produced by the reaction of 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanol with deoxyguanosine and DNA. Chem Res Toxicol. 2003;16:180–190. doi: 10.1021/tx0256376. [DOI] [PubMed] [Google Scholar]
- 23.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht SS, Spratt TE, Trushin N. Absolute configuration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol formed metabolically from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 1997;18:1851–1854. doi: 10.1093/carcin/18.9.1851. [DOI] [PubMed] [Google Scholar]
- 27.Sturla SJ, Scott J, Lao Y, Hecht SS, Villalta PW. Mass spectrometric analysis of relative levels of pyridyloxobutylation adducts formed in the reaction of DNA with a chemically activated form of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 2005;18:1048–1055. doi: 10.1021/tx050028u. [DOI] [PubMed] [Google Scholar]
- 28.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem Res Toxicol. 2006;19:319–324. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta RC. Enhanced sensitivity of 32P-postlabeling analysis of aromatic carcinogen:DNA adducts. Cancer Res. 1985;45:5656–5662. [PubMed] [Google Scholar]
- 30.Schuller HM, Orloff M. Tobacco-specific carcinogenic nitrosamines. Ligands for nicotinic acetylcholine receptors in human lung cancer cells. Biochem Pharmacol. 1998;55:1377–1384. doi: 10.1016/s0006-2952(97)00651-5. [DOI] [PubMed] [Google Scholar]
- 31.Schuller HM. Nitrosamines as nicotinic receptor ligands. Life Sci. 2007;80:2274–2280. doi: 10.1016/j.lfs.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuller HM, Tithof PK, Williams M, Plummer H., 3rd The tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone is a beta-adrenergic agonist and stimulates DNA synthesis in lung adenocarcinoma via beta-adrenergic receptor-mediated release of arachidonic acid. Cancer Res. 1999;59:4510–4515. [PubMed] [Google Scholar]
- 33.Peterson LA, Hecht SS. O6-methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenesis in A/J mouse lung. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
- 34.Pauly GT, Peterson LA, Moschel RC. Mutagenesis by O6-[4-oxo-4-(3-pyridyl)butyl]guanine in Escherichia coli and human cells. Chem Res Toxicol. 2002;15:165–169. doi: 10.1021/tx0101245. [DOI] [PubMed] [Google Scholar]
- 35.Hecht SS, Chen CB, Ohmori T, Hoffmann D. Comparative carcinogenicity in F344 rats of the tobacco-specific nitrosamines, N′-nitrosonornicotine and 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 1980;40:298–302. [PubMed] [Google Scholar]
- 36.Rivenson A, Hoffmann D, Prokopczyk B, Amin S, Hecht SS. Induction of lung and exocrine pancreas tumors in F344 rats by tobacco-specific and Areca-derived N-nitrosamines. Cancer Res. 1988;48:6912–6917. [PubMed] [Google Scholar]
- 37.Reed CJ. Drug metabolism in the nasal cavity: relevance to toxicology. Drug Metab Rev. 1993;25:173–205. doi: 10.3109/03602539308993975. [DOI] [PubMed] [Google Scholar]
- 38.Jalas JR, Hecht SS, Murphy SE. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem Res Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 39.Su T, Sheng JJ, Lipinskas TW, Ding X. Expression of CYP2A genes in rodent and human nasal mucosa. Drug Metab Dispos. 1996;24:884–890. [PubMed] [Google Scholar]
- 40.Thornton-Manning JR, Dahl AR. Metabolic capacity of nasal tissue interspecies comparisons of xenobiotic-metabolizing enzymes. Mutat Res. 1997;380:43–59. doi: 10.1016/s0027-5107(97)00126-7. [DOI] [PubMed] [Google Scholar]
- 41.Peterson LA, Ng DK, Stearns RA, Hecht SS. Formation of NADP(H) analogs of tobacco-specific nitrosamines in rat liver and pancreatic microsomes. Chem Res Toxicol. 1994;7:599–608. doi: 10.1021/tx00041a003. [DOI] [PubMed] [Google Scholar]
- 42.Murphy SE, Spina DA, Nunes MG, Pullo DA. Glucuronidation of 4-((hydroxymethyl)nitrosamino)-1-(3-pyridyl)-1-butanone, a metabolically activated form of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, by phenobarbital-treated rats. Chem Res Toxicol. 1995;8:772–779. doi: 10.1021/tx00047a018. [DOI] [PubMed] [Google Scholar]
- 43.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, France: 2004. Tobacco smoke and involuntary smoking. [PMC free article] [PubMed] [Google Scholar]
- 44.Prokopczyk B, Hoffmann D, Bologna M, Cunningham AJ, Trushin N, Akerkar S, Boyiri T, Amin S, Desai D, Colosimo S, Pittman B, Leder G, Ramadani M, Henne-Bruns D, Beger HG, El-Bayoumy L. Identification of tobacco-derived compounds in human pancreatic juice. Chem Res Toxicol. 2002;15:677–685. doi: 10.1021/tx0101088. [DOI] [PubMed] [Google Scholar]
- 45.Prokopczyk B, Leder G, Trushin N, Cunningham AJ, Akerkar S, Pittman B, Ramadani M, Straeter J, Beger HG, Henne-Bruns D, El-Bayoumy K. 4-Hydroxy-1-(3-pyridyl)-1-butanone, an indicator for 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced DNA damage, is not detected in human pancreatic tissue. Cancer Epidemiol Biomarkers Prev. 2005;14:540–541. doi: 10.1158/1055-9965.EPI-04-0626. [DOI] [PubMed] [Google Scholar]
- 46.Prokopczyk B, Rivenson A, Hoffmann D. A study of betel quid carcinogenesis. IX Comparative carcinogenicity of 3-(methylnitrosamino)propionitrile and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone upon local application to mouse skin and rat oral mucosa. Cancer Lett. 1991;60:153–157. doi: 10.1016/0304-3835(91)90222-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.