Abstract
Mammalian polarity proteins have been studied predominantly in cell culture systems, and little is known about their functions in vivo. To address this issue, we used a shRNA lentiviral system to manipulate gene expression in mouse mammary stem/progenitor cells. Transplantation of Par3-depleted stem/progenitor cells into the mammary fat pad severely disrupted mammary development, and glands were characterized by ductal hyperplasia, luminal filling, and highly disorganized end bud structures that were unable to remodel into normal ductal structures. Unexpectedly, Par3-depleted mammary glands also had an expanded progenitor population. We identified a novel function for the atypical protein kinase C (aPKC)-binding domain of Par3 in restricting Par3 and aPKC to the apical region in mammary epithelia in vivo, and found that mammary morphogenesis is dependent on the ability of Par3 to directly bind aPKC. These results reveal a new function for Par3 in the regulation of progenitor differentiation and epithelial morphogenesis in vivo and demonstrate for the first time an essential requirement for the Par3–aPKC interaction.
Keywords: Mammary gland, progenitor, end bud, polarity, Par3, lentivirus
Par3 is an evolutionarily conserved polarity protein that was originally identified as an essential factor for the asymmetric cell division of the Caenorhabditis elegans zygote (Goldstein and Macara 2007). It was later found to be required for neuroblast and epithelial polarization during Drosophila embryogenesis, and in vertebrates regulates various modes of polarization during migration, neuronal development, and tight junction formation, as well as tissue organization during heart and brain development (Mertens et al. 2005; Hirose et al. 2006; Goldstein and Macara 2007; Pegtel et al. 2007; Costa et al. 2008). Par3 and Par6 can function independently of one another or as part of the Par complex together with atypical protein kinase C (aPKC). Each component of this complex can interact directly with the other two components. However, the regulation of the complex is not fully understood. Par6 binds to the N-terminal regulatory domain of aPKC and inhibits its kinase activity (Joberty et al. 2000; Yamanaka et al. 2001). This inhibition is relieved by the binding of Par6 with Cdc42-GTP (Yamanaka et al. 2001; Atwood et al. 2007). Par3 also binds aPKC, through the kinase domain, and can act as an inhibitor or a substrate of aPKC (Lin et al. 2000; Nagai-Tamai et al. 2002). Phosphorylation of Par3 by aPKC within the aPKC-binding domain (on Ser827) causes the two proteins to dissociate (Nagai-Tamai et al. 2002). The interaction between Par3 and aPKC is likely dynamic since protein phosphatase 1 can dephosphorylate Ser827 (Traweger et al. 2008). Despite evolutionary conservation of the aPKC-binding domain in Par3 orthologs, its biological function remains unknown. No role has been ascribed to this domain of Par3 in cell polarization or the symmetric cell divisions of Drosophila or C. elegans. In mammalian cells, the overexpression of Par3 that lacks the aPKC phosphorylation site slightly reduces tight junction formation in MDCK monolayers, as compared with the wild-type Par3 (Hirose et al. 2002; Nagai-Tamai et al. 2002). However, the mechanism was not investigated.
In general, the establishment and regulation of polarity in mammalian epithelial morphogenesis has been studied predominantly in two-dimensional (2D) and three-dimensional (3D) cultures (Zegers et al. 2003; Debnath and Brugge 2005) but remains poorly understood in vivo, where more complex cell–cell and cell–matrix interactions are present. The ability of the mammary gland to completely regenerate following the transplantation of mammary cells into a cleared fat pad (Deome et al. 1959) makes it an ideal model for studying epithelial polarization and morphogenesis in vivo. Mammary gland stem cells are necessary for the regenerative potential of the mammary gland, and the transplant mammary model has become an excellent tool for understanding stem cell renewal and differentiation (Shackleton et al. 2006; Stingl et al. 2006). The mammary gland is a bilayered epithelial ductal system that consists of a luminal epithelial layer surrounded by a contractile myoepithelial layer. The luminal layer is characterized by expression of cytokeratin 8 (K8) and cytokeratin 18 (K18), whereas the myoepithelial layer expresses cytokeratin 14 (K14) and smooth muscle actin (SMA). After the onset of puberty in rodents, terminal end bud structures form at the tips of growing ducts and consist of two major cell types: multilayered inner body cells and an outer layer of cap cells that are thought to be progenitors for the myoepithelial layer (Hinck and Silberstein 2005). The end bud serves as a stem/progenitor cell niche during development (Kenney et al. 2001). Ductal elongation and branch bifurcation are the result of the coordination of many processes including proliferation, apoptosis, differentiation and cell motility. However, the cellular mechanisms that control epithelial organization during branching morphogenesis are not well understood.
To determine the role of Par3 during epithelial morphogenesis in vivo, we used shRNA lentiviruses to deplete Par3 from mammary stem/progenitor cells and assessed subsequent outgrowth after transplantation into host mice.
Results
Knockdown of Par3 in mouse mammary cells
We first tested two shRNA sequences for their ability to deplete Par3 in mouse NIH 3T3 cells (Fig. 1A,B). ShPar3-1 was more effective and was used to further define the conditions for Par3 depletion in primary mouse mammary epithelial cells (MECs).
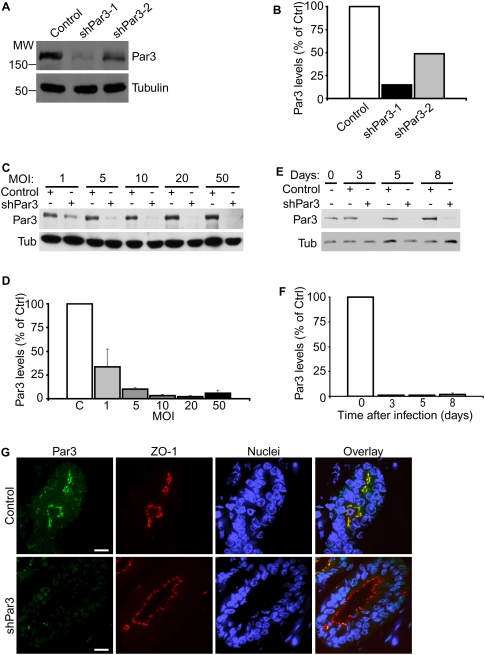
Figure 1.
Knockdown of Par3 in primary mouse mammary epithelial and stem cells. (A) NIH 3T3 cells were transduced with two different shRNA lentiviruses against mouse Par3. Par3 and tubulin levels were assayed by immunoblot 7 d after transduction. (B) Quantification of Par3 knockdown in A. Data represent means from two independent experiments. Tubulin was used as an internal control. (C) Primary mouse MECs were transduced with control or shPar3 lentivirus at MOIs equal to 1, 5, 10, 20, or 50. Par3 and tubulin protein levels were assayed by immunoblot 10 d after lentiviral transduction. (D) Quantification of Par3 knockdown in C. Data represent means from two independent experiments. Tubulin was used as an internal control. Error bars, SD. (E) Time course for lentiviral knockdown of Par3 in primary mouse MECs. Cells were transduced at a MOI = 20 with shPar3-1 lentivirus. Par3 protein levels were assayed by immunoblot at the indicated times after lentiviral transduction. (F) Quantification of Par3 knockdown in E. Points represent means from two independent experiments. Tubulin was used as an internal control. Error bars, SD. (G) Paraffin sections of control and shPar3 mammary glands 8 wk after transplantation. Sections were immunostained for Par3 and ZO1. Bars, 20 μm.
To determine the optimal multiplicity of infection (MOI) for Par3 silencing, we infected primary MECs at MOIs ranging from 1 to 50 (Fig. 1C,D). Relative to controls, Par3 levels were diminished at all MOIs tested and were maximally depleted at a MOI between 10 and 20, resulting in >95% loss of Par3 (Fig. 1D). Importantly, we did not observe toxic effects of lentivirus on primary MECs, even at a very high MOI (MOI > 100) (data not shown).
Next, we examined the time course for lentiviral-mediated knockdown of Par3 by transducing primary MECs with control or shPar3-1 lentiviruses and culturing the cells for up to 10 d. Strikingly, Par3 levels were reduced by >95% by 3 d after transduction (Fig. 1E,F). Par3 silencing was persistent in culture for at least 8 d (Fig. 1) and was maintained for at least 8 wk in vivo, as assessed by immunostaining outgrowths of shPar3-transduced mammary stem/progenitor cells (Fig. 1G). These data demonstrate that lentiviral-mediated RNAi is a fast and efficient tool for studying loss-of-gene functions in vivo, even when knockdown results in a severe growth disadvantage (see below; Fig. 2).
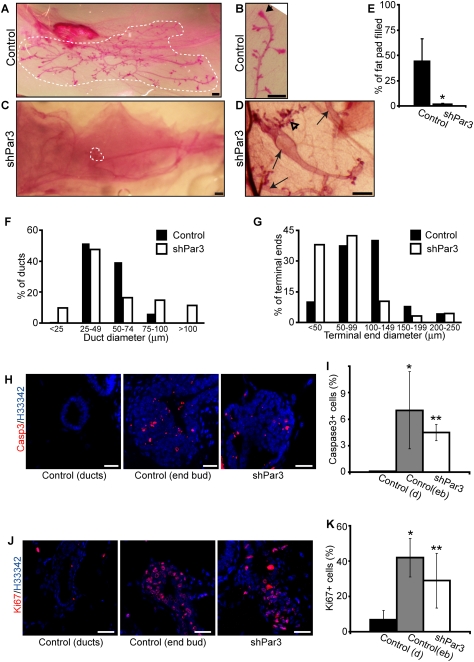
Figure 2.
Par3 is essential for normal mammary gland development. (A,B) Carmine alum-stained whole-mount images of control mammary glands after 8 wk. Arrowhead in B shows an end bud in the developing gland. Bars, 0.5 mm. (C,D) Carmine alum-stained whole-mount images of shPar3 mammary glands after 8 wk of regeneration. Arrows in D show multiple small independent outgrowths in a single fat pad. The open arrowhead in D shows regions of small disorganized ducts that lack prominent end buds. Bars, 0.5 mm. (E) The percentage of the fat pad filled by control (n = 6) and Par3-depleted (n = 8) mammary glands was quantified. Error bars, SD. (*) P = 0.00004. (F) Distribution of duct diameters in control (n = 6) and Par3-depleted (n = 8) mammary glands. The diameters of all ducts from control and Par3-depleted glands were measured and categorized in 20-μm increments. (G) Distribution of end bud sizes of control and Par3-depleted mammary glands. The diameters of all terminal ends from control and Par3-depleted glands were measured and categorized in 50-μm increments. (H) Tissue sections from control and Par3-depleted glands were stained for cleaved Caspase-3 (red) and Hoechst 33342 (blue). Bars, 20 μm. (I) Quantification of cleaved Caspase-3-positive cells in control ducts (black bar, n = 5), control end buds (gray bar, n = 5), and Par3-depleted (white bar, n = 5) glands. Error bars, SD. (*) P = 0.034 control duct versus control end bud; (**) P = 0.001 shPar3 versus control duct; (**) P = 0.37 shPar3 versus control end bud. (J) Tissue sections from control and Par3-depleted glands were stained for Ki67 (red) and Hoechst 33342 (blue). Bars, 20 μm. (K) Quantification of Ki67+ cells in control ducts (black bar, n = 5), control end buds (gray bar, n = 4), and Par3-depleted (white bar, n = 5) glands. Error bars, SD. (*) P = 0.0004, control duct versus control end bud; (**) P = 0.016, shPar3 versus control duct; (**) P = 0.20, shPar3 versus control end bud.
Par3 depletion in progenitors inhibits mammary gland growth
To examine a possible role for Par3 in mammary gland morphogenesis in vivo, single-cell suspensions of freshly isolated MECs were transduced by centrifugation at 300g for 3 h. Prior to injection into the cleared fat pad of 4-wk-old hosts, the transduced cells were grown for 2–3 d as suspension mammospheres, which has been shown to enrich for mammary progenitors (Dontu et al. 2003; Liao et al. 2007; Sansone et al. 2007). We transduced 10,000 cells for each mammary fat pad injection, which resulted in outgrowths in 61% of control transplants (n = 23) and 74% of transplants from Par3-depleted progenitor cells (n = 27) (Supplemental Table 1). YFP marked transduced cells and was expressed uniformly throughout the ducts (Supplemental Fig. S1). Using this method, as few as 1000 cells were sufficient for outgrowths in 33% of control (n = 6) or 60% of shPar3 (n = 10) transplants (Supplemental Table 1). The ability to transduce and transplant a small number of cells is important because it circumvents the problem of using low-titer viruses, such as those that encode large cDNAs (Proia and Kuperwasser 2006; Welm et al. 2008), and has enabled us to perform rescue experiments by expressing a bicistronic lentivirus that contains both shPar3 and cDNAs for human Par3, a 180-kDa protein (see below, Fig. 3).
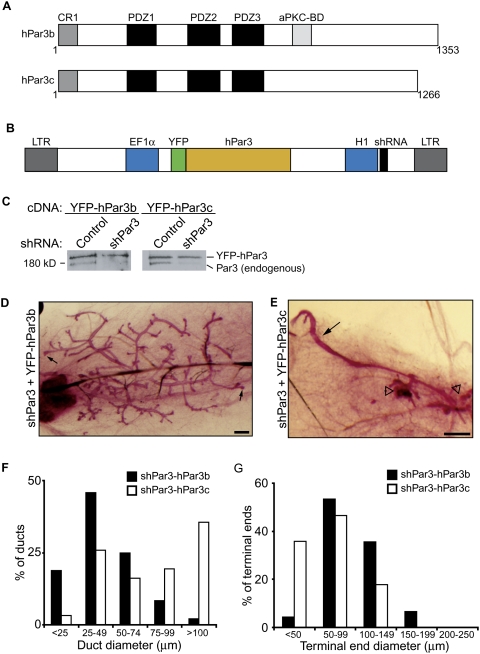
Figure 3.
Mammary development requires aPKC-binding domain of Par3. (A) Schematic representation of human Par3b and human Par3c. Domains shown are the conserved region (CR), PSD95/Dlg/ZO1 domain (PDZ), and atypical protein kinase C-binding domain (aPKC-BD). Numbers indicate amino acid number. (B) Schematic representation of bicistronic lentiviral plasmid for expression of shRNA and cDNA. (LTR) long terminal repeat; (EF1α) EF1α promoter; (H1) H1 RNA polymerase III promoter. (C) Immunoblots of mammary epithelial lysates from cells expressing control/YFP-hPar3b, shPar3/YFP-hPar3b, control/YFPhPar3c, and shPar3/YFPhPar3c bicistronic lentiviruses. Blots were probed using anti-Par3 antibodies. The top band is YFP-hPar3 and the bottom band is endogenous Par3. (D,E) Carmine alum-stained whole-mount images of shPar3-YFPhPar3b (D) and shPar3/YFPhPar3c (E) mammary glands after 8 wk. Small arrows show end buds in D. Open arrowheads show disorganized ducts in E. Bars, 0.5 mm. (F) Distribution of duct diameters in shPar3–hPar3b and shPar3–hPar3c mammary glands (n = 3). The diameters of ducts were measured and categorized in 25-μm increments. (G) Distribution of terminal end sizes from shPar3–hPar3b and shPar3–hPar3c mammary glands (n = 3). The diameters of terminal ends were measured and categorized in 50-μm increments.
Regenerated glands were examined by whole-mount staining and by subsequent immunostaining of paraffin sections. After 8 wk, control mammary glands filled 46% ± 20.3% (SD, n = 6) of the fat pad, whereas Par3-depleted glands filled 2% ± 0.8% (SD, n = 8) of the fat pad (Fig. 2 A,C,E). Several of the shPar3 glands had multiple small outgrowths in a single fat pad (Fig. 2D, arrows), whereas control glands appeared as single outgrowths (Fig. 2A).
Control mammary glands contained regularly branched ducts with a uniform diameter (92% were 25–74 μm wide) (Fig. 2A,F) that penetrated into the mammary fat pad. In contrast, Par3-depleted mammary glands contained ducts with nonuniform diameters and some outgrowths had enlarged primary ducts (26% were >75 μm) (Fig. 2D,F) with few branches. The enlarged ducts in Par3-depleted glands were multilayered (see below, Fig 4A, panel c, arrow) and sometimes contained cells within the lumen (Fig. 5A, panel b, arrows), features that resemble low-grade carcinoma in situ (Feeley and Quinn 2008). Par3-depleted glands also had hyperbranched regions at the ends of the ducts that consisted of several short disorganized ducts with normal widths (Fig. 2D, open arrowhead).
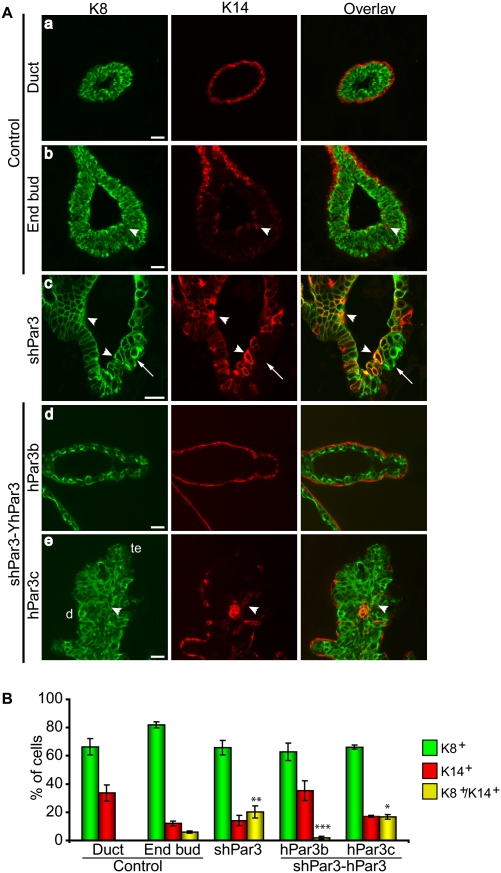
Figure 4.
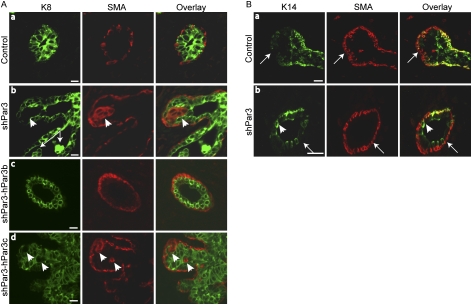
Loss of Par3 perturbs mammary progenitor differentiation and disrupts the myoepithelial layer. (A) Paraffin sections of control and shPar3 mammary glands 8 wk after transplantation were immunostained for K8 (green) and K14 (red). (Panel c) The arrow shows a multilayered duct. In panel e: (te) Terminal end; (d) duct. Bars, 10 μm. (B) Quantification of K8+, K14+, and K8+K14+ cells. A uniform threshold was applied to all images and the proportion of single-positive and dual-positive epithelial cells were counted. Error bars, SD (n = 5). (*) P = 0.00003; (**) P = 0.009; (***) P = 0.02 versus control end bud; (***) P = 0.1 versus control ducts.
Figure 5.
Par3-depleted mammary glands have disorganized basal layers and are surrounded by cap cells. (A) Paraffin sections of control or Par3-depleted mammary glands 8 wk after transplantation were immunostained for K8 (green) and SMA (red). (Panels b,d) Arrowheads indicate SMA+ cell clusters inside the luminal compartment. (Panel b) The arrow shows cells within the lumen. Bars, 10 μm. (B) Paraffin sections of control or Par3-depleted mammary glands 8 wk after transplantation were immunostained for K14 (green) and SMA (red). Arrows show K14−/SMA+ cap cells. The arrowhead shows luminal K14+ cells. Bars, 20 μm.
In addition to disrupted duct structures, Par3-depleted mammary glands lacked the normal end bud structures that are typical of developing mouse mammary glands (Fig. 2B,D,G). As a measure of terminal end morphology, we measured the size distribution of all termini from control (n = 6) and shPar3 (n = 8) mammary glands (Fig. 2G) after 8 wk of regeneration. The majority (79%) of terminal ends in control mammary glands were 50–149 μm, whereas shPar3 glands had fewer terminal ends in this range (52%) and several were very small (38% were <50 μm), corresponding to the short hyperbranched regions of the glands (Fig. 2D, open arrowhead). In control mammary glands, end buds were eight to 12 cells thick, whereas the terminal end structures from shPar3 mammary glands were only one to four cell layers thick (Supplemental Fig. S2A). Together, these results demonstrate that Par3 is necessary for the development of normal end bud and ductal structures during mammary morphogenesis.
To confirm that the defects in mammary morphogenesis were specific to Par3 depletion rather than off-target effects, we examined mammary glands from the second hairpin sequence, shPar3-2 (Supplemental Fig. S3). This hairpin sequence reduced Par3 levels by >80% in primary MECs (Supplemental Fig. S3A) and produced glands that were similar to those observed using shPar3-1 with disorganized ducts that lack normal end buds (Supplemental Fig. S3C).
To further test the specificity of the Par3 shRNA, we designed rescue experiments using a wild-type human Par3 cDNA (hPar3b) that is not recognized by the murine-specific shRNA (Fig. 3A,C). Human Par3 variants were expressed as YFP fusion proteins from bicistronic lentiviral plasmids that contain both YFP-hPar3 and the murine Par3 shRNA sequences (Fig. 3B). Transduction of primary MECs with these viruses demonstrated that endogenous Par3 was effectively knocked down, while the YFP-hPar3 fusions were each expressed at near-endogenous levels and were not targets for the mouse-specific shRNA (Fig. 3C). Expression of shPar3-YFP-hPar3b (the wild-type, full-length variant) completely restored mammary gland development and the development of end bud structures (Fig. 3D [arrow], G), which further confirms the specificity of the shPar3 knockdown. While some of the ducts in shPar3-YFP-hPar3b glands were dilated, they were uniform in diameter and formed normal bilayers of luminal and myoepithelial cells (Fig. 5A, panel c).
We next examined the requirement for the interaction between aPKC and Par3 in mammary gland development by expressing hPar3c, a naturally occurring splice variant that lacks the aPKC-binding domain (Fig. 3A; Gao et al. 2002). In contrast to YFP-hPar3b, the expression of YFP-hPar3c did not restore normal mammary outgrowth. The transduced glands contained unbranched ducts of varying diameters (Fig. 3E, arrow) and regions with several small disorganized ducts (Fig. 3E, open arrowheads), similar to the outgrowths observed in Par3-depleted glands. Collectively, these results demonstrate that Par3 is essential for regulating the epithelial organization required for ductal elongation during mammary morphogenesis. Furthermore, these data provide the first evidence of a key biological function for the aPKC-binding domain of Par3.
Proliferation and apoptosis in Par3-depleted mammary glands
In the mammary gland, both proliferation and apoptosis have been shown to be highest in end buds compared with mature ducts, and are necessary for mammary morphogenesis (Humphreys et al. 1996; Mailleux et al. 2007). We speculated that the small size of shPar3 mammary glands could result from either decreased proliferation or increased cell death. To investigate these possibilities, we stained control and Par3-depleted mammary gland sections for cleaved Caspase-3, a marker for apoptosis, and Ki67, a proliferation marker. Consistent with previous reports, cleaved Caspase-3+ cells were detected only within the end buds of control glands (Humphreys et al. 1996; Mailleux et al. 2007). However, cleaved Caspase-3+ cells were seen throughout the Par3-depleted mammary glands at levels (4.5% ± 0.9% [SD]) similar to those detected in control end buds (7.0% ± 4.4% [SD]) (Fig. 2H,I). To examine whether proliferation was altered in Par3-depleted mammary glands, we stained for Ki67. Surprisingly, although Par3-depleted mammary glands are very small, they had a relatively high percentage of Ki67+ cells (27% ± 12.4% [SD]), which was significantly greater than the proliferation levels in control ducts (7% ± 5.3% [SD]), but similar to the percentage of Ki67+ cells found in control end buds (42% ± 10.9% [SD]) (Fig. 2J,K). Therefore, Par3-depleted mammary glands show high levels of both proliferation and apoptosis. The high proliferation rate of shPar3 mammary glands is offset by apoptosis, expansion of the duct diameter, generation of multilayered ducts, and luminal filling, which collectively will reduce gland size.
Expansion of K8+K14+ dual-positive cells in Par3-depleted mammary glands
To examine the architecture of Par3-depleted mammary glands, we stained for K8 for luminal epithelial cells and for K14 for the myoepithelial layer. K8+ cells were uniformly present within the luminal layer of control ducts, as expected, and were surrounded by a single layer of K14+ myoepithelial cells along the ducts, but not around the end buds (Fig. 4A, panels a,b). In contrast, the keratin staining profile in Par3-depleted mammary glands was severely disrupted. K14+ single-positive cells were frequently absent from the basal layer of shPar3-depeleted mammary ducts (Fig. 4A, panel c, arrow) but were present within the luminal compartment as part of an unusually large K8+K14+ dual-positive population (20% ± 4%) (Fig. 4A [panel c, arrowhead], B).
The end bud represents a potential stem/progenitor cell niche in the developing rodent mammary gland (Kenney et al. 2001), and several groups have correlated bipotential progenitor identity with K8+K14+ dual expression (Woodward et al. 2005; Shackleton et al. 2006; Villadsen et al. 2007; Wang et al. 2008). In support of this idea, dual-positive K8+K14+ cells were only found in the end buds (6.0% ± 0.8%) of control glands, while none were detected in mature ducts (Fig. 4A,B). In Par3-depleted glands, there was a twofold expansion of K8+K14+ dual-positive cells compared with control end buds (Fig. 4). This expansion appears to be at the expense of myoepithelial cells, because there is a concomitant decrease in the proportion of K14+ single-positive cells, with no change in K8+ cells. Importantly, Par3-depleted glands that expressed hPar3b, but not hPar3c, restored the normal phenotype and exhibited normal K8 and K14 expression profiles in mammary glands (Fig. 4A [panels d,e], B). This result indicates that the binding of aPKC to Par3 normally limits the expansion of K8+K14+ dual-positive cells.
Par3-depleted mammary glands have disorganized myoepithelial layers
To determine whether distinct basal and luminal layers were present in Par3-depleted mammary glands, we examined SMA as a myoepithelial marker that, unlike K14, is also present in cap cells. As expected, SMA+ cells were restricted to the single outer layer of myoepithelial cells in mature ducts and end buds in control mammary glands (Fig. 5A [panel a], B [panel a]). Strikingly, the Par3-depleted mammary ducts also possessed a layer of SMA+ cells outside of the luminal cells, confirming that distinct luminal and basal layers are present in Par3-depleted mammary glands (Fig. 5A, panel b). However, in some glands there were small gaps in the SMA+ basal layer (Supplemental Fig. S4), but this was much less severe than the gaps between K14+ cells in the basal layer. Furthermore, clusters of SMA+ cells were intermixed with luminal K8+ cells (Fig. 5A, panel b, arrowhead), but none of these SMA+ cells coexpressed K8, arguing that they are distinct from the population of K8+K14+ dual-positive cells. Expression of shPar3 plus YFP-hPar3b was able to restore the wild-type duct morphology, with a normal distribution of SMA+ cells, whereas mammary glands expressing shPar3 plus YFP-hPar3c resembled Par3-depleted glands, with a disorganized basal layer of SMA+ cells (Fig. 5A, panel d, arrowhead). Therefore, although there are distinct luminal and basal layers in Par3-depleted glands, the layers are disrupted and disorganized. Moreover, Par3 requires the aPKC-binding domain for normal mammary gland organization.
In control mammary glands, ducts are surrounded by a K14+SMA+ myoepithelial layer, whereas end buds are surrounded by K14−SMA+ cap cells (Fig. 5B; Mailleux et al. 2007). Double-staining for SMA and K14 shows that Par3-depleted mammary ducts are also surrounded by a K14−SMA+ cap cell layer (Fig. 5B). Taken together, these data all argue that in the absence of Par3, mammary end buds are defective in cell fate determination, which prevents the generation of mature ducts.
Direct association between Par3 and aPKC is required for their correct subcellular distribution
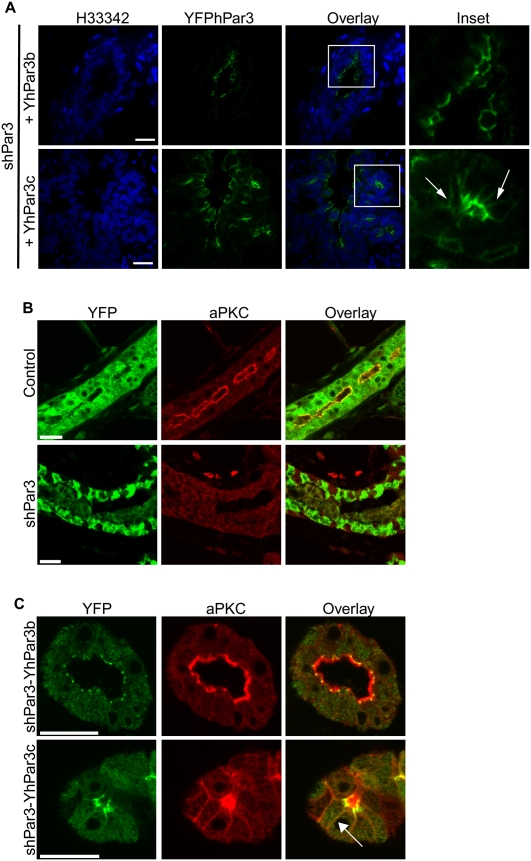
We demonstrated that full-length hPar3 is able to restore normal mammary gland development and that hPar3c is unable to restore any aspect of mammary development or prevent progenitor expansion in Par3-depleted mammary glands, demonstrating for the first time an essential role for the aPKC-binding domain of Par3. To address the mechanism underlying the requirement for aPKC binding to Par3 in mammary development, we examined the subcellular distribution of YFP-hPar3b and YFP-hPar3c. Endogenous Par3 and YFP-hPar3b were both apical in luminal cells, and concentrated at tight junctions (Figs. 1G, 6A). In contrast, although YFP-hPar3c is present at the apical surface of luminal cells, it was also visible on the lateral surfaces (Fig. 6A, inset). This result suggests that binding of aPKC helps restrict Par3 apically, which is necessary for its function in regulating mammary morphogenesis. The localization of hPar3c on lateral membranes is unlikely to be an artifact of overexpression because hPar3b and hPar3c were expressed at similar levels, and were expressed at near-endogenous levels in MECs transduced with our bicistronic lentiviruses (Fig. 3C). In Drosophila and C. elegans embryos, aPKC localization is dependent on Par3 (Tabuse et al. 1998; Harris and Peifer 2005). We asked, therefore, whether aPKC localization was disrupted in Par3-depleted mammary glands. Indeed, while aPKC was concentrated at the apical surface in control mammary glands, the kinase was completely absent from membranes in Par3-depleted mammary glands (Fig. 6B). We further investigated the subcellular distribution of aPKC in the Par3-depleted glands expressing YFP fusions of hPar3b or hPar3c. The expression of hPar3b completely restored aPKC localization to the apical surface in luminal epithelial cells (Fig. 6C). In contrast, expression of hPar3c partially restored aPKC localization to the apical surface, but aPKC was also mislocalized to the lateral membranes along with hPar3c (Fig. 6C, arrow). Unexpectedly, therefore, Par3 and aPKC are mutually dependent for their restricted apical localization, which requires the aPKC-binding domain of Par3.
Figure 6.
Par3 and aPKC are mutually dependent for their subcellular distribution. (A) Tissue sections of shPar3-YFPhPar3b and shPar3-YFPhPar3c mammary glands were stained for YFP (green) and with Hoechst 33342 (H33342, blue). Arrows show hPar3c localized on the lateral membrane. (B) Control and Par3-depleted mammary glands were stained for YFP (which marks transduced cells, green) and aPKC (red). (C) shPar3-YFPhPar3b and shPar3-YFPhPar3c mammary glands were stained for YFP (green) and aPKC (red). The arrow shows mislocalization of aPKC and hPar3c on the lateral membranes. Bars, 20 μm.
Par3 and aPKC regulate mammary progenitor differentiation
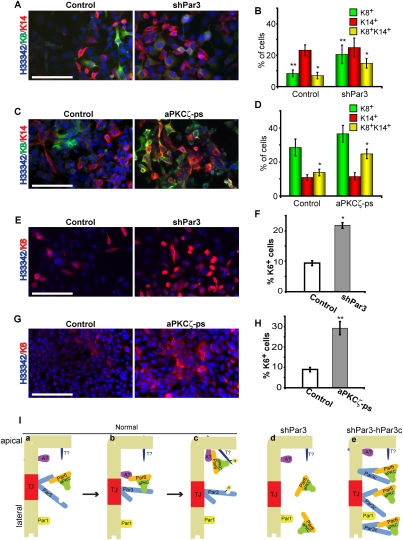
Since the aPKC-binding domain of Par3 is necessary for restricting the expansion of K8+K14+ progenitors, we tested directly whether aPKC is involved in the differentiation pathway of MECs. To address this question, we performed in vitro differentiation assays using Comma-1D cells. Comma-1D cells are a mammary stem/progenitor cell line that is capable of regenerating a complete mammary gland when injected into cleared mammary fat pads, and also undergoes consistent differentiation in vivo and in vitro (Danielson et al. 1984; Wang et al. 2008; Zhan et al. 2008). Similar to what we observed in vivo, depletion of Par3 resulted in a twofold increase in the number of K8+K14+ dual-positive cells (Fig. 7A,B). There was also an increase in the number of luminal (K8+) cells in Par3-depleted cultures at the expense of K8−K14− dual-negative cells, but the relevance of this increase is unclear since we did not observe a similar elevation in the luminal population in vivo. Importantly, inhibition of aPKC using a cell-permeable pseudosubstrate inhibitor also caused a twofold increase in the percentage of K8+K14+ dual-positive progenitors (Fig. 7C,D). To further examine differentiation of mammary progenitor cells, we stained for a second putative mammary progenitor marker, keratin 6 (K6) (Li et al. 2003; Woodward et al. 2005). Depletion of Par3 or inhibition of aPKC each caused a significant increase in the percentage of K6+ cells by twofold or threefold, respectively (Fig. 7E–H). Therefore, depletion of Par3 or inhibition of aPKC disrupts the normal differentiation pathway and causes the accumulation of mammary progenitors in vitro.
Figure 7.
Par3 and aPKC regulate progenitor differentiation. (A) Control or Par3-depleted COMMA-1D cells were grown for 7 d and stained for K8 and K14. (B) Quantification of A. (*) P = 0.005 (control vs. shPar3); (**) P = 0.01 (control vs. shPar3). (C) COMMA-1D cells were treated with 40 μM myristoylated aPKCζ pseudosubstrate every 48 h for 7 d, then stained for K8 and K14. (D) Quantification of C. (*) P = 0.003 (control vs. shPar3). (E) Control or Par3-depleted COMMA-1D cells were grown for 7 d and stained for K6. (F) Quantification of E. (*) P = 1.2 × 10−13 (control vs. shPar3). (G) COMMA-1D cells were treated with 40 μM myristoylated aPKCζ pseudosubstrate every 48 h for 7 d, then stained for K6. (H) Quantification of G. (**) P = 5.8 × 10−7 (control vs. aPKCζ-ps). Bars, 100 μm;error bars, SD; n = 4. (I) Model for Par3/aPKC function in MECs. Par3 permits the recruitment of the constitutive aPKC/Par6 complex to tight junctions (panel a), where the complex is handed off to an apical protein (panel b; apical protein [A?]) that is required for phosphorylating an apical target (panel c; apical target [T?]). (Panel d) In the absence of Par3, aPKC is not recruited to the plasma membrane and cannot phosphorylate apical targets. (Panel e) The Par3c variant can recruit aPKC to the plasma membrane, but cannot hand off aPKC appropriately, so both proteins remain bound and diffuse onto the apical and lateral surfaces, but aPKC cannot phosphorylate its normal targets. See the Discussion for additional details.
Discussion
Organ regeneration from transplanted stem cells that have been transduced with lentiviruses provides a powerful approach to studying gene function during morphogenesis (Siwko et al. 2008). Not only can gene expression be silenced easily using shRNAs, but the same lentivirus can be used to rescue expression by a knock-in strategy, using wild-type or mutant genes that are resistant to the shRNA. We took this approach to address the question of polarity protein function during mammary gland morphogenesis, and identified a novel function for Par3 and aPKC in regulating mammary differentiation and morphogenesis.
We discovered that silencing of Par3 severely inhibits ductal elongation of mammary glands. The ducts that do form are disorganized and lack normal end bud structures. Some of the ducts are multilayered, with cells inside the lumen, which resembles hyperplastic tissue found in ductal carcinoma in situ (Feeley and Quinn 2008). We propose that Par3-depleted glands are composed predominantly of an expanded end bud cell population that does not form normal bulbous end bud structures and is unable to differentiate and organize into normal mammary epithelial ducts. This idea is supported by several lines of evidence. First, the Par3-depleted glands, like end buds, are highly proliferative and apoptotic (Fig. 2). In normal glands, end bud proliferation is necessary to produce cells to form ducts as they elongate and invade through the fatty stroma, and apoptosis is also important for lumen formation (Humphreys et al. 1996; Mailleux et al. 2007). Second, Par3-depleted glands contain K8+K14+ dual-positive cells throughout the glands. In normal mouse glands, K8+K14+ dual-positive cells were located exclusively in end buds, although a small number of dual-positive cells have been reported by others in ducts from human breast tissue (Villadsen et al. 2007). Third, both end buds and Par3-depleted glands are surrounded by K14−SMA+ cap cells.
How does Par3 regulate mammary morphogenesis? Using bicistronic lentiviruses that express both a short hairpin to mouse Par3 and human Par3 cDNAs, we found that the aPKC-binding domain of Par3 is essential for mammary development. Full-length Par3 was able to restore all aspects of mammary gland defects from Par3-depleted mammary glands, whereas a Par3 variant that lacks the aPKC-binding domain did not rescue mammary gland development.
The relationship between Par3 and aPKC is context-dependent, and is not fully understood. For example, in the C. elegans zygote, Par3, Par6, and aPKC colocalize in a mutually dependent fashion (Goldstein and Macara 2007), but in the embryonic epithelium of Drosophila, Par3 localization is independent of aPKC, and is positioned below aPKC and Par6, while aPKC localization requires Par3 (Harris and Peifer 2005). In mammalian epithelial cells, Par3 is associated with the tight junctions, and aPKC and Par6 are predominantly apical. However, the interdependencies of positioning have not been elucidated in mammalian cells. Moreover, the roles of direct binding of aPKC to Par3 versus indirect association through Par6 are not understood. Here we demonstrated for the first time that mammalian aPKC localization is dependent on a direct interaction with Par3. In the absence of Par3, the apical distribution of aPKC is completely lost, but can be rescued by the expression of a human YFP-Par3 fusion. Strikingly, however, the expression of YFP-Par3c, a splice variant that cannot directly bind aPKC, resulted in the abnormal codistribution of aPKC and the YFP-Par3c along the lateral membranes of the luminal epithelial cells. We conclude that a direct Par3- aPKC association is needed both to restrict Par3 to the tight junctions, and to retain aPKC at the apical surface. What mechanism might account for this behavior? We propose the following model (Fig. 7I): (1) Par3 permits the recruitment of the constitutive aPKC/Par6 complex to tight junctions, where the complex is transferred to an apical binding partner, such as Pals1 or Crumbs (Hurd et al. 2003b; Harris and Peifer 2005). (2) In the absence of Par3, aPKC is not recruited to the plasma membrane (Fig. 6B), and therefore cannot phosphorylate targets at the apical surface. (3) The Par3c variant can recruit aPKC to the plasma membrane, but cannot appropriately release aPKC to the apical surface, and both proteins remain in a complex that can spread onto the apical and lateral surfaces (Fig. 6C). Since Par3 can change the target specificity of aPKC (Wirtz-Peitz et al. 2008), Par3c may both mislocalize aPKC and prevent the phosphorylation of its normal targets. (4) Par3 is normally excluded from the lateral membranes through phosphorylation by Par1, a serine/threonine kinase located at the lateral cortex of epithelial cells (Benton and St Johnston 2003; Hurd et al. 2003a). However, if Par1-mediated phosphorylation of Par3 is dependent on the absence of aPKC, then the Par3c variant will not dissociate from the lateral domain, and will accumulate there. The basis for this last part of the model is that aPKC can phosphorylate and inhibit Par1 (Suzuki et al. 2004). Thus, Par3c, which can associate only indirectly with aPKC, would recruit an active kinase to the lateral membrane, which would inhibit Par1 and prevent Par1 from releasing Par3 into the cytoplasm. Further studies will be required to definitively test this speculative model.
We found that Par3 depletion from stem/progenitor cells caused the accumulation of both K8+K14+ dual-positive cells as well as K6+ progenitors. Bipotential progenitors that express both luminal keratins (K8, K18, or K19) and K14 have been identified in the prostate (Wang et al. 2006) and mammary gland (Woodward et al. 2005; Shackleton et al. 2006; Villadsen et al. 2007; Wang et al. 2008). In cultured organoids, K8+K14+ cells are detected in the distal end bud (Fata et al. 2007), a potential stem cell niche (Williams and Daniel 1983; Kenney et al. 2001), and are also rarely found in ducts from human mammary tissue (Villadsen et al. 2007). In the mouse cortex, overexpression of Par3 or Par6 promotes self-replicating progenitor divisions (Costa et al. 2008). In the mammary gland, we show that depletion or inhibition of aPKC activity promotes accumulation of progenitors. Therefore, the Par complex may have a general role in regulating progenitor cell fate determination. This result supports the idea in the model described above, that silencing of Par3—or expression of the Par3c variant—prevents the normal phosphorylation of aPKC targets (Fig. 7I).
One function of Par3 in Drosophila stem cells is to orient the plane of mitosis to ensure correct segregation of cell fate determinants (Schober et al. 1999). However, it is not known whether Par3 has a similar function in mammals. Oriented divisions are important for mammary morphogenesis, at least during ductal extension, where cells divide parallel to the plane of the duct (Taddei et al. 2008), but it is unlikely that cells within the multilayered terminal end bud possess specific orientations during mitosis, and it is not yet known whether mammary progenitor or stem cells undergo asymmetric cell divisions. We were unable to measure spindle orientation in Par3-depleted glands because the mitotic cells were all within multilayered regions and did not contact the lumen. By analogy to Drosophila, loss of Par3 might disrupt asymmetric cell division of the mammary stem/progenitor cells, which would result in an increased population of such progenitors. Indeed, we found that Par3-depleted stem/progenitor cells were able to initiate mammary gland repopulation more frequently than control glands, indicating that Par3 might regulate mammary stem cell self-renewal. However, directly testing this hypothesis will require the ability to track the fate of individual stem cells during mammary gland morphogenesis.
Materials and methods
Plasmids and shRNAs
GFP in pLVTHM (Wiznerowicz and Trono 2003) was replaced with seYFP (Venus) with a C-terminal multiple cloning site (BamHI, NotI, and EcoRI) inserted for generating YFP fusion proteins (pL-Venus). Additional BamHI, NotI, and EcoRI sites were removed using the QuikChange XL kit (Stratagene) according to the manufacturer's instructions, leaving unique restriction sites within the multiple cloning site. shRNA complementary DNA strands (19mer sequences: shPar3, 5′-ACAAGCGTGGCATGATCCA-3′; shPar3-2, 5′-GTAGGCAAGAGGCTCAACA-3′) with flanking sequences were annealed and ligated into the MluI/ClaI sites of pLVTHM or pLV-Venus. All shRNAs were verified by DNA sequencing. Lentiviral vectors for bicistronic expression of human Par3b and human Par3c with shPar3 were made in pLV-Venus. Control lentiviruses were virus alone, or contained shRNA to Luciferase.
Lentivirus transduction of mammary epithelial/stem cells
Lentivirus was produced by calcium phosphate transfecting 293LT cells in 10-cm dishes with 20 μg of lentiviral plasmid, 10 μg of packaging plasmid (psPAX2), and 5 μg of VSVG coat protein plasmid (pMD2.G). Viral supernatants were collected after 48 h, filtered (0.45 mm), and concentrated 10×–20× with 100-k ultrafilters (Millipore). Concentrated virus was aliquoted and frozen at −80°C, then titered using 293LT cells. Typical titers were 106–108 transducing units (TU) per milliliter for shRNA lentiviruses, and 105–106 TU/mL for cDNA lentiviruses.
The third through fifth mammary gland pairs were removed from 8-wk-old C3H female mice, minced with scissors, and digested in Digestion media (DMEM/F12, 2 mg/mL Collagenase I [Roche], 5 μg/mL insulin [Sigma], 600 U/mL Nystatin [Sigma], 100 U/mL penicillin/streptomycin) for 3 h at 37°C with periodic mixing. Epithelial organoids were collected by centrifugation at 1000 rpm for 5 min. The cell pellet was resuspended in 5 mL of DMEM/F12 and centrifuged at 1000 rpm for 5 min. The cell pellet was resuspended in DMEM/F12 containing DNaseI and centrifuged at 1000 rpm for 10 min. The pellet was resuspended in 10 mL of 5% calf serum in PBS and cells were collected by centrifugation five times at 1500 rpm for 15 sec. The cells were resuspended in 1 mL of trypsin/EDTA (Invitrogen) for 12 min, dissociated into a single-cell suspension, and passed through a 45-μm filter (Fisher) to obtain a single-cell suspension of MECs. Freshly isolated MECs or NIH 3T3 cells were transduced by centrifugation at 300g for 3 h, without polybrene, in a final volume of 200 μL, and containing 5 ng/mL insulin. After transduction, cells were mechanically dissociated and transferred to six-well low-attachment dishes (Corning) and grown as mammosphere cultures for 2–5 d (Dontu et al. 2003). NIH 3T3 cells were plated directly in six-well tissue culture dishes and were grown in DMEM with 10% calf serum. Primary mammary cells for immunoblots were grown as mammospheres for 2 d, then transferred to six-well tissue culture plates and grown as monolayers in DMEM/F12 containing 5% FBS, 5 ng/mL EGF, 100 U/mL Pen/strep, 2 μg/mL hydrocortisone, and 1× insulin/transferring/selenium.
In vitro differention assays
COMMA-1D cells, generously provided by D. Medina (Baylor College of Medicine, Houston, TX), were maintained in DMEM/F12 containing 10 μg/mL insulin, 5 ng/mL EGF, and 2% fetal calf serum and penicillin/streptomycin. Cells were plated at a density of 2200 cells per square centimeter in four-well chamber slides. After 1 wk, cells were fixed in 4% paraformaldehyde and stained for K6, K8, and K14. The myristoylated aPKCζ pseudosubstrate was used at a concentration of 40 μM and was added to cultures every 48 h.
Antibodies and immunoblots
Rabbit anti-Par3 sera raised against a Par3 C-terminal peptide were affinity-purified and frozen in aliquots at −80C.
Cells were lysed in RIPA buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 2 μM aprotinin, 1 mM PMSF, 1 μM leupeptin) for 30 min on ice, and 30 μg of whole-cell lysate were separated by SDS-PAGE using 10% gels. Blots were probed using rabbit anti-Par3 antibody (0.6 μg/mL) and mouse anti-α tubulin (1:5000; Sigma). Protein was detected using HRP-conjugated secondary antibodies (Jackson ImmunoResearch) and PicoWest Chemiluminescent reagent (Pierce). Primary antibodies used for immunostaining were rat anti-K8 (Univeristy of Iowa Hybridoma Bank, TROMA-1); rabbit anti-GFP (1:1000; Abcam, ab6556); rabbit anti-K14 (1:500; Covance, PRB-155P); mouse anti-SMA (1:50; Abcam, ab18147); rabbit anti-Ki67 (1:75; Abcam, ab15580); rabbit anti-cleaved Caspase-3 (1:100; Covance, 9664); and rabbit anti-K6 (1:100; Covance, PRB-169P).
Mammary transplants
For transplants, mammospheres from 1000 or 10,000 transduced mammary epithelial cells in 7.5 μL of Injection Medium (DMEM/F12, 10% Matrigel [BD Biosciences] 5ng/mL EGF [Invitrogen], 20 ng/mL FGF2 [R&D Systems], 0.01% trypan blue) were injected into the cleared #4 fat pads of 4-wk-old C3H female mice (Charles River Laboratories) using a Hamilton syringe with a 26-guage needle.
Mammary gland whole mounts and immunostaining
After 7–10 wk of mammary ductal outgrowth, mammary fat pads were removed and fixed in Carnoy's fixative (60% ethanol, 30% chloroform, 10% acetic acid) and stained with Carmine alum. For immunofluorescence, 5-μm paraffin sections were deparaffinized and boiled in TE buffer (20 mM Tris, 1mM EDTA, 0.05% Tween-20 at pH 9.0) for 20 min in a microwave oven. Sections were blocked in 10% normal goat serum for 1 h followed by overnight incubation of primary antibody at 4°C. Sections were incubated for 1 h with the appropriate secondary antibody conjugated to Alexa488 (1:1000 dilution), Alexa546 (1:1000 dilution), or Alexa633 (1:200 dilution). Confocal images were taken on a Nikon (Eclipse TE2000-E) with a Yokogawa spinning disk confocal system (Solamere Technologies) using 40× (NA = 0.95) or 60× (NA = 1.40) objective lenses (Solamere Technology) or with a Zeiss 510 META with a 60× (NA = 1.40) objective lens.
Contrast enhancement was applied equally for control and shPar3 images using ImageJ.
Statistical analysis
All measurements were made using ImageJ. To calculate knockdown efficiencies, Par3 protein levels were normalized to α-tubulin as an internal control and were reported as means. Fat pad filling was measured by drawing a shape around the fat pad using the Freehand tool. Ductal areas were measured by drawing a shape that connected tips of ducts. The percentage fat pad filled was measured by the equation (ductal area/fat pad area) × 100. Measurements are reported as mean (±SD). For measurements of terminal ends and ductal diameters, the diameter of all terminal ends and ducts were measured at the thickest point using the Straight line tool.
To measure K8+/K14+ cells, a uniform threshold for each channel (K8, green; K14, red) was applied to control and shPar3 sections, and the number of green, red, and yellow cells was counted. All epithelial regions were counted from the indicated number of glands.
Measurements are reported as mean (±SD). All statistical analyses were performed using Excel with unpaired, two-tailed t-tests.
Acknowledgments
This work was supported by a research grant from the National Institutes of Health (GM090702) to I.G.M. L.M.M. is the recipient of a Post-doctoral fellowship from the Canadian Institutes of Health Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1795909.
Supplemental material is available at http://www.genesdev.org.
Note added in proof
While this manuscript was in press, a study by Horikoshi et al. (2009) shows that the Par3/aPKC interaction is necessary for apical domain formation in MDCK monolayers and 3D cycsts.
References
- Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14–3–3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Danielson KG, Oborn CJ, Durban EM, Butel JS, Medina D. Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci. 1984;81:3756–3760. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata JE, Mori H, Ewald AJ, Zhang H, Yao E, Werb Z, Bissell MJ. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFα and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley L, Quinn CM. Columnar cell lesions of the breast. Histopathology. 2008;52:11–19. doi: 10.1111/j.1365-2559.2007.02890.x. [DOI] [PubMed] [Google Scholar]
- Gao L, Macara IG, Joberty G. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene. 2002;294:99–107. doi: 10.1016/s0378-1119(02)00681-9. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: Fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L, Silberstein GB. Key stages in mammary gland development: The mammary end bud as a motile organ. Breast Cancer Res. 2005;7:245–251. doi: 10.1186/bcr1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, Yonemura S, Ohno S. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci. 2009;122:1595–1606. doi: 10.1242/jcs.043174. [DOI] [PubMed] [Google Scholar]
- Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T, Suzuki Y, Yamanaka T, Suzuki A, Mizuno K, et al. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J Cell Sci. 2002;115:2485–2495. doi: 10.1242/jcs.115.12.2485. [DOI] [PubMed] [Google Scholar]
- Hirose T, Karasawa M, Sugitani Y, Fujisawa M, Akimoto K, Ohno S, Noda T. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 2006;133:1389–1398. doi: 10.1242/dev.02294. [DOI] [PubMed] [Google Scholar]
- Humphreys RC, Krajewska M, Krnacik S, Jaeger R, Weiher H, Krajewski S, Reed JC, Rosen JM. Apoptosis in the terminal endbud of the murine mammary gland: A mechanism of ductal morphogenesis. Development. 1996;122:4013–4022. doi: 10.1242/dev.122.12.4013. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Liu CJ, Kweon HK, Hakansson K, Margolis B. Phosphorylation-dependent binding of 14–3–3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003a;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003b;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Kenney NJ, Smith GH, Lawrence E, Barrett JC, Salomon DS. Identification of stem cell units in the terminal end bud and duct of the mouse mammary gland. J Biomed Biotechnol. 2001;1:133–143. doi: 10.1155/S1110724301000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MJ, Zhang CC, Zhou B, Zimonjic DB, Mani SA, Kaba M, Gifford A, Reinhardt F, Popescu NC, Guo W, et al. Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity. Cancer Res. 2007;67:8131–8138. doi: 10.1158/0008-5472.CAN-06-4493. [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev Cell. 2007;12:221–234. doi: 10.1016/j.devcel.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens AE, Rygiel TP, Olivo C, van der Kammen R, Collard JG. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol. 2005;170:1029–1037. doi: 10.1083/jcb.200502129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Ellenbroek SI, Mertens AE, van der Kammen RA, de Rooij J, Collard JG. The Par–Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front–rear polarity. Curr Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Proia DA, Kuperwasser C. Reconstruction of human mammary tissues in a mouse model. Nat Protocols. 2006;1:206–214. doi: 10.1038/nprot.2006.31. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafe M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Siwko SK, Bu W, Gutierrez C, Lewis B, Jechlinger M, Schaffhausen B, Li Y. Lentivirus-mediated oncogene introduction into mammary cells in vivo induces tumors. Neoplasia. 2008;10:653–662. doi: 10.1593/neo.08266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Taddei I, Deugnier MA, Faraldo MM, Petit V, Bouvard D, Medina D, Fassler R, Thiery JP, Glukhova MA. β1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat Cell Biol. 2008;10:716–722. doi: 10.1038/ncb1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traweger A, Wiggin G, Taylor L, Tate SA, Metalnikov P, Pawson T. Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par-3. Proc Natl Acad Sci. 2008;105:10402–10407. doi: 10.1073/pnas.0804102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F, Aguet M, de Sauvage FJ, Gao WQ. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol. 2008;28:3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJP, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Daniel CW. Mammary ductal elongation: Differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–3594. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Suzuki A, Sugiyama Y, Kitamura K, Maniwa R, Nagai Y, Yamashita A, Hirose T, Ishikawa H, et al. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells. 2001;6:721–731. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
- Zegers MM, O'Brien LE, Yu W, Datta A, Mostov KE. Epithelial polarity and tubulogenesis in vitro. Trends Cell Biol. 2003;13:169–176. doi: 10.1016/s0962-8924(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]