Abstract
Cell growth is an essential requirement for cell cycle progression. While it is often held that growth is independent of cell cycle position, this relationship has not been closely scrutinized. Here we show that in budding yeast, the ability of cells to grow changes during the cell cycle. We find that cell growth is faster in cells arrested in anaphase and G1 than in other cell cycle stages. We demonstrate that the establishment of a polarized actin cytoskeleton—either as a consequence of normal cell division or through activation of the mating pheromone response—potently attenuates protein synthesis and growth. We furthermore show by population and single-cell analysis that growth varies during an unperturbed cell cycle, slowing at the time of polarized growth. Our study uncovers a fundamental relationship whereby cell cycle position regulates growth.
Keywords: Actin polarization, cdc28, cell cycle, cell growth, cell size, Saccharomyces
Growing cells and organisms must increase in biomass as well as in cell number. The coordinated production of new cells (the cell cycle) and growth (the increase in biomass of individual cells) is essential for both development of multicellular organisms and fitness of microorganisms (Stocker and Hafen 2000; Hall et al. 2004; Jorgensen and Tyers 2004). In order to maintain long-term size homeostasis, cells must on average double in size before they divide. In most organisms, this is accomplished by establishing a dependency on growth for cell cycle entry. Cells typically initiate a new cell cycle in late G1 phase only once a critical cell size, a function of protein synthesis rate, is achieved (Hartwell et al. 1974; Johnston et al. 1977; Jorgensen and Tyers 2004). At least in simpler eukaryotes, nutrient conditions determine both growth rate and the critical size threshold, such that slower growing cells are smaller than faster growing cells (Johnston et al. 1979). Growth and cell cycle entry can be genetically separated by altering the expression of limiting cell cycle regulators that control the activity of cyclin-dependent kinase (CDK), which drives cell cycle progression. For example, increasing G1 cyclin dosage while maintaining a constant growth rate causes a reduction in cell size known as a “Whi” phenotype (Cross 1988; Nash et al. 1988; Jorgensen et al. 2002).
In budding yeast, the mechanisms whereby growth controls cell cycle progression are partly understood. The protein kinase A (PKA) and target of rapamycin (TOR) pathways sense nutrient status directly or indirectly and, when ample nutrients are available, promote macromolecular synthesis (Thevelein and de Winde 1999; Hall et al. 2004; Inoki et al. 2005; Santangelo 2006). The rate of ribosome production and, in turn, the rate of protein synthesis affect the rate of accumulation of the G1 cyclins, which are among the most unstable proteins in the cell (Schneider et al. 1998; Jorgensen et al. 2002; Jorgensen and Tyers 2004). A minimum rate of G1 cyclin synthesis is required to initiate early events that promote entry into the cell cycle (Schneider et al. 2004). The accumulation of the G1 cyclin Cln3 is in particular controlled at the level of CLN3 mRNA translation (Barbet et al. 1996; Polymenis and Schmidt 1997; Hall et al. 1998). Once sufficient Cln3 levels have been achieved, Cln3-Cdc28 induces a positive feedback loop in which two downstream G1 cyclins, Cln1 and Cln2, stimulate their own synthesis to ensure coherent entry into the cell cycle (Cross and Tinkelenberg 1991; Dirick and Nasmyth 1991; Murray and Hunt 1993; Tyers et al. 1993; Nasmyth 1996; Di Talia et al. 2007; Skotheim et al. 2008).
In order to grow in size, cells must fuse new lipid vesicles with the cell membrane. The vesicles are transported to sites of fusion on actin cables (Pruyne et al. 2004; Moseley and Goode 2006; Park and Bi 2007). In budding yeast, the actin cytoskeleton undergoes periodic polarization during the cell cycle, and as a consequence the areas of vesicle deposition change as a function of cycle position (Kilmartin and Adams 1984; Amberg 1998). In G1 phase, actin cables are evenly distributed throughout the cytoplasm, resulting in uniform deposition, isotropic growth, and spherical cell morphology. As cells transition into S phase, the actin cytoskeleton becomes polarized, and vesicle deposition occurs apically at the site of bud emergence. After initial bud emergence, growth remains limited to the developing daughter cell, but becomes isotropic to create a spherical bud. After anaphase and nuclear division, the actin cytoskeleton reorients toward the site of division to enable cytokinesis (Pruyne et al. 2004; Moseley and Goode 2006; Park and Bi 2007).
Polarization of the actin cytoskeleton requires the concerted action of many proteins. Cortical cues established in the prior cell cycle cause the Rho-like GTPase Cdc42 to accumulate at the site of bud emergence where it activates the formins Bni1 and Bnr1 to nucleate actin cable formation (Evangelista et al. 1997; Sagot et al. 2002; Park and Bi 2007). Cell cycle position dictates the pattern of actin polarization and growth. The Cln-CDKs phosphorylate and activate components of the actin polarizing machinery in late G1 phase (Lew and Reed 1993; Moffat and Andrews 2004; McCusker et al. 2007). The B-type cyclin (Clb)-CDKs, which are activated subsequently to Cln-CDKs and promote S phase and mitosis, mediate the switch from polarized growth to isotropic growth, likely through down-regulation of the Cln-CDK activity (Lew and Reed 1993; Amon et al. 1994). Whether these dramatic changes in cell architecture affect growth has not been studied.
Previous studies in budding yeast suggest that growth rate is constant throughout the cell cycle (Elliott and McLaughlin 1978, 1979; Di Talia et al. 2007); however, growth rates do appear to change during the cell cycle in fission yeast (Mitchison and Nurse 1985). When either budding or fission yeast cells are arrested at different cell cycle stages using temperature-sensitive cell division cycle (cdc) mutants, cells continue to synthesize proteins and increase in volume (Johnston et al. 1977; Adams et al. 1990; Su and O'Farrell 1998). Similarly, cell cycle arrest in tissue culture cells also results in abnormally large cells due to ongoing growth (Hemerly et al. 1995; Weigmann et al. 1997; Neufeld et al. 1998; Su and O'Farrell 1998). These observations led to the notion that cell growth is unabated in the absence of cell division.
Here we analyze cell growth during various cell cycle arrests in budding yeast and find that cell growth varies markedly with cell cycle position. The capacity to grow is higher in anaphase- and G1-arrested cells than cells arrested in S phase and early mitosis. Unexpectedly, pheromone treatment of G1-arrested cells overrides this capacity for extended growth. This growth limitation by pheromone and at the G1/S transition is mediated by actin polarization; many genes involved in protein synthesis are down-regulated in response to pheromone treatment. Polarization of the actin cytoskeleton in general is sufficient to limit protein synthesis and growth in all cell cycle stages examined. Finally, we present evidence to suggest that growth varies during an unperturbed cell cycle slowing at the time of polarized growth. Our results demonstrate that cell cycle position can directly affect cell growth in part through changes is actin polarization.
Results
cdc mutants arrest with different terminal cell sizes
Hartwell and colleagues (Johnston et al. 1977) reported that temperature-sensitive cdc mutants continue to increase in size when incubated at the restrictive temperature. However, as these mutants do cease to grow eventually, cell cycle progression must also affect cell growth in some way. To investigate this effect, we examined the growth properties of cells terminally arrested in different cell cycle stages using the α-factor mating pheromone, overexpression of a stable version of the B-type cyclin Clb2 (Clb2dbΔ), and isogenic cdc mutants. In all cases, cdc mutants were grown to exponential phase at the permissive temperature of 20°C and shifted to the restrictive temperature of 37°C (with the exception of GAL1-CLB2dbΔ strains, which were grown at 30°C due to the diminished activity of the GAL1 promoter at 37°C). As a measure of growth, modal cell volume was determined and plotted for various times after temperature shift (see the Materials and Methods). Cell number was monitored to ensure that cells remained arrested during the course of the experiment.
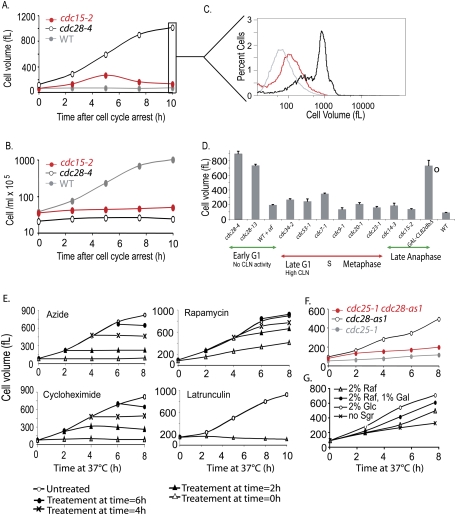
Consistent with previous observations (Johnston et al. 1977), all tested cdc mutants continued to grow after cell cycle arrest. However, 2–4 h after temperature shift, most cdc mutants (cdc15-2 mutants are shown as an example) dramatically slowed their growth (Fig. 1A,B; see also Figs. 2B, 6C [below]). Wild-type cells exhibited a modal cell volume of 70 fL, while most cdc mutants arrested with a cell volume between 100 and 300 fL (Fig. 1D). Two cdc mutant arrests were, however, notable exceptions: cdc28 and GAL1-CLB2dbΔ. Both temperature-sensitive cdc28 alleles that we analyzed continued to grow for at least 8 h and reached a terminal cell volume of 700 fL or more (Fig. 1A–D; Supplemental Fig. 1A). Likewise, a strain that overexpressed a nondegradable form of Clb2 arrested with a similar large terminal phenotype (Fig. 1D). The large size of the Clb2dbΔ strain was not due to growth at 30°C because the strain was still larger compared with metaphase-arrested Cdc20-depleted cells arrested at 30°C (Supplemental Fig. 1B).
Figure 1.
The ability of cells to grow is increased in G1 and anaphase-arrested cells. (A–C) Comparison of the sizes of wild-type (A2587), cdc28-4 (A17132), and cdc15-2 (A2596) cells. Cells were shifted to 37°C, and cell size (A) and cell number (B) were determined at the indicated times. C shows the cell size distributions of the strains in A after 10 h of temperature shift. (D) Exponentially growing yeast cells were arrested, and growth was monitored over a period of 10 h. cdc mutants carrying the indicated alleles were arrested at 37°C in YEPD. Pheromone arrest was maintained by repeated addition of α-factor to 20 μg/mL every 2 h. Cells carrying the GAL-CLB2dbΔ fusion (marked by ○ to indicate different growth temperature) were grown in YEP supplemented with 2% raffinose at 30°C. At the time of galactose addition (1%), cells were also shifted to 30°C. Wild-type cells were likewise shifted to 37°C, although the mode cell volume is similar at 30°C. The average peak (mode) cell volume for the 10 h time point of at least three independent replicates is shown (error bars represent standard deviation). The following strains were used for this analysis: wild type (A2587), cdc28-4 (A17132 or A2594), cdc28-13 (A4370), wild type with α-factor (A2589, bar1∷HisG), cdc34-2 (A1467), cdc53-1 (A1469), cdc7-1 (A344), cdc9-1 (A2599), cdc20-1 (A937), cdc23-1 (A785), cdc14-3 (A5321), cdc15-2 (A2596), and GAL-CLB2dbΔ (A17873). (E) The large size of cdc28-4 mutants is due to cell growth. cdc28-4 cells (A17132) were shifted to 37°C (0 h time point). At the indicated times, 5-mL aliquots were removed and treated with 0.1% azide, 200 μM latrunculin A, 10 μM rapamycin, or 0.33 mg/mL cycloheximide. Mode cell volume was measured for all subsequent time points in each drug-treated culture. (F) The RAS pathway is required for growth. Exponentially growing cdc25-1 (A20752), cdc28-as1 (A4370), and cdc25-1 cdc28-as1 double mutants (A21093) were shifted to 37°C and treated with 5 μM 1-NM-PP1 CDK inhibitor at time 0 h. Cell volume was determined at indicated times. (G) The volume increase of cdc28-4 mutants responds to carbon source identity. cdc28-4 cells (A17132) were grown to exponential phase in YEP supplemented with 2% raffinose. Upon shift to 37°C, cells were transferred into YEP medium either lacking any sugars, containing 2% raffinose, 2% raffinose with 1% galactose, or 2% glucose, and mode cell size distribution was determined.
Figure 2.
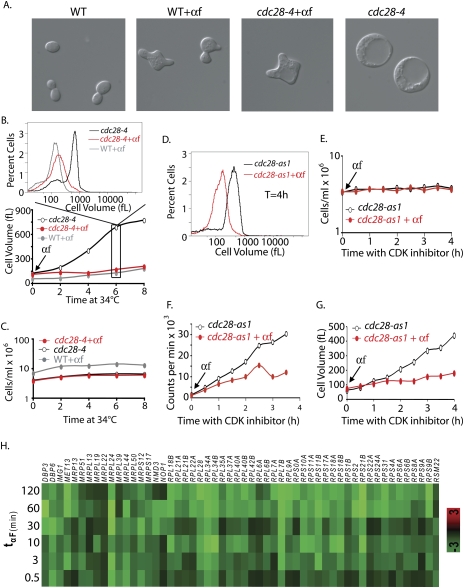
Polarization limits growth in cdc28-4 mutants. The micrographs in A show wild-type (A2589) and cdc28-4 (A22928) cells at 6 h after the temperature shift with or without α-factor treatment. (B,C) Wild-type (A2589, bar1∷HisG; gray circles) and cdc28-4 bar1∷HisG (A22928) cells were shifted to 34°C and either treated with 20 μg/mL α-factor (black circles) or left untreated (red circles). Modal cell volume (B) and cell number (C) were determined at the indicated times. The cell volume distributions for the 6-h time point are shown above the graph in B. (D–G) cdc28-as1 (A4370) cells were shifted to 30°C and arrested with 5 μM CDK inhibitor, and 35S methionine and 35S cysteine (20 μCi/mL final each) were added. (F) The culture was split and either treated with mating pheromone (20 μg/mL; red circles) or mock-treated (black circles) and 35S incorporation into total protein was determined as described in Materials and Methods. A parallel culture treated identically but with unlabeled amino acids was used to determine cell number (E) and cell volume (G). The cell volume distribution of the two cultures at 4 h after temperature shift is shown in D. (H) Gene expression profile of 54 down-regulated RPs during mating in the wild-type strain (MT1567). Genome-wide gene expression was examined at different time points after pheromone addition. Down-regulated genes were significantly enriched for only one MIPS functional category: RPs (54 genes; P < 1.198e-06). Log2 (treated/untreated) values are indicated for each gene (y-axis) at various times (min) after pheromone addition (x-axis).
Figure 6.
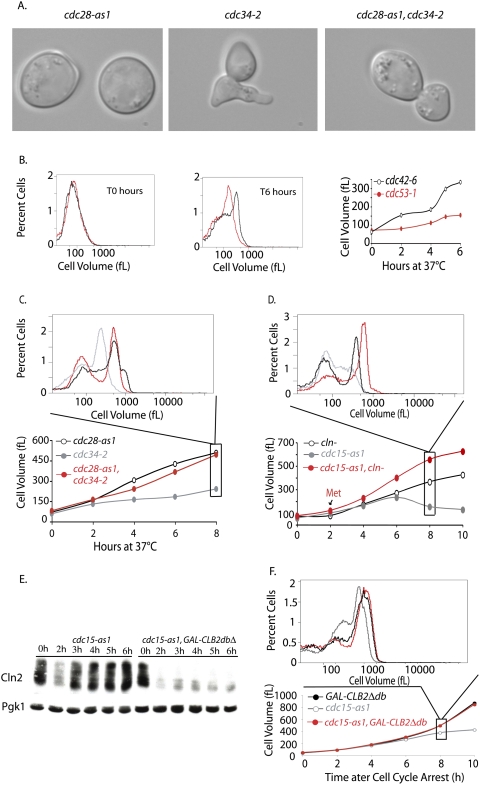
The limited growth of SCF and MEN mutants is due to Cln-CDK activity. (A,C) cdc34-2 (A1467), cdc28-as1 (A4370), and cdc34-2 cdc28-as1 (A17188) cells were shifted to 37°C (0 h time point). Single mutants were treated with 5 μM 1-NM-PP1 inhibitor at the time of temperature shift. The double mutant was grown for 2 h at 37°C, by which time ∼80% of the double mutants had budded, which was followed by addition of CDK inhibitor. (A) Micrographs of mutants after 4 h of growth at 37°C. (B) Comparison of the growth of cdc42-6 (A18221) and cdc53-1 (A1469) mutants. Exponentially growing cells were shifted from room temperature to 37°C, and growth was monitored using a Coulter counter. Cell volume distribution for both strains is shown for the T0 h and T6 h time points, and the mode cell volume for the complete time course is plotted. (C) Inactivation of Cdc28 rescues the growth inhibition of cdc34-2 mutants. Cell size distributions were measured at the indicated time points after shift to 37°C. The histogram above displays the cell volume distribution 8 h after temperature shift. Cells were grown as described in A. (D) Depletion of Cln-CDKs allows cdc15-2 mutants to grow in size. MET-CLN2 cln3Δ cln1Δ (A2624; red circles), cdc15-as1 (A17872; gray circles), and MET-CLN2 cln3Δ cln1Δ cdc15-as1 (A18316; black circles) cells were grown to exponential phase in synthetic complete medium lacking methionine. Cells were transferred into YEPD at 37°C in the presence of 10 μM 1-N-PP1 inhibitor to inhibit the cdc15-as1 allele (0 h time point). The single mutants were treated with 8 mM methionine at the time of temperature shift and cdc15-as1 inhibition, to repress Cln2 production. The double mutant was grown in the absence of methionine for 2 h to allow cells to accumulate in the cdc15-as1 block prior to methionine addition. Subsequently, 8 mM methionine was readded every 2 h to all cultures. Cell size distributions were measured at the indicated time points after shift to 37°C. The histogram above displays the cell volume distribution 8 h after temperature shift. (E) CLNs accumulate in cdc15-as1-arrested cells, but not in cdc15-as1-arrested cells overexpressing CLB2dbΔ. cdc15-as1 (A18982) and cdc15-as1 GAL-CLB2dbΔ (A18986) strains, both expressing a tagged version of CLN2 (Cln2-3xHA), were pregrown at 30°C in YPE supplemented with 2% raffinose. At time 0 h, 10 μM 1-N-PP1 inhibitor and 1% galactose were added to both strains to inhibit cdc15-as1 and to induce CLB2dbΔ, respectively. Samples were withdrawn at the indicated times. Samples were run by 10% SDS-PAGE and prior to transfer to a membrane and blotting. The membrane was blotted with anti-HA antibody to detect Cln2-3X-HA. The same membrane was stripped and reprobed with anti-Pgk1 antibody. (F) Expression of a stable form of Clb2 rescues the growth phenotype of MEN mutants. cdc15-as1 (A17873), GAL-CLB2 dbΔ (A17872), and cdc15-as1 GAL-CLB2 dbΔ double mutant (A17874) were arrested in YPE supplemented with 2% rafinose media with α-factor for 90 min after which 1% galactose was added to induce the expression of Clb2Δdb. Cells were further incubated with α-factor for 45 min and were released into fresh YPE media supplemented with 2% raffinose and 1% galactose at 30°C. At this time, Cdc15-as1 inhibitor, 1-N-PP1, was added to all strains at 10 μM concentration. The volume of all strains was monitored to 10 h. The mode cell volume for each time point is plotted: cdc15-as1 (gray circles), GAL-CLB2dbΔ (black circles), and double mutant (red circles).
To ascertain whether the cdc28 effect was a function of the cell cycle stage (G1) in which they arrest, we examined cells arrested in G1 with mating pheromone (Figs. 1D, 2B). Whereas cdc28 mutants reached a volume of 800 fL or more, the pheromone-treated cells grew to a much smaller volume of ∼195 fL. This difference was independent of the initial cell volume prior to onset of arrest (Supplemental Fig. 1A). We observed a similar situation in late anaphase. Cells overexpressing Clb2dbΔ arrested in late anaphase, as either budded or unbudded cells (Amon et al. 1994), with a terminal cell size of ∼700 fL (Fig. 1D). In contrast, cells defective in the mitotic exit network (MEN) such as cdc14-3 and cdc15-2 mutants (henceforth MEN mutants), which also arrest in late anaphase, achieve a maximal size of <200 fL (Fig. 1A,D). Note that the apparent decrease in the size of MEN mutants (Fig. 1A) is due to cell lysis and cells undergoing septation, which is more pronounced at 37°C than at 30°C (Fig. 6D,F, below). Our results indicate that either a growth promoting factor(s) is present at the cdc28 and GAL-CLB2dbΔ arrests or that a growth inhibitor(s) is present in other cdc arrests and pheromone-treated cells.
The cdc28-4 arrest is associated with continued cell growth
We first explored the molecular basis for the different growth capabilities of cdc28 mutants and α-factor-treated cells, but in what follows below, it will become evident that one of the reasons for the growth difference between the two G1 arrests is also responsible for the growth difference observed among the mutants that arrest in late anaphase.
To ensure that the increase in volume of cdc28 mutants observed during the prolonged incubation times was due to cell growth rather than osmotic swelling, we examined whether the volume increase was associated with protein synthesis and whether it was energy-dependent. To test this, cells were shifted to the restrictive temperature, and at various times, either azide (an ATP synthesis inhibitor) or cycloheximide (a protein synthesis inhibitor) was added. The cell volume increase ceased shortly after addition of either inhibitor regardless of the time of addition (Fig. 1E). Addition of rapamycin, an inhibitor of the TOR pathway and protein synthesis, or latrunculin A, which depolymerizes the actin cytoskeleton disrupting vesicle trafficking, also severely restricted the growth of cdc28-4 mutants (Fig. 1E). We also observed that inactivating Cdc25, the upstream GTP exchange factor for the Ras/PKA pathway (Murray and Hunt 1993; Santangelo 2006; Zaman et al. 2008), concomitantly with inactivating Cdc28 drastically limited increases in cell size (Fig. 1F). The growth observed in cdc28-4 mutants was also responsive to carbon source identity as the volume increase was faster in the presence of a sugar than in its absence and was faster in the presence of glucose than in the presence of the poorer carbon source raffinose (Fig. 1G). Consistent with continued macromolecular synthesis, the cdc28-4 mutant strain incorporated radiolabeled amino acids into cellular proteins for at least 4 h after temperature shift (Fig. 5B, below). We conclude that the continuous cell volume increase of cdc28-4 mutants is due to intact and unbridled macromolecular biosynthesis.
Figure 5.
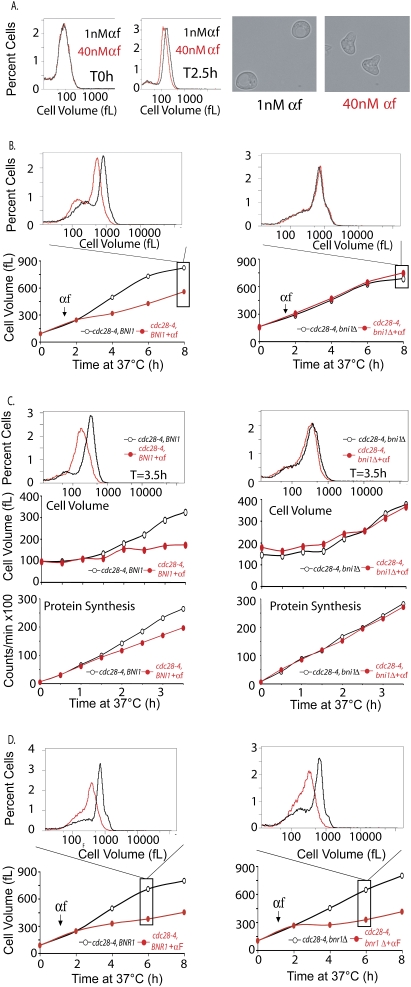
BNI1 but not BNR1 is required for the growth inhibitory effect of pheromone. (A) Mutants that are hypersensitive to mating pheromone, bar1 (A4370), were grown in YPE media supplemented with 2% glucose at 30°C. The culture was split and treated with a final concentration of either 1 nM pheromone (black) or40 nM pheromone (red). After 2.5 h, aliquots were collected and analyzed for budding, the percent of cells with shmoos, and cell volume distributions (see text for details). Images of cells were taken 2.5 h after pheromone addition. (B,D) cdc28-4 (A17132) and cdc28-4 bni1Δ (A18238) cells (B) or cdc28-4 (A17132) and cdc28-4 bnr1Δ (A19911) cells (D) were grown as described in Figure 3A, and the cell volume distributions were determined at indicated times points. The histograms above each plot display the cell volume distribution 6 h (D) or 8 h (B) after temperature shift. (C) Cell volume (middle) and protein synthesis (bottom) rates were determined for cdc28-4 (A17132) and cdc28-4 bni1Δ (A18238) cells in the presence and absence of pheromone as described in Figure 2F. (Top) The cell volume distribution of each culture 3.5 h after temperature shift is shown in the histogram.
Mating pheromone inhibits growth in cdc28-4 mutants
We next determined why arrested cdc28-4 mutants grow much larger than α-factor-treated cells, despite the fact that both arrests occur in G1 phase. We performed a simple epistasis test by adding mating pheromone to cdc28-4 cells concurrently with the shift to 37°C. The untreated cdc28-4 mutant grew continuously to reach a modal cell volume of >800 fL after 8 h of growth at 37°C, while the mating-pheromone-treated cells reached a volume of only 195 fL under the identical conditions (Fig. 2A,B). Both untreated and pheromone-treated cells remained arrested throughout the experiment (Fig. 2C). Pheromone reduced the rate of volume increase of cdc28-4 mutants from 120 fL/h in untreated cells to 12 fL/h, which was close to the rate of 16 fL/h in wild-type cells treated with pheromone (Fig. 2B). Similar results were obtained with an analog-sensitive cdc28-as1 allele (Bishop et al. 2000) and the cdc28-13 allele (Fig. 2D–G; Supplemental Fig. 1). As expected, mating pheromone also down-regulated protein synthesis in cdc28-as1 and cdc28-4 cells (Figs. 2D–G, 5C [below]). To exclude the possibility that pheromone causes a G1 arrest that is entirely distinct from that of the cdc28 arrest, we prearrested a cdc28-4 mutant at the restrictive temperature for 90 min prior to pheromone addition. Pheromone also restricted the growth of the cdc28-4 mutant under these conditions (Fig. 3). Consistent with a growth inhibitory role of pheromone, we observed that 54 ribosomal proteins (RPs) were down-regulated by pheromone treatment (P < 1.198 × 10−06) (Fig. 2H). This down-regulation occurred within 10 min of pheromone addition. The velocity of this response is comparable with that observed in response to rapamycin treatment (Jorgensen and Tyers 2004), raising the possibility that the effects of pheromone on cell growth are direct. We conclude that even though cells continue to grow in the presence of pheromone, this growth is limited compared with that seen in cdc28 mutants (Fig. 1D). Importantly, the growth inhibition caused by pheromone is epistatic to the unrestricted growth in a cdc28 arrest and is associated with the rapid down-regulation of a core set of RP genes that are critical for cell growth.
Figure 3.
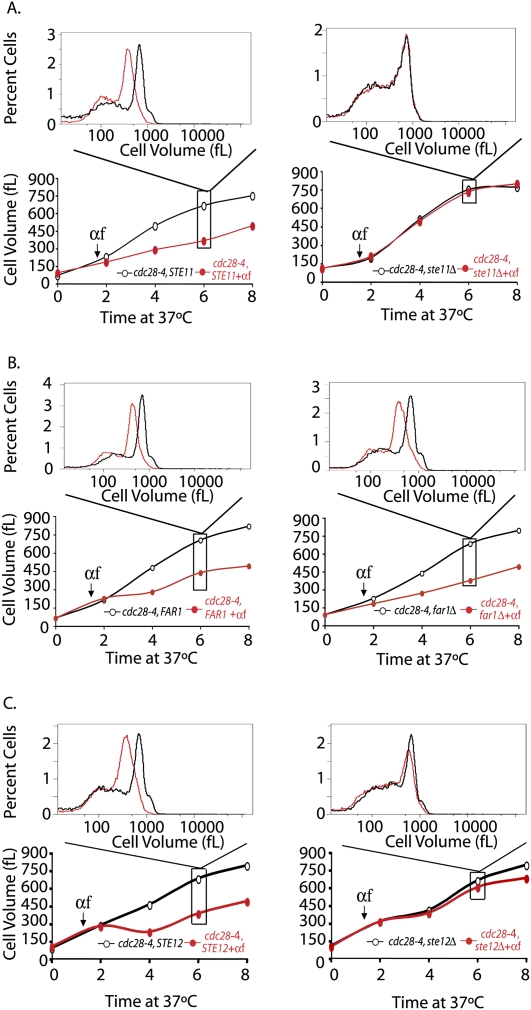
The mating pheromone pathway, but not the mating pheromone effector FAR1, is required for the growth inhibitory effects of mating pheromone. (A–C) Parallel cultures of cdc28-4 (A17132) and cdc28-4 ste11Δ (A18205) (A), cdc28-4 (A17132) and cdc28-4 far1Δ (A18204) (B), and cdc28-4 (A17132) and cdc28-4 ste12Δ (A18434) (C) were shifted to 37°C (0-h time point). After 90 min, cells were either treated with mating pheromone (20 μg/mL; red circles) or left untreated (black circles). Pheromone was readded every 2 h. Mode cell volume was determined at the indicated times. The histograms above each plot display the cell volume distribution 6 h after temperature shift.
The mating pheromone pathway limits growth
Pheromone signaling is initiated upon binding of pheromone to a G-protein-coupled seven transmembrane receptor, which in turn activates a mitogen-activated protein (MAP) kinase signaling cascade (Fig. 4C; Dohlman and Thorner 2001). Deletion of STE11, which encodes the MAP kinase kinase kinase in the pathway, abolished the growth inhibitory effects of mating pheromone on a cdc28-4 strain (Fig. 3A). Deletion of the gene encoding the MAP kinase Fus3, but not of the gene encoding the partially redundant MAP kinase Kss1, abrogated the growth inhibition by pheromone (Supplemental Fig. 2A,B). The suppression caused by deleting FUS3 was, however, incomplete, suggesting that KSS1 does contribute to promoting pheromone-mediated growth inhibition (Herskowitz 1995; Breitkreutz et al. 2001).
Figure 4.
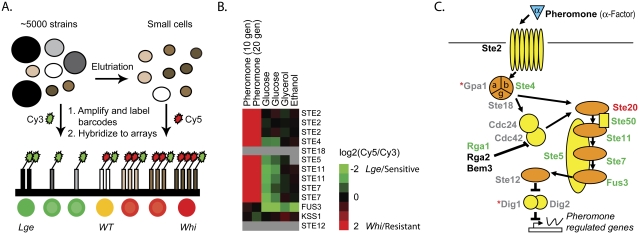
The pheromone pathway affects cell size during exponential growth. (A) Mixed yeast haploid deletion pools were elutriated to enrich for small cells. Unique bar codes from yeast pools collected immediately before and after elutriation were amplified and differentially labeled with Cy3 (before) and Cy5 (after) and hybridized to bar code microarrays. Green indicates depletion by elutriation (lge cells) while red indicates enrichment (whi cells). (B) Elutriation bar code microarrays experiments were performed with different rich (glucose) (Cook et al. 2008) or poor (glycerol, ethanol) carbon sources. Fitness experiments were performed in the presence of mating pheromone. A number of pheromone pathway gene deletions display a large cell size in the absence of pheromone, and in a manner dependent on the carbon source. The degree of enrichment (small = whi; pheromone-resistant; red) or depletion (large = lge; pheromone sensitive; green) for both upstream and downstream bar codes is expressed as log2(elutriated/total) or log2(treated/untreated) and is indicated by the color bar. Gray indicates no available data either due to insufficient signal or the absence of the strain from the deletion pool. (C) Schematic of the mating pathway. The cell size of gene deletions, relative to the deletion pool, is indicated by the text color: lge (green), whi (red), wild type (black), and no viable data (gray). gpa1Δ and dig1Δ (red asterisks) were both found to be lge; however, they displayed the incorrect pheromone sensitivity/resistance phenotype, likely due to secondary mutations, and were thus excluded.
Collectively, the two pheromone MAPKs phosphorylate and activate several effectors of the pheromone response: Far1, which mediates cell cycle arrest in response to pheromone treatment (Elion et al. 1993; Peter et al. 1993; Tyers and Futcher 1993); Ste12, the transcription factor required for the expression of pheromone-responsive genes (Song et al. 1991; Elion et al. 1993); and Bni1, a formin responsible for inducing polarized growth and formation of a mating projection (Matheos et al. 2004). Mating pheromone still substantially limited the cell volume increase in a cdc28-4 far1Δ double-mutant strain. As expected, given the requirement for basal level transcription for expression of several pheromone pathway genes, a cdc28-4 ste12Δ double mutant was resistant to the growth inhibitory effect of pheromone. Unexpectedly, however, deletion of BNI1 in a cdc28-4 strain also abolished the growth inhibitory effect of pheromone (Fig. 5B,C).
Importantly, the mating pheromone pathway appeared to not only limit growth during pheromone response, but its effect on cell growth was also apparent during logarithmic growth. We performed a sensitive genome-wide cell size screen in which a pooled culture of all ∼5000 viable yeast gene deletion strains was separated into different size fractions by centrifugal elutriation (Fig. 4A). The different size fractions were then probed for the relative enrichment or depletion of all strains by bar code microarray analysis of sequence tags specific to each deletion cassette (Giaever et al. 2002; Cook et al. 2008). This quantitative method is able to detect more subtle variations in cell size than previous screens performed using direct size analysis (Jorgensen et al. 2002; Cook et al. 2008). As a control, we treated the pool with a low concentration of mating pheromone to identify the ste gene deletion strains, which fail to arrest in G1 and hence acquire a smaller size than the rest of the pool (Fig. 4B). Strikingly, when grown in the absence of pheromone in rich medium, nearly all strains deleted for components of the mating pheromone pathway displayed a large cell size, suggesting that basal signaling through the pathway normally antagonizes growth (Fig. 4B,C; Supplemental Fig. 3; Supplemental Table 1). These effects were dependent on nutrient conditions as the large relative size of the ste mutants was eliminated by culture in the poor carbon sources glycerol or ethanol (Fig. 4B; Supplemental Fig. 3). A poor nitrogen source similarly reduced the size effect of the pheromone pathway mutations (data not shown). The growth inhibitory effects of pheromone in a cdc28-4 arrest were also more pronounced under conditions of fast growth than under conditions of slowed growth (data not shown). Taken together, these results support a previously unnoticed specific role for the pheromone pathway in the attenuation of growth in wild-type cells in both pheromone-treated cells and to a modest extent even during vegetative growth.
Pheromone-mediated actin polarization limits growth
How does the pheromone signaling pathway limit growth? Pheromone-treated cells differ from cdc28-arrested cells in that they form a mating projection (aka shmoo), the formation of which depends on actin polarization. However, only treatment of cells with high concentrations of pheromone leads to projection formation; treatment of cells with low concentrations of pheromone causes a transient G1 arrest in the absence of shmoo formation (Farley et al. 1999). We exploited the different responses to low and high doses of pheromone to interrogate the relationship between actin polarization and growth inhibition. All cells treated with 1 nM α-factor for 2.5 h arrested in G1 phase, but only 12% of these formed shmoos; in contrast, 40 nM pheromone caused 79% of cells to form shmoos. Furthermore, shmoo morphology was distinct in the two conditions. In the presence of high levels of pheromone, cells formed narrow and highly polarized mating projections, whereas in the presence of low pheromone concentrations, shmoos were less polarized (Fig. 5A, pictures on the right). Strikingly, cells treated with 1 nM pheromone grew larger than cells with 40 nM pheromone (Fig. 5A). The extent of projection formation and growth also correlated in other experiments. Treatment of a cdc28-4 far1Δ strain with pheromone caused 61% of cells to form highly polar shmoos with concomitant growth suppression (Fig. 3B; data not shown), whereas only 19% of cdc28-4 ste12Δ cells formed shmoos with no significant growth inhibition (Fig. 3C; data not shown). Deletion of STE11 and FUS3 produced effects similar to that of STE12 deletion (Fig. 2A; Supplemental Fig. 2A; data not shown). Conversely, deletion of BAR1, a gene that encodes a pheromone protease (Hicks and Herskowitz 1976), renders cells supersensitive to mating pheromone and to the formation of highly polarized mating projections (Sprague and Herskowitz 1981; Chan and Otte 1982). The growth restriction of pheromone was much more dramatic in bar1Δ cells than in wild-type cells (Supplemental Fig. 2C).
If pheromone-induced actin polarization, indeed, restricts growth, preventing actin polarization in cells with a functional mating-pheromone pathway should restore the ability of pheromone-treated cells to grow. The pheromone-pathway kinase Fus3 phosphorylates the formin Bni1, and that is thought to be essential for polarization of growth and shmoo formation (Matheos et al. 2004). Indeed, Bni1, but not its homolog Bnr1, appears to have a role in α-factor signaling (Qi and Elion 2005). However, unlike ste12Δ cells, bni1Δ cells are not sterile (Matheos et al. 2004; data not shown). As reported previously for wild-type cells (Jorgensen et al. 2002), deletion of BNI1 led to an increase in cell size of cdc28-4 cells at the permissive temperature (Fig. 5B). However, more interestingly, inactivation of BNI1 in a cdc28-4 strain completely abolished the growth inhibitory effects of mating pheromone (Fig. 5B,C). Deletion of SPA2, another gene involved in establishing cell polarity (Park and Bi 2007), also had a dramatic effect on the ability of pheromone to limit growth (Supplemental Fig. 4C). In contrast, deletion of BNR1, which is dispensable for actin polarization in response to pheromone (data not shown), had no effect (Fig. 5D). We conclude that one of the mechanisms whereby α-factor limits the growth of cdc28-4 mutants is through its ability to induce actin polarization.
Actin polarization limits growth after passage through G1 phase
Our results suggest that during G1 phase, actin polarization can limit growth. To test whether actin polarization can limit growth in other stages of the cell cycle, we examined the growth properties of strains defective in the ubiquitin ligase SCF (cdc34-2, cdc53-1; henceforth SCF mutants) and a strain carrying a cdc42-6 allele. When SCF mutants are shifted to the restrictive temperature, they grow in a highly polarized fashion due to high levels of Cln-CDK activity (Tyers et al. 1992; Lew and Reed 1993), but growth is limited (Figs. 1D, 6A). In contrast, a cdc42-6 mutant, which fails to maintain a polarized actin cytoskeleton and thus grows isotropically (Gladfelter et al. 2001), grew significantly larger than SCF mutants (Fig. 6B). To test whether actin polarization accounted for the limited growth in SCF mutants, we examined the consequences of inactivating Cln-CDKs in this background. A cdc34-2 cdc28-as1 double-mutant strain was first arrested for 2 h at 37°C to arrest cells in a state of high Cln-CDK activity and hyperpolarized growth. CDK activity was then inhibited by addition of the adenosine analog 1-NM-PP1 after which cells grew isotropically and reached cell sizes nearly identical to that of the cdc28-as1 single mutant (Fig. 6A,C). Inactivation of Cdc42 in a SCF mutant, thus causing istotropic growth, also restored the ability of cells to grow more than SCF single mutants (data not shown). Deletion of STE12 in a cdc34-2 strain had no effect, indicating that elevated pheromone pathway signaling did not account for the observed growth suppression (Supplemental Fig. 4A). These results indicate that Cln-CDK activity and actin polarization limit cell growth in SCF mutants.
Actin polarization restricts growth in MEN mutants
Given the above results, we postulated that differences in actin polarization might be responsible for the marked difference in growth capacity of MEN mutants and cells that overexpress a nondegradable form of Clb2. Both MEN mutants and GAL-CLB2dbΔ cells arrest in anaphase due to the inability of the cells to sufficiently reduce the level of B-type cyclins (Stegmeier and Amon 2004). However, GAL-CLB2dbΔ strains arrest as large budded or unbudded cells with a depolarized actin cytoskeleton (Amon et al. 1994; Monje-Casas and Amon 2009), whereas MEN mutants polarize their actin cytoskeleton and form an elongated bud after prolonged incubation at 37°C (Jimenez et al. 1998). To directly test the possibility that actin polarization limits growth in MEN mutants, we examined the consequences of inactivating Cln-CDKs in a cdc15-as1 strain, which mimics the cdc15-2 allele in all respects upon treatment with 1-NM-PP1 (D'Aquino et al. 2005). To allow conditional expression of Cln-CDK activity, the cdc15-as1 strain was also deleted for all CLN genes (CLN1, CLN2, and CLN3) and rendered viable by expression of CLN2 under the methionine-repressible MET3 promoter. Cells were first arrested in anaphase by the addition of NM-PP1, and then CLN2 transcription was repressed by the addition of methionine (Fig. 6D). The depletion of G1 cyclin activity allowed cdc15-as1 cln1Δ cln2Δ cln3Δ cells to grow to a significantly larger size than control cdc15-as1 cells (Fig. 6D). Finally, we found that cdc15-2 mutants overexpressing a stabilized version of Clb2 (Amon et al. 1994), which also represses Cln-CDK activity (Fig. 6E), were able to grow to the same extent as GAL-CLB2dbΔ strains (Fig. 6F). Either depletion of CLNs or expression of Clb2dbΔ minimized the formation of polarized projections seen in MEN mutants arrested for prolonged periods of time (data not shown), further extending the correlation between polarization of the actin cytoskeleton and limitation of growth. In addition, deletion of STE12 in a cdc15-2 strain had no effect on the growth properties of the MEN mutant, indicating that elevated pheromone pathway signaling in this context also did not account for the observed growth suppression (Supplemental Fig. 4B). We conclude that Cln-CDK activity restricts growth in both SCF and MEN mutants via effects on actin polarization. Interestingly, inactivation of Cdc28 activity did not allow cdc20-1 and cdc23-1 to grow larger (data not shown). This finding demonstrates that cell cycle events other than Cdc28-mediated actin polarization restrict growth in these metaphase-arrested cells.
Growth rates change during the cell cycle
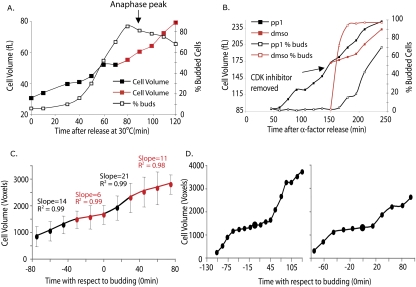
To address whether cell cycle position affects growth under normal physiological conditions, we examined rates of cell volume increase in wild-type cells synchronized in early G1 by centrifugal elutriation. Consistent with our cell cycle arrest studies, we observed a subtle slowing of growth rate around the time of bud formation (30–40 min after release) (Fig. 7A) and a significantly slower growth rate as cells progressed through mitosis until cells entered anaphase, at which time growth rate accelerated (Fig. 7A; Supplemental Fig. 5A).
Figure 7.
Growth rates change during the cell cycle. (A) Wild-type cells (A2587) grown in YEP + 2% glucose were elutriated at 4°C to collect small G1 cells. The collected cells were transferred to YEP supplemented with 2% glucose at 30°C, and cell volume (black or red filled squares), budding (black, open squares), and percent anaphase cells (anaphase peak indicated with an arrow) were determined at the indicated times. Similar observations were made in at least three independent experiments (Supplemental Fig. 5B). (B) cdc28-as1 cells (A18367) were arrested at 22°C with α-factor for 2.5 h to allow accumulation of cells in G1. Subsequently, cells were released from arrest in the presence of 20 μM 1-NM-PP1 inhibitor at 30°C, and the cell volume and budding were determined at 15-min intervals. After 135 min, the culture was washed to remove the CDK inhibitor and resuspended in medium containing either CDK inhibitor (black) or DMSO (red). Cell volume (filled squares) and budding (open squares) were determined at the indicated times following the 135-min time point. Similar observations were made in at least three independent experiments (Supplemental Fig. 5A). (C) Wild-type cells (A2597) were grown in a microfluidic device in YPD media at room temperature. Cells were synchronized with pheromone for 1.5 h and released in fresh YPD media, and the growth properties of their decedents were analyzed. Pheromone-treated cells were not included in the analysis to eliminate effects of pheromone treatment on growth in the ensuing cell cycle. Images of cells were acquired every 15 min by automated time-lapse microscopy. Image processing algorithms were used to calculate the cell volumes as described in Materials and Methods. Calculated cell volumes of individual cells (volume of mother + volume of bud) were plotted versus the time of bud emergence (T = 0 min). The data plotted are the average of the growth patterns of 20 individual cells. The plot was segmented into four linear parts, and the fit to a linear for each segment (slope and R 2) is indicated. (D) The growth of two individual cells from the experiment, which were used to produce the average in C.
The slowing in growth rate at the time of bud formation after elutriation was subtle. We therefore wished to examine growth rates during the cell cycle using other synchronization methods. We arrested cdc28-as1 cells in G1 phase with NM-PP1 and cell volume was monitored upon release from the G1 block (Fig. 7B; Supplemental Fig. 5B). Upon removal of the inhibitor and concomitant with polarized growth and bud formation, we observed a decrease in cell growth (red filled squares in Fig. 7B; Supplemental Fig. 5B). A control culture that was manipulated in the same manner but in which the CDK inhibitor was reapplied during the release (black filled squares in Fig. 7B) did not slow cell growth until cells began to escape the G1 arrest (Fig. 7B; Supplemental Fig. 5B).
To exclude the possibility that the growth rate changes observed upon release from a cdc28-as1 block were a result of the chemical–genetic perturbation, we measured growth in wild-type cells using a single-cell approach. Increase in cell size was monitored by time-lapse video microscopy, and the volume of mother and daughter cells was estimated by image processing algorithms (Taylor et al. 2009; D Falconnet, A Niemistö, RJ Taylor, M Ricicova, and T Galitski, in prep.). Cells grew in the microfluidic device at room temperature in YPD with a doubling time of ∼120 min, indicating that the proliferation timing in the microfluidics device was comparable with that of cells grown in flasks. Analysis of single cells demonstrated that cell growth slowed prior to and during bud emergence. Although cell-to-cell variation exists, we often observed cells that decreased growth for 45–60 min around the time of budding (Fig. 7D; Supplemental Fig. 6). Furthermore, averaging of the data showed that growth was reduced at around the time of bud emergence (Fig. 7C). This period of reduced growth was followed by a 30-min window of accelerated growth, followed again by attenuated growth most likely when cells are in mitosis (note that the analysis did not allow us to examine cell cycle parameters other than budding). The volume measurements were best fit by four linear growth rates with R 2 values of 0.984 or above. A single exponential growth curve fit represented the data less well (R 2 = 9.71). Based on the results obtained with synchronized and asynchronous cell cultures and two different measurement methods, we conclude that growth is not constant throughout an unperturbed cell cycle. Rather, the transitions between isotropic and apical patterns of growth inherent to cell cycle progression appear to dictate changes in the rate of cell growth. The subtlety of the slowing in growth at the time of budding in the elutriated cells could be the result of stress caused by the elutriation procedure, or perhaps because elutriation selects for newborn cells that are subject to additional growth regulation in the first cell cycle (Di Talia et al. 2007).
Discussion
It is well-established that cell growth controls cell division in budding and fission yeast (Jorgensen and Tyers 2004). We show here that in budding yeast, the reciprocal relationship also holds: Cell cycle stage dictates cell growth. In particular, the ability of cells to grow is higher in anaphase- and G1-arrested cells than in other cell cycle stages. Through manipulation of actin dynamics, either by mutation or by pheromone treatment, we demonstrate that polarization of the actin cytoskeleton markedly attenuates growth and protein synthesis rates throughout the cell cycle. During the normal cell cycle, passage through the G1/S-phase boundary drives a transition from an isotropic to an apical pattern of growth; correspondingly, we find a decrease in growth rate at this boundary, consistent with the notion that actin polarization regulates growth. Our data also indicate that polarization is not the only mechanism whereby cell cycle progression affects growth. Mutants that arrest during S (cdc9-1, cdc7-1) and M phase (cdc20-1, cdc23-1) grow isotropically, but their growth capacity is reduced (Fig. 1D). These findings add a new dimension to the coordination of cell growth and cell division.
Growth during the cell cycle
The relationship between cell growth and division has remained a central question in cell biology since Boveri and others first described the phenomenon of cell size homeostasis (Wilson 1925). Johnston, Pringle, and Hartwell measured protein synthesis and cell diameter in cdc mutants upon shift to the restrictive temperature and found that in most cdc mutants, these parameters continued to increase for 6–8 h after temperature shift; these findings led to the proposal that growth is independent of cell division (Johnston et al. 1977). Subsequent observations suggesting constant protein synthesis rates in synchronous cycling cells or individual cells further substantiated this model (Elliott and McLaughlin 1978, 1979; Woldringh et al. 1993; Di Talia et al. 2007). In contrast, our results indicate that while growth does continue in arrested cells, the rate and extent to which growth occurs are not the same in all cell cycle states. We initially noticed this effect in the markedly different growth rates of cells arrested by different cdc mutations. Importantly, we also observe a consistent reduction in growth rate as synchronized cells pass through the G1/S–phase boundary, as well as in single-cell analysis. Superficially, these observations appear at odds with a recent study that concluded growth rate increases exponentially throughout the cell cycle (Di Talia et al. 2007). However, upon closer analysis, these data are fully compatible with our observations. Di Talia et al. (2007) described their measurements using a simple exponential fit, and as a corollary a constant growth rate, because this approach assumes fewer variables. However, Di Talia et al. (2007) also noted that growth rates are equally well-described by two linear growth components. In our analysis, the imposition of multiple linear fits more accurately matched the growth rates observed in individual cell measurements than a single exponential fit. It is also notable that Di Talia et al. (2007) reported a decrease in growth rate at or near the time of budding (Fig. 2A; Figs. S1A–C, S2A,B in Di Talia et al. 2007). It is worth noting that Woldringh et al. (1993), who measured growth rates in elutriated single cells, also observed a change in growth similar to what we observe in elutriated cells: Volume increase accelerated after bud emergence. All salient observations are thus consistent with the model that growth varies not only between different cell cycle arrests but also during an unperturbed cell cycle.
The pheromone pathway represses growth
Our results indicate that pheromone treatment limits growth in G1-arrested cdc28 mutants. This growth inhibition is mediated by the MAP kinase pathway through a mechanism that is distinct from the FAR1-dependent cell cycle arrest in G1 phase but that requires pheromone-induced actin polarization. We also identified the main elements of the pheromone signaling pathway as large mutants in an unbiased cell size screen. We emphasize that this later observation runs contrary to the naive prediction that ablation of the cell cycle delay caused by basal signaling of the pheromone pathway should produce smaller, not larger cells. Coupled with the dependence of this size effect on carbon and nitrogen source, the genome-wide elutriation screen results suggest that basal pheromone signaling serves, in fact, to attenuate growth. This inhibition of growth by the active mating pheromone pathway may have been selected to help prevent susceptibility to cell lysis during a prolonged G1 arrest as cells seek a mating partner.
Actin polarization inhibits growth in all cell cycle phases
Pheromone exerted a very strong growth inhibitory effect. This is at least in part mediated through its stimulation of actin polarization. However, we note that the effect of pheromone on RP gene expression is very rapid, raising the possibility that pheromone affects growth by more than one pathway. It is nevertheless clear that actin polarization plays a key role in attenuating growth. The extent of growth inhibition correlated with the degree of actin polarization and depended on the formin Bni1, which is essential for pheromone-induced polarization, but is not restricted to mating per se (Matheos et al. 2004). Actin polarization dominantly inhibits growth in cell cycle stages other than G1 phase; for example, in the cdc34-2 and cdc15-2 arrests. In contrast, the pheromone pathway did not appear to exert any obvious effect outside of G1 phase, presumably because signaling activity is stringently restricted to G1 phase, in large part by CDK activity (Strickfaden et al. 2007).
It is possible that the actin cytoskeleton impinges on growth regulatory pathways via the Protein Kinase C (PKC) pathway, also known as the cell-wall integrity pathway (Levin 2005). The PKC pathway has been shown previously to inhibit protein synthesis in mutants defective in vesicular transport and secretion (Mizuta and Warner 1994; Nierras and Warner 1999). Furthermore, the pathway is activated both during an unperturbed cell cycle (Zarzov et al. 1996) and during α-factor treatment (Zarzov et al. 1996; Buehrer and Errede 1997). PKC1 mutants have weakened cell walls and lyse at the bud tip or shmoo (Levin et al. 1994; Roemer et al. 1994). These observations suggest that the PKC pathway ameliorates cell wall stress under adverse conditions in part by attenuating growth. Polarization of the actin cytoskeleton is also likely to activate the PKC response; indeed, bni1 strains show a delayed and decreased induction of the PKC pathway when treated with pheromone (Buehrer and Errede 1997). In an unperturbed cell cycle, stress caused by polarized growth is likely limited to late G1 and early S phase when bud formation occurs. However, as our analysis indicates, polarized growth can limit protein synthesis at any point in the cell cycle.
It is important to note that actin polarization is not the only cell cycle event controlling growth. Cells arrested in S phase and metaphase grow isotropically, yet their growth abilities are reduced compared with cdc28 mutants. Furthermore, inactivation of CDC28 and hence actin cytoskeleton depolymerization in these mutants does not lead to increased growth (A Goranov, unpubl.). Thus, other growth inhibitory factors are present in these cell cycle arrests that antagonize cell growth. Identifying this other growth inhibitory pathway will be an important next step.
Why does growth rate change during the cell cycle?
Our data suggest that in budding yeast, growth is maximal during anaphase and G1, and more limited in S phase and mitosis. Cells may have evolved this mechanism in order to stockpile the necessary cellular resources prior to the critical and irrevocable phases of DNA replication and chromosome segregation. This notion of resource accumulation is consistent with the concept of cell cycle commitment, called Start in yeast and the Restriction Point in mammalian cells (Hartwell et al. 1974; Pardee 1974). Once cell cycle entry has occurred, actin polarization may help ensure that less energy is directed to protein synthesis and hence available for duplication and segregation of the genetic material. In addition, cell cycle-dependent decreases in growth rate may also have been selected to allow the cell to better cope with cell membrane and cell wall stress that is inherent to bud growth, as discussed above in the case of activating the PKC pathway.
In fission yeast, growth occurs at the old end of the cell during the first one-third of the cell cycle and subsequently switches to growth at both ends. This phenomenon is called NETO (new-end take-off) and is cell cycle-controlled (Mitchison and Nurse 1985). At the NETO transition there is also a change of growth rate (Mitchison and Nurse 1985; Sveiczer et al. 1996), further supporting the notion that growth rate varies during the cell cycle. In addition, the growth of Schizosaccharomyces pombe cdc25 mutants, which arrest with low CDK activity somewhat analogous to the Saccharomyces cerevisiae cdc28 mutants, is limited by mating pheromone (Stern and Nurse 1997). These observations raise the possibility that this principle of growth regulation may be conserved across divergent fungal species.
In multicellular organisms, the relationship between cell growth and division is flexible and under tight developmental control (Su and O'Farrell 1998; Stocker and Hafen 2000; Saucedo and Edgar 2002; Edgar 2006). We speculate that in these organisms too, growth is influenced by cell cycle state. This principle may in part explain the longstanding enigma of how normal organ size can be maintained in the face of drastic alterations in the size of constituent cells (Fankhauser 1945; Weigmann et al. 1997; Neufeld et al. 1998). For example, reduction of CDK activity in Drosophila imaginal discs results in fewer cells, and yet overall disk size is not altered (Weigmann et al. 1997; Neufeld et al. 1998). Increased growth in the absence of CDK activity could help explain this phenomenon. Developing methodologies to exactly measure growth rates in vivo will be essential to further address this question. Similarly, it will be interesting to determine whether cell morphology affects cell growth and whether the cytoskeleton influences cell growth, proliferation, and fate decisions in multicellular organisms (Chen et al. 1997; Ingber 1997; Nelson et al. 2005).
Materials and methods
Yeast strains, plasmids, and growth conditions
All yeast strains are derivatives of W303 and are listed in Supplemental Table 2. Yeast strains were generated and manipulated as described previously (Gutherie and Fink 1991). α-Factor at a stock concentration of 20 mg/mL in water was used at a concentration of 20 μg/mL for cell cycle arrests. For reversible G1 synchronization, α-factor was used at 5 μg/mL. To decrease the chance of wild-type pheromone-treated cells escaping the arrest, these cells carried a deletion of the α-factor-inactivating protease, BAR1, rendering cells hypersensitive to pheromone and allowing for prolonged cell cycle arrest. The following drugs were used at the indicated concentrations: cycloheximide, 330 μg/mL in water; sodium azide, 0.1% (w/v) in water; latrunculin A, 200 μM in DMSO; and rapamycin, 100 nM in ethanol. 1-NM-PP1 inhibitor was used at 5 μM to inhibit Cdc28-as1 and arrest cells in G1 (Bishop et al. 2000), and 1-N-PP1 was used at 10 μM to inhibit Cdc15-as1 (D'Aquino et al. 2005).
Cell volume determination
To determine the cell volume distribution of a culture, 500 μL of cells was collected, briefly sonicated (1–3 sec), and diluted into Isoton II solution (Beckman) to give <12% saturation when processed with a Beckman Coulter Multisizer 3 (usually 1:100 dilution). Usually, 20,000–60,000 cells were counted and sized. In rare instances, as few as 15,000 were counted and sized. To decrease background signal and chances of counting dead yeast, particles smaller than 3 μm in diameter were excluded from the data analysis. In wild-type cells, >99% of the population is larger than 3 μm in diameter. Cell volume distributions were smoothed with a rolling window of three bins. When cell size distributions were determined and smoothed, we obtained median, mean, and mode statistical descriptions for the population. In populations of cdc mutants grown for prolonged periods of time at the restrictive temperature, a fraction of cells undergo lysis or escape the cell cycle arrest, resulting in the accumulation of smaller cells in culture. This subpopulation of lysed or nongrowing cells affects the median and mean descriptions but not mode. We therefore chose to plot the mode of cell populations as this most accurately represents the mean size of arrested and growing cells. Indeed, in growing subpopulations of cells, the mode is nearly identical to the mean and median cell size (Supplemental Fig. 7). Integration of the area under the curve revealed that usually 50%–80% of cells fall within these growing subpopulations.
Protein synthesis analysis
Cells were grown to exponential phase in YEP + 2% glucose or YEP + 2% raffinose + 2% galactose that was supplemented with 12 μL of S35-Cys (120 μCi final) and 12 μL of S35-Met (120 μCi final) and incubated at 37°C. Radioactively labeled amino acids were obtained from Perkin Elmer at 10 mCi/mL stock concentrations. Time points were collected when appropriate by collecting two 250-μL aliquots per sample. Aliquots were immediately mixed with 250 μL of cold 10% trichloroacetic acid (TCA) and were incubated for at least 20 min at 4°C. 100-μL glass beads were added to each sample, and cells were lysed using a Biopulverizer 101 by three cycles of 45 sec each at max speed. To destroy charged amino acids, samples were incubated for 10 min at 95°C. Samples were cooled to room temperature, extracts were cleared, and 400 μL of the supernatant was spotted on a glass fiber filter (company) prewetted with 5% TCA. Samples were washed with ∼10 mL of 5% TCA and with 100% ethanol. Filters were air-dried for an additional 20 min and mixed with scintillation fluid, and incorporation was determined with a scintillation counter (company) for 1 min per sample. The two duplicate aliquots per sample were averaged and reported.
Microscopy
Five hundred microliters of live cells grown at the appropriate conditions were collected and sonicated briefly to disperse clumps. Five microliters was spotted on a slide and imaged with a Zeiss Axioplan 2 microscope equipped with Hamamatsu ORCA-ER C4742-80 digital CCD camera and processed with Openlab (Improvision) software. For assessing spindle morphology, cells were collected at the indicated times, fixed in 3.7% formaldehyde (v/v) overnight, and processed for indirect immunodetection of tubulin structures as described previously (Visintin et al. 2003).
Elutriation
Elutriations were performed essentially as described (Amon 2002). Strains were grown in 1 L of YEP supplemented with either 2% raffinose or 2% glucose at room temperature to OD600 of 1.0–2.0. Cells were collected, chilled to 4°C, and resuspended in 30 mL of YPE without sugar (Supplemental Fig. 3B) or in the presence of 2% glucose (Fig. 6B). Cells were sonicated to break clumps and were kept at 4°C for the duration of the elutriation. A Beckman elutriation rotor JE 5.0 was chilled to 4°C and equilibrated with YEP (no sugar) at 2400 rpm. Cells were loaded into the rotor using a pump speed of 15 mL/min and filled ∼80% of the chamber. Cells were allowed to equilibrate for 30 min at a rotor speed of 2400 rpm and pump speed of 15 mL/min. Pump speed was increased until small unbudded cells were elutriated. One liter was collected at that pump speed (usually ∼20 mL/min) and kept on ice. The elutriated cells were concentrated with a filtration apparatus and resuspended in YEPD supplemented with 2% glucose at 30°C to initiate the cell cycle.
Western blot analysis
Cells were collected at the indicated times, pelleted to remove media, and resuspended in 5 mL of 5% cold TCA. After more than 15 min in TCA, cells were pelleted, supernatant was removed, and cells were resuspended in 1 mL of acetone. Cells were vigorously vortexed and spun down to remove acetone, and dried for 3 h. Cells were lysed in 150 μL of TE (pH 7.5) in the presence of 2.75 mM DTT. To detect Cln2-HA, a 1° monoclonal, mouse, anti-HA antibody (Covance 16B112) at 1:500, and a 2° rabbit anti-mouse HRP-conjugated antibody (Molecular Probes) at 1:1000 dilution were used. The membrane was reprobed with monoclonal 1° mouse anti-Pgk1 antibody (Invitrogen) at 1:20,000 dilution and probed with a 2° rabbit anti-mouse HRP- conjugated antibody (Molecular Probes) at 1:1000 dilution.
Genome-wide screen to identify deletions that affect cell size
Bar code elutriation experiments were performed as described (Cook et al. 2008). Briefly, 1.5 × 107 yeast cells from a pool of the complete haploid MATa deletion collection were inoculated into 1 L of XY liquid media (YEPD + 100 mg/L adenine + 200 mg/L tryptophan) supplemented with 100 μg/mL G418 and containing 2% of glucose (Cook et al. 2008), ethanol, or glycerol, and grown to a density of ∼1 × 107 to 3 × 107 cells per milliliter. Yeast cells were elutriated, and samples were collected immediately before and after elutriation for subsequent analysis. Pheromone fitness screens were performed in the presence of 0.8 μg/mL α-factor (Chi Scientific). Yeast cells (1.5 × 107) from the deletion pool were diluted into 50 mL of XY + 100 μg/mL G418 in the presence or absence of 0.8 μg/mL α-factor and grown to saturation at 30°C (∼10 generations). 1.5 × 107 yeast cells from the saturated culture were rediluted in 50 mL of fresh XY + 100 μg/mL G418 in the presence or absence of 0.8 μg/mL α-factor and grown to saturation (∼20 generations total). Yeast cells were collected from the 10-generation and 20-generation samples from both pheromone-treated and -untreated samples for subsequent analysis.
Genomic DNA was extracted and used for amplification of unique bar codes with Cy5- and Cy3-labeled primers (Operon), which were hybridized to in-house printed 13K short unmodified bar code arrays (SUBarrays); experimental methods and data analysis were performed as described (Cook et al. 2008). LOWESS normalized and filtered microarray data for the indicated deletion strains were visualized using Java TreeView version 1.1.1 (Saldanha 2004).
Expression microarrays
The sample labeling, hybridizations, and normalization of the expression experiments were done by The University Health Network Microarray Center (Toronto, Ontario, Canada), details of which are available at http://www.microarrays.ca. Briefly, total RNA was reverse-transcribed using SuperScript II (Invitrogen), oligo-dT20-dVdN as a primer and AA-dUTP. Amino allyl cDNA was purified using the CyScribe GFX purification kit (GE Healthcare). Labeling was done by incubating samples with monoreactive cyanine dyes for 1 h. The fluorescently labeled cDNAs were combined and purified. A cocktail containing digoxygenin Easy Hyb (Roche) solution and salmon sperm DNA was added to the fluorescently labeled cDNAs, and the solution was heated for 2 min to 65°C, cooled to 50°C, and applied to the spotted microarray slides. Slides then were incubated overnight at 37°C in a sealed humidified chamber, washed successively with 1× SSC/0.1% SDS at 50°C (three times for 10 min), 0.1× SSC at room temperature (three times for 10 min), dried by centrifugation, and scanned.
Single-cell volume estimation using microfluidic technology and image analysis algorithms
Cells were grown to exponential phase in YPD media, spun down, and concentrated in YPD supplemented with 20 mg/mL BSA (Sigma Aldrich) to an OD600 = 2. Cell suspensions were mixed at a ratio of 1:1 with a 2.8% gel of low melting agarose (Sigma A2576, Type IX-A) that was dissolved in YPD, heated to boiling temperature, and cooled down to 30°C. The vigorously vortexed cell suspension gel was transferred to a microfluidic device designed and fabricated as described previously (Taylor et al. 2009; D Falconnet, A Niemistö, RJ Taylor, M Ricicova, and T Galitski, in prep.) using multilayer soft lithography (Unger et al. 2000; Hansen et al. 2004). The cell suspension gel was isolated in individual chambers of the microfluidic chip. The whole device was then cooled to 4°C in order to polymerize the agarose gel that leads to cell immobilization. Cell imaging chambers were designed to be in diffusive contact with perfusion channels to allow for constant growth conditions by diffusive media exchange throughout the experiment (D Falconnet, A Niemistö, RJ Taylor, M Ricicova, and T Galitski, in prep.). After an initial 90-min perfusion with YPD media, cells were synchronized with 10 μM α-factor (in YPD) for 90 min and afterward released from G1 phase by supplying YPD for 5 h. Only daughters and their progeny of pheromone-treated cells were analyzed to exclude effects of pheromone arrest on cell growth. Bright field images of each chamber were taken every 15 min by automated time-lapse microscopy (D Falconnet, A Niemistö, RJ Taylor, M Ricicova, and T Galitski, in prep.). Automated image processing algorithms were used to track individual cells in selected chambers after G1 release and to calculate cell volumes over time (Taylor et al. 2009; D Falconnet, A Niemistö, RJ Taylor, M Ricicova, and T Galitski, in prep.). Image processing steps include segmentation to identify cell walls and volume calculations based on the “conical method” (Gordon et al. 2007). Volumes were calculated in cubic pixels (voxels), where 1 voxel corresponds to ∼0.26 μm3. An automated tracking algorithm assigned a number to each cell with its computed volume and tracked individual cells in all images taken during the examined period. New buds were allocated to appropriate mother cells using time-lapse imaging videos. The total cell volume estimation of each cell and its progeny was calculated over multiple generations as a sum of all volumes in each time point. The segmentation was manually verified, and if a cell was erroneously segmented by the algorithm at a particular time, this time point was excluded from the analysis.
Acknowledgments
We thank D. Pellman, G. Fink, S. Bell, and members of their laboratories for reagents. We thank Fred Cross, Bruce Futcher, Paul Nurse, Frank Solomon, and members of the Amon laboratory for their critical reading of the manuscript. This work was supported by the National Institutes of Health GM056800 to A.A., by grants from the Canadian Institutes of Health Research and the Wellcome Trust to M.T., by Post-doctoral Fellowship from the American Cancer Society to A.G., and by an Ontario Student Opportunity Trust Fund Award to M.C. A.A. is an investigator of the Howard Hughes Medical Institute, and M.T. is a Research Professor of the Scottish Universities Life Sciences Alliance as supported by the Scotish Funding Council, with additional support from a Royal Society Wolfson Research Merit Award.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1777309.
Supplemental material is available at http://www.genesdev.org.
References
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. Synchronization procedures. Methods Enzymol. 2002;351:457–467. doi: 10.1016/s0076-6879(02)51864-4. [DOI] [PubMed] [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Boucher L, Tyers M. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol. 2001;11:1266–1271. doi: 10.1016/s0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- Buehrer BM, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Cook MA, Chan CK, Jorgensen P, Ketela T, So D, Tyers M, Ho CY. Systematic validation and atomic force microscopy of non-covalent short oligonucleotide barcode microarrays. PLoS One. 2008;3:e1546. doi: 10.1371/journal.pone.0001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- D'Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: Genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Elion EA, Satterberg B, Kranz JE. FUS3 phosphorylates multiple components of the mating signal transduction cascade: Evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SG, McLaughlin CS. Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 1978;75:4384–4388. doi: 10.1073/pnas.75.9.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SG, McLaughlin CS. Synthesis and modification of proteins during the cell cycle of the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1185–1190. doi: 10.1128/jb.137.3.1185-1190.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fankhauser G. Maintenance of normal structure in heteroploid salamander larvae through compensation of changes in cell size by adjustment in cell number. J Exp Zool. 1945;100:445–455. doi: 10.1002/jez.1401000310. [DOI] [PubMed] [Google Scholar]
- Farley FW, Satterberg B, Goldsmith EJ, Elion EA. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Moskow JJ, Zyla TR, Lew DJ. Isolation and characterization of effector-loop mutants of CDC42 in yeast. Mol Biol Cell. 2001;12:1239–1255. doi: 10.1091/mbc.12.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- Gutherie C, Fink GR. Guide to yeast genetics and molecular biology. Academic Press; San Diego: 1991. [Google Scholar]
- Hall DD, Markwardt DD, Parviz F, Heideman W. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 1998;17:4370–4378. doi: 10.1093/emboj/17.15.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MN, Raff M, Thomas G. Cell growth: Control of cell size. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Hansen CL, Sommer MO, Quake SR. Systematic investigation of protein phase behavior with a microfluidic formulator. Proc Natl Acad Sci. 2004;101:14431–14436. doi: 10.1073/pnas.0405847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler Jde A, Bergounioux C, Van Montagu M, Engler G, Inze D, Ferreira P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: For mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hicks JB, Herskowitz I. Evidence for a new diffusible element of mating pheromones in yeast. Nature. 1976;260:246–248. doi: 10.1038/260246a0. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Tensegrity: The architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Li Y, Guan KL. Signaling by target of rapamycin proteins in cell growth control. Microbiol Mol Biol Rev. 2005;69:79–100. doi: 10.1128/MMBR.69.1.79-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Cid VJ, Cenamor R, Yuste M, Molero G, Nombela C, Sanchez M. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J Cell Biol. 1998;143:1617–1634. doi: 10.1083/jcb.143.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Ehrhardt CW, Lorincz A, Carter BL. Regulation of cell size in the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Bowers B, Chen CY, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: Regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheos D, Metodiev M, Muller E, Stone D, Rose MD. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J Cell Biol. 2004;165:99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR, III, Gygi SP, Kellogg DR. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–515. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Warner JR. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Andrews B. Late-G1 cyclin–CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell. 2009;16:132–145. doi: 10.1016/j.devcel.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: From cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A, Hunt T. The cell cycle. Oxford University Press; New York: 1993. [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Nierras CR, Warner JR. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- Pardee AB. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes & Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Qi M, Elion EA. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J Cell Sci. 2005;118:2837–2848. doi: 10.1242/jcs.02418. [DOI] [PubMed] [Google Scholar]
- Roemer T, Paravicini G, Payton MA, Bussey H. Characterization of the yeast (1 → 6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA. Why size matters: Altering cell size. Curr Opin Genet Dev. 2002;12:565–571. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Patton EE, Lanker S, Mendenhall MD, Wittenberg C, Futcher B, Tyers M. Yeast G1 cyclins are unstable in G1 phase. Nature. 1998;395:86–89. doi: 10.1038/25774. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Zhang J, Markwardt J, Tokiwa G, Volpe T, Honey S, Futcher B. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start. Mol Cell Biol. 2004;24:10802–10813. doi: 10.1128/MCB.24.24.10802-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Dolan JW, Yuan YL, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes & Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- Sprague GF, Jr, Herskowitz I. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J Mol Biol. 1981;153:305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stern B, Nurse P. Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B-p34cdc2 kinase. EMBO J. 1997;16:534–544. doi: 10.1093/emboj/16.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Hafen E. Genetic control of cell size. Curr Opin Genet Dev. 2000;10:529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, O'Farrell PH. Size control: Cell proliferation does not equal growth. Curr Biol. 1998;8:R687–R689. doi: 10.1016/S0960-9822(98)70436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveiczer A, Novak B, Mitchison JM. The size control of fission yeast revisited. J Cell Sci. 1996;109:2947–2957. doi: 10.1242/jcs.109.12.2947. [DOI] [PubMed] [Google Scholar]
- Taylor RJ, Falconnet D, Niemisto A, Ramsey SA, Prinz S, Shmulevich I, Galitski T, Hansen CL. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc Natl Acad Sci. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Visintin R, Stegmeier F, Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol Biol Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigmann K, Cohen SM, Lehner CF. Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development. 1997;124:3555–3563. doi: 10.1242/dev.124.18.3555. [DOI] [PubMed] [Google Scholar]
- Wilson EB. The cell in development and heredity. Macmillan; New York: 1925. [Google Scholar]
- Woldringh CL, Huls PG, Vischer NO. Volume growth of daughter and parent cells during the cell cycle of Saccharomyces cerevisiae a/α as determined by image cytometry. J Bacteriol. 1993;175:3174–3181. doi: 10.1128/jb.175.10.3174-3181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]