Abstract
The resection of DNA double-strand breaks (DSBs) in bacteria is a motor-driven process performed by a multisubunit helicase–nuclease complex: either an Escherichia coli-type RecBCD enzyme or a Bacillus-type AddAB enzyme. Here we identify mycobacterial AdnAB as the founder of a new family of heterodimeric helicase–nucleases with distinctive properties. The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal nuclease module. The AdnAB ATPase is triggered by dsDNA with free ends and the energy of ATP hydrolysis is coupled to DSB end resection by the AdnAB nuclease. The mycobacterial nonhomologous end-joining (NHEJ) protein Ku protects DSBs from resection by AdnAB. We find that AdnAB incises ssDNA by measuring the distance from the free 5′ end to dictate the sites of cleavage, which are predominantly 5 or 6 nucleotides (nt) from the 5′ end. The “molecular ruler” of AdnAB is regulated by ATP, which elicits an increase in ssDNA cleavage rate and a distal displacement of the cleavage sites 16–17 nt from the 5′ terminus. AdnAB is a dual nuclease with a clear division of labor between the subunits. Mutations in the nuclease active site of the AdnB subunit ablate the ATP-inducible cleavages; the corresponding changes in AdnA abolish ATP-independent cleavage. Complete suppression of DSB end resection requires simultaneous mutation of both subunit nucleases. The nuclease-null AdnAB is a helicase that unwinds linear plasmid DNA without degrading the displaced single strands. Mutations of the phosphohydrolase active site of the AdnB subunit ablate DNA-dependent ATPase activity, DSB end resection, and ATP-inducible ssDNA cleavage; the equivalent mutations of the AdnA subunit have comparatively little effect. AdnAB is a novel signature of the Actinomycetales taxon. Mycobacteria are exceptional in that they encode both AdnAB and RecBCD, suggesting the existence of alternative end-resecting motor–nuclease complexes.
Keywords: DNA repair, helicase, double-strand breaks, ATP-dependent nuclease, molecular ruler
DNA double-strand breaks (DSBs) are an acute threat to cell survival. The major pathway of DSB repair in bacteria is via homologous recombination (HR). In HR, an intact copy of the broken chromosome segment, typically a newly replicated sister chromatid, serves as a template for DNA synthesis across the break. In this mechanism, one or both of the DSB ends is resected by an exonuclease to leave a 3′ single-stranded tail, which then invades the sister chromatid and pairs with its complementary DNA sequence. The RecA protein performs the critical steps of locating and pairing the homologous sequences and promoting strand invasion (Chen et al. 2008). A DNA polymerase at the invading 3′-OH end copies the sequence information from the sister chromatid onto the broken end and drives the formation of a displaced single-strand loop. Resolution of the recombination intermediates ultimately results in a faithfully restored chromosome with no mutations. RecA-dependent HR provides bacteria with a means of rescuing collapsed DNA replication forks, a frequent and potentially lethal event caused by impediments to fork progression (Lusetti and Cox 2002), as well as a defense against exogenous DNA-damaging agents.
The resection of DSB ends in bacteria is an elegantly orchestrated motor-driven process (Spies et al. 2007) performed by a multisubunit helicase–nuclease complex exemplified by Escherichia coli RecBCD (Wright et al. 1971; Goldmark and Linn 1972; Singleton et al. 2004; Dillingham and Kowalczykowski 2008). RecBCD is a heterotrimer composed of two helicase subunits, RecB and RecD, that unwind the DSB ends with opposite polarities (Dillingham et al. 2003; Taylor and Smith 2003) and one nuclease module, located at the C terminus of RecB, that incises the single strands displaced by the helicases. The Bacillus subtilis ATP-dependent DNase enzyme AddAB provides a different way of solving the bacterial DNA end resection problem. AddAB is a functional analog of RecBCD but is sui generis structurally (Chedin and Kowalczykowski 2002). AddAB, a heterodimer, consists of a single helicase motor in the AddA subunit, plus two distinct nuclease modules—one each in AddA and AddB (Haijema et al. 1996a,b; Yeeles and Dillingham 2007). Bacterial species encoding RecBCD/AddAB enzymes fall into two clades: those that have an E. coli-type RecBCD enzyme but not AddAB, and those that have a Bacillus-type AddAB helicase–nuclease but not RecBCD (Rocha et al. 2005). Included in the AddAB bacterial clade is the gastric ulcer-causing human pathogen Helicobacter pylori, which relies on AddAB activity to survive in the stomach (Amundsen et al. 2008).
We are interested in the role of DSB repair in the physiology and pathophysiology of mycobacteria. Mycobacterium tuberculosis, the agent of human TB, and its avirulent cousin Mycobacterium smegmatis can avail themselves of two separate pathways to repair DSBs: (1) a RecA-dependent HR system (Pitcher et al. 2007a; Stephanou et al. 2007), and (2) a nonhomologous end-joining (NHEJ) system that depends on dedicated DNA ligases and the DNA end-binding protein Ku (Gong et al. 2004, 2005; Pitcher et al. 2006; Aniukwu et al. 2008). NHEJ helps protect the bacterial chromosome against DSBs during quiescent states, when there is no sister chromatid available to direct HR (for review, see Pitcher et al. 2007b; Shuman and Glickman 2007). Unlike NHEJ in budding yeast, the mycobacterial NHEJ mechanism is conspicuously mutagenic, even when repairing complementary 5′ overhangs or “clean” blunt DSBs (Gong et al. 2005). The repair infidelity reflects two alternative means of end processing: (1) the addition of templated or nontemplated nucleotides to the DSB ends prior to ligation, or (2) nucleolytic resection of one or both DSB ends prior to sealing (Aniukwu et al. 2008).
The identity of the end-resecting enzyme in the mycobacterial HR and NHEJ pathways is uncertain, notwithstanding that M. smegmatis encodes a homolog of E. coli RecBCD. An M. smegmatis ΔrecBCD mutant is viable, as is a ΔrecA mutant (Stephanou et al. 2007). Whereas the ΔrecA strain is hypersensitive to killing by UV irradiation, as expected, the ΔrecBCD strain is no more sensitive than wild-type M. smegmatis (Stephanou et al. 2007). Moreover, deletion of recBCD has virtually no effect on the fidelity of mycobacterial NHEJ (Aniukwu et al. 2008). The spectrum of unfaithful NHEJ outcomes at blunt and 5′ overhang DSBs in the ΔrecBCD mutant is similar to wild-type mycobacteria. In particular, there is no decline in the prevalence or size of the deletion tracts, indicating that RecBCD is not the principal catalyst of deletion formation during mycobacterial NHEJ in vivo (Aniukwu et al. 2008). These findings suggest that there are other mycobacterial helicase–nucleases available to process DSB ends for HR and NHEJ when RecBCD is absent. Because mycobacteria do not have a Bacillus-type AddAB enzyme (Rocha et al. 2005), we surmised that they might possess a novel end-resecting machinery.
Mycobacterial helicase-like proteins are plausible candidate components of this hypothetical system. We had previously characterized two mycobacterial UvrD-like helicases. UvrD1 was identified based on its interaction with mycobacterial Ku and then shown to be a Ku-dependent 3′-to-5′ DNA helicase (Sinha et al. 2007). In contrast, its paralog UvrD2 is a vigorous 3′-to-5′ helicase per se (Sinha et al. 2008). However, neither UvrD1 nor UvrD2 has an associated nuclease activity. In a broader survey of mycobacterial proteomes, we identified a paralogous pair of uncharacterized UvrD-like proteins encoded by adjacent co-oriented genes suggestive of an operon. The M. smegmatis version of this operon, which we named “adn” (ATP-dependent nuclease), encodes a 1045-amino-acid AdnA polypeptide and a 1095-amino-acid AdnB polypeptide (Fig. 1). The compelling feature of these two large proteins is that they both consist of an N-terminal motor-like domain fused to a C-terminal nuclease-like domain (Supplemental Fig. S1). The adnAB gene cluster is present and conserved in M. tuberculosis (Fig. 1). Here we show that mycobacterial AdnAB is a heterodimeric DSB end processing machine with distinctive properties vis a vis RecBCD and AddAB. We suggest that mycobacteria resemble eukarya (and certain other bacteria) in having functionally redundant DSB processing systems with helicase and nuclease components (Sanchez et al. 2006; Mimitou and Symington 2008; Zhu et al. 2008).
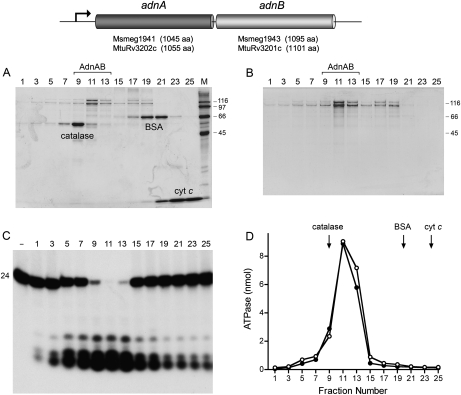
Figure 1.
AdnAB is a heterodimeric ATPase/nuclease. The M. smegmatis and M. tuberculosis adnAB operons comprise two adjacent ORFs encoding AdnA and AdnB polypeptides of the sizes specified. (A) Glycerol gradient sedimentation of recombinant M. smegmatis AdnAB with internal standards. Aliquots (20 μL) of odd-numbered glycerol gradient fractions were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The internal standards catalase, BSA, and cytochrome c are indicated. The positions and sizes (in kilodaltons) of molecular-weight markers included in lane M are indicated on the right. (B) Glycerol gradient sedimentation of AdnAB alone. Aliquots (20 μL) of odd-numbered glycerol gradient fractions were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. (C) AdnAB-associated nuclease. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 2.4 nM 5′ 32P-labeled 24-mer DNA oligonucleotide (see Fig. 4A), and 1 μL of the indicated glycerol gradient fraction were incubated for 5 min at 37°C. Products were analyzed by Urea-PAGE and visualized by autoradiography. A control reaction lacking added enzyme is included in lane −. (D) AdnAB-associated ATPase. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 1 mM [γ32P]ATP, 1 μg of salmon sperm DNA, and 1 μL of the indicated glycerol gradient fractions from A (○) or B (●) were incubated for 5 min at 37°C. The positions of the internal standards from A are indicated by arrows.
Results
AdnAB is a heterodimeric DNA-dependent ATPase/nuclease
The N-terminal motor-like domains of M. smegmatis AdnA and AdnB are homologous to one another and to the motor domains of E. coli UvrD and Bacillus PcrA, which are structurally well-characterized exemplars of superfamily I DNA repair helicases (Velankar et al. 1999; Lee and Yang 2006). An amino acid sequence alignment highlights 156 positions of side chain identity/similarity in AdnA, AdnB, and UvrD within a 630-amino-acid segment of the UvrD motor domain, including nearly all of the UvrD side chains that comprise the ATPase active site and many of the UvrD side chains that comprise the binding site for ssDNA (shaded in yellow in Supplemental Fig. S1). The presence of two seemingly complete motor-like domains distinguishes mycobacterial AdnAB from Bacillus AddAB.
To assess what activities, if any, are associated with mycobacterial AdnA and AdnB, we coexpressed the two polypeptides in E. coli with an N-terminal His10Smt3 tag appended to the AdnA subunit only. The tagged mycobacterial protein was isolated from a soluble E. coli extract by Ni-agarose affinity chromatography and then passed over a DEAE-Sephacel column to remove residual nucleic acid. The His10Smt3 tag was then cleaved from AdnA with the Smt3-specific protease Ulp1 (Mossessova and Lima 2000) and the tagless protein was separated from the tag by a second round of Ni-agarose chromatography. The second Ni-agarose fraction, which contained recombinant AdnA and AdnB polypeptides, was then subjected to zonal velocity sedimentation in a glycerol gradient, with and without internal standards catalase (a 248-kDa tetramer of a 62-kDa polypeptide), bovine serum albumin (BSA; a 66-kDa monomer), and cytochrome c (a 12-kDa monomer) (Fig. 1). SDS-PAGE analysis of the gradient fractions revealed two resolved forms of the 110-kDa AdnA and 118-kDa AdnB polypeptides: one sedimenting in fraction 11 on the “light” side of the catalase peak at a position consistent with an AdnAB heterodimer, and a second sedimenting in fractions 17–19 on the “heavy” side of BSA, consistent with a mixture of AdnA and AdnB monomers (Fig. 1A). The sedimentation profile of the Adn polypeptides was the same in the absence of the internal standards (Fig. 1B). Based on staining intensity, the AdnA protein (the one originally bearing the affinity tag) appeared to be in slight excess, and this excess portion comprised a diffusely sedimenting component evident in fractions 1–19 (Fig. 1A,B), suggesting that AdnA tends to aggregate when not complexed with AdnB (vide infra).
An assay of the glycerol gradient fractions for hydrolysis of [γ32P]ATP in the presence magnesium and salmon sperm DNA revealed a single peak of ATPase activity in fractions 9–13 coincident with the AdnAB heterodimer (Fig. 1D). The fractions were also assayed for single-strand deoxyribonuclease activity with a 5′ 32P-labeled 24-mer DNA oligonucleotide substrate (Fig. 1C). A single symmetrical peak of ssDNase activity that converted the labeled 24-mer to a mixture of 5-mer and 6-mer 32P-labeled oligonucleotide products was detected in fractions 9–13 (Fig. 1C). We surmise that mycobacterial AdnA and AdnB form a heterodimeric ATPase/nuclease in the absence of any other mycobacterial polypeptides.
dsDNA-dependent NTPase activity of AdnAB requires free DNA ends
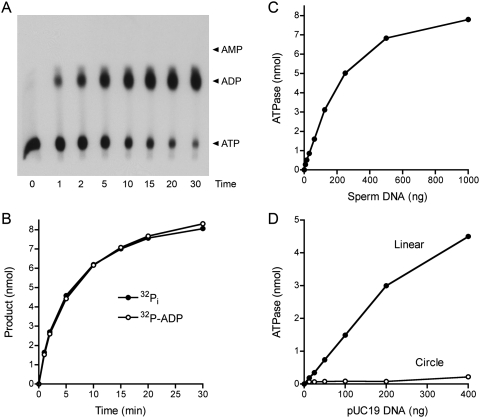
Further characterization of the phosphohydrolase activity was performed using the glycerol gradient-purified AdnAB heterodimer. The rate of release of 32Pi from 1 mM [γ32P]ATP in the presence of salmon sperm DNA was identical to the rate of conversion of [α32P]ATP to [α32P]ADP in a parallel reaction mixture containing the same amount of AdnAB (Fig. 2B). We detected no formation of [α32P]AMP during the reaction (Fig. 2A). We conclude that AdnAB catalyzes the hydrolysis of ATP to ADP and Pi. From the initial rates, we estimated a turnover number of 300 sec−1. ATP hydrolysis by AdnAB was strictly dependent on a DNA cofactor; the extent of product formation was proportional to the amount of salmon sperm DNA added up to 250 ng/10-μL reaction and began to level off at 1 μg of input DNA (Fig. 2C). Whereas linear pUC19 plasmid dsDNA was an effective cofactor for the AdnAB ATPase, circular pUC19 dsDNA was not (Fig. 2D). We conclude that AdnAB requires a free end for dsDNA-dependent ATP hydrolysis.
Figure 2.
AdnAB is a DNA-dependent ATPase. (A,B) Reaction mixtures (60 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM [α32P]ATP or [γ32P]ATP, 1 mM MgCl2, 10 μg of salmon sperm DNA, and AdnAB (60 ng AdnB) were incubated at 37°C. Aliquots (5 μL, containing 5 nmol input ATP) were withdrawn at the times specified and quenched with formic acid. Reaction products were analyzed by PEI-cellulose TLC and visualized by autoradiography. The analysis of products from the reaction containing [α32P]ATP is shown in A, with the positions of cold ATP, ADP, and AMP standards indicated on the right. The extent of conversion of [γ32P]ATP to 32Pi (●) or [α-32P]ATP to [α-32P]ADP (○) is plotted as a function of time in B. (C) DNA dependence of ATP hydrolysis. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 1 mM [γ32P]ATP, 1 mM MgCl2, AdnAB (10 ng AdnB), and salmon sperm DNA as specified were incubated for 5 min at 37°C. 32Pi release is plotted as function of the amount of DNA added. (D) ATP hydrolysis requires free DNA ends. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM [γ32P]ATP, 1 mM MgCl2, AdnAB (10 ng AdnB), and either closed circular pUC19 DNA or blunt-ended linear pUC19 DNA (SmaI-digested) were incubated for 5 min at 37°C. 32Pi release is plotted as function of the amount of DNA added.
AdnAB is an ATP-dependent dsDNA exonuclease
The capacity of AdnAB to resect DSB ends was tested by incubating the enzyme with a blunt-end linear duplex DNA (SmaI-digested pUC19) in the presence of magnesium and ATP. AdnAB converted the 2.7-kb linear duplex, which migrated as a discrete ethidium-bromide-stained species during native agarose gel electrophoresis, into a diffuse smear of more rapidly migrating products (Fig. 3A). The extent of decay of the linear substrate was proportional to the amount of AdnAB added. AdnAB was also adept at resecting linear pUC19 with either a 5′ single-stranded overhang (BamHI-treated) or a 3′ single-stranded tail (KpnI-treated) (Fig. 3B, left panel). In contrast, closed circular pUC19 DNA was resistant to digestion by AdnAB (Fig. 3A), signifying that AdnAB is an obligate exonuclease on a dsDNA substrate.
Figure 3.
AdnAB is an ATP-dependent dsDNA exonuclease. (A) dsDNA exonuclease. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP (where indicated by +), 200 ng of circular pUC19 or blunt-ended linear pUC19 (SmaI-digested), and AdnAB were incubated for 5 min at 37°C. AdnAB-containing reactions indicated by + received 10 ng of AdnB polypeptide. The reactions in the AdnAB titration series received (from left to right) 0.15, 0.31, 0.63, 1.25, 2.5, 5, and 10 ng of AdnB polypeptide. The reaction products were analyzed by electrophoresis through a 0.7% native agarose gel and were visualized by staining with ethidium bromide. The positions and sizes (kilobases) of linear DNA markers are indicated on the right. (B, left panel) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0); 2 mM MgCl2; 1 mM ATP; 200 ng of linear pUC19 DNA prepared by digestion with KpnI, BamHI, or SmaI as specified; and AdnAB (10 ng AdnB) were incubated for 5 min at 37°C. A control reaction containing SmaI-cut DNA without AdnAB is included in lane −. (Right panel) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM ATP, 200 ng of linear SmaI-cut pUC19 DNA, AdnAB (10 ng AdnB), and MgCl2 as specified were incubated for 5 min at 37°C. The DNase reaction products were analyzed by agarose gel electrophoresis. (C) ATP dependence. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 200 ng of SmaI-cut pUC19 DNA, AdnAB (5 ng of AdnB), and ATP as specified were incubated for 5 min at 37°C. (D) Ku protects DSB ends from AdnAB. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng of SmaI-cut pUC19 DNA, AdnAB (10 ng of AdnB, where indicated by +), and purified M. tuberculosis Ku (either 75, 150, or 300 ng, where indicated by +, ++, and +++, respectively) were incubated for 5 min at 37°C. Ku was prepared according to Sinha et al. (2007). (E) Nucleotide specificity in NTP hydrolysis. Reaction mixtures (20 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 1 mM NTP/dNTP as indicated, 2 μg of salmon sperm DNA, and AdnAB (20 ng of AdnB) were incubated for 5 min at 37°C. The reactions were quenched by adding 1 mL of malachite green reagent (Biomol Research Laboratories). Phosphate release was determined by measuring A620 and interpolating the value to a phosphate standard curve. (F) Nucleotide specificity in DSB end resection. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM NTP/dNTP as indicated, 200 ng of linear SmaI-cut pUC19 DNA, and AdnAB (5 ng of AdnB) were incubated for 5 min at 37°C. The DNase reaction products were analyzed by agarose gel electrophoresis.
The AdnAB dsDNA exonuclease was dependent on magnesium (Fig. 3B, right panel) and ATP (Fig. 3C). The extent of substrate consumption was proportional to ATP concentration and saturated at ≥125 μM ATP (Fig. 3C). Partially digested intermediates were detectable at limiting concentrations of ATP (8–16 μM) or magnesium (31–62 μM) (Fig. 3B,C). The dsDNA exonuclease was active over a broad pH range in Tris buffers, from pH 6.0 to pH 9.6; this activity was suppressed at pH ≤5.0 (Supplemental Fig. S2).
The NTP substrate specificity of AdnAB was investigated by a malachite green colorimetric assay of the release of Pi from 1 mM ATP, GTP, CTP, UTP, dATP, dGTP, dCTP, and dTTP. AdnAB readily hydrolyzed all of the purine rNTPs and dNTPs, but was poorly active with the pyrimidine NTPs, in the following order: UTP > TTP > dCTP > CTP (Fig. 3E). The ability of the various NTPs to support DSB end resection correlated nicely with their relative efficacy in NTP hydrolysis (Fig. 3F). We surmise that DSB end processing by AdnAB is coupled to an NTPase motor that unwinds the DNA duplex from the free ends and thereby exposes the displaced single strands to the single-strand nuclease activity associated with the AdnAB complex (Fig. 1C).
Novel properties of the AdnAB single-strand nuclease
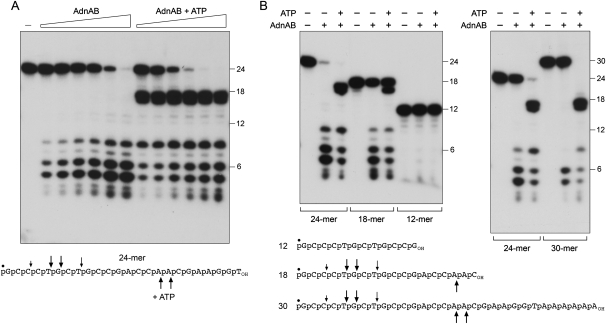
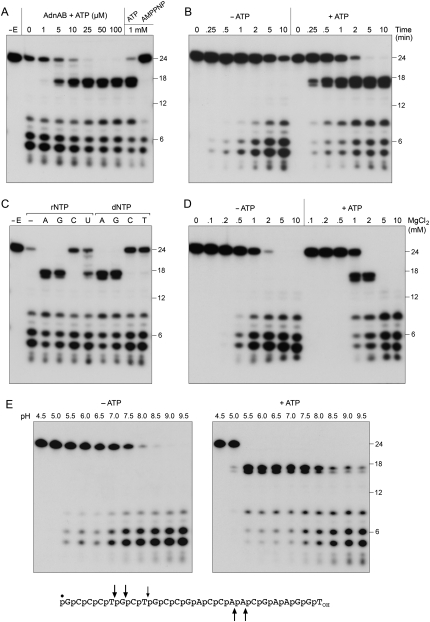
AdnAB incised a 5′ 32P-labeled 24-mer ssDNA oligonucleotide substrate in the absence of ATP to release a mixture of 32P-labeled oligonucleotide products, predominantly 5-mer, 6-mer, and 8-mer species resulting from cleavage of the phosphodiester bonds denoted by the arrows above the oligonucleotide sequence in Figure 4A. The extent of DNA cleavage was proportional to input AdnAB and all of the oligonucleotide substrate was digested at saturating enzyme (Fig. 4A). Inclusion of 1 mM ATP in the AdnAB nuclease reaction mixture had two noteworthy effects. First, ATP increased the nuclease-specific activity by about fourfold, as gauged by the consumption of the 24-mer as a function of input AdnAB. Second, ATP triggered a shift in the distribution of the 5′ 32P-labeled cleavage products, now dominated by a doublet of 16-mer and 17-mer species that were not observed in the absence of ATP (Fig. 4A). Induction of cleavage at the novel sites was dependent on the concentration of ATP and was fully manifest at ≥25 μM ATP (Fig. 5A). In contrast, 1 mM AMPPNP failed to trigger these cleavages (Fig. 5A), suggesting that they required ATP hydrolysis.
Figure 4.
ssDNase activity of AdnAB with 5′ end-labeled DNA. (A) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA substrate (depicted at bottom), either no ATP or 1 mM ATP, and AdnAB (in the amount of 0.63, 1.25, 2.5, 5, 10, and 20 ng of AdnB, from left to right in each titration series) were incubated for 5 min at 37°C. Enzyme was omitted from the control reaction in lane −. The reaction products were analyzed by Urea-PAGE and visualized by autoradiography. 5′ 32P-labeled 18-mer, 12-mer, and 6-mer oligonucleotides of the same 5′-terminal sequence as the 24-mer substrate were analyzed in parallel; the positions of the size markers are indicated on the right. (B) Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0); 2 mM MgCl2; 0.5 mM DTT; 0.1 μM 5′ 32P-labeled 30-mer, 24-mer, 18-mer, or 12-mer DNA substrates (depicted at bottom); 1 mM ATP (where indicated by +); and AdnAB (10 ng of AdnB, where indicated by +) were incubated for 5 min at 37°C. The principal sites of AdnAB incision of the 30-mer, 24-mer, and 18-mer substrates in the absence of ATP are indicated by arrows above the DNA sequences; the cleavage sites induced by ATP are indicated below the DNA sequences.
Figure 5.
Characterization of the ssDNase. (A) ATP dependence of cleavage site choice. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA, AdnAB (5 ng of AdnB), and ATP or AMPPNP as specified were incubated for 5 min at 37°C. Enzyme was omitted from the control reaction in lane −E. (B) Kinetics. Reaction mixtures (100 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA, AdnAB (60 ng of AdnB), and either no nucleotide or 1 mM ATP were incubated at 37°C. Aliquots (10 μL) were withdrawn at the times specified and quenched immediately with formamide/EDTA. (C) Nucleotide specificity. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA, AdnAB (5 of ng AdnB), and either no nucleotide (lane −) or 0.1 mM of the indicated NTP or dNTP were incubated for 5 min at 37°C. Enzyme was omitted from the control reaction in lane −E. (D) Magnesium dependence. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA, either no nucleotide or 1 mM ATP, AdnAB (5 ng of AdnB), and MgCl2 as specified were incubated for 5 min at 37°C. (E) pH dependence. Reaction mixtures (10 μL) containing 20 mM Tris buffer (either Tris-acetate pH 4.5–7.0 or Tris-HCl pH 7.5–9.5), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA, either no nucleotide or 1 mM ATP, and AdnAB (10 ng of AdnB) were incubated for 5 min at 37°C. The principal sites of AdnAB incision of the 24-mer DNA in the absence of ATP are indicated by arrows above the sequence; the cleavage sites induced by ATP are indicated below.
The nucleotide specificity of the cleavage site switching effect was investigated by performing the nuclease reaction in the presence of 0.1 mM ATP, GTP, CTP, UTP, dATP, dGTP, dCTP, and dTTP. Induction of the 16-mer and 17-mer cleavage products was triggered by any of the purine rNTPs and dNTPs (Fig. 5C). Among the pyrimidine nucleotides, UTP had a feeble inductive effect and the others were ineffective (Fig. 5C). Thus, the nucleotide specificity in cleavage site switching mirrored the nucleotide preference of AdnAB for NTP hydrolysis.
ATP elicited an approximately fivefold increase in the rate of consumption of the input 24-mer (Fig. 5B). Each of the radiolabeled cleavage products seen in the ATP-containing reaction accumulated with similar kinetic profiles and persisted at the same level even after all of the 24-mer had been consumed. Thus, there was no evidence for a precursor–product relationship between the ATP-dependent 16-mer/17-mer doublet and the 5-mer/6-mer doublet that predominated in the absence of ATP.
Having seen that hydrolysable ATP affected the rate and mode of cleavage of a 24-mer ssDNA, we queried whether the 24-mer ssDNA could trigger ATP hydrolysis by AdnAB (Supplemental Fig. S3). Indeed it did, and the extent of ATP hydrolysis displayed a hyperbolic dependence on the amount of input 24-mer; the apparent K m for 24-mer ssDNA was 1 μM (Supplemental Fig. S3).
AdnAB nuclease has a molecular ruler that measures from the 5′ end of ssDNA
Insights to the nuclease mechanism and the basis of cleavage site selection emerged from a comparison of the products generated by AdnAB with 32P-labeled DNA oligonucleotides of different lengths with the same nucleotide sequence at their 5′ ends (Fig. 4B). AdnAB cleaved 18-mer, 24-mer, and 30-mer DNA substrates in the absence of ATP to yield an identical cluster of 5-mer, 6-mer, and 8-mer products, signifying that the initial incision was dictated by distance from the DNA 5′ end. In the presence of ATP, AdnAB cleaved the 24-mer and the 30-mer to form an identical 16-mer/17-mer doublet (Fig. 4B, right panel) and it incised the 18-mer to yield a 16-mer product corresponding to the lower band in that doublet (Fig. 4B, left panel). These results attest that ATP increases the measured distance from the 5′ end to the cleavage site. The cleavage patterns are incompatible with a mechanism for measuring cleavage from the 3′ end. AdnAB was unable to digest a 12-mer DNA oligonucleotide, whether or not ATP was added (Fig. 4B), indicating that the enzyme required a minimal length of DNA 3′ of the scissile phosphodiester. (Note also that the 24-mer was a better substrate than the 18-mer when tested in parallel.) We infer that the length effects on cleavage efficiency account for why the 32P-labeled 16-mer/17-mer excised from the 24-mer and 30-mer in the presence of ATP was not processed further (by cleavage 5–6 nucleotides [nt] from its 5′ end) once all the input substrate had been consumed.
The conclusion that AdnAB measures the distance from the 5′ end to its cleavage site was fortified by an experiment using a 25-mer DNA oligonucleotide (of the same sequence as the 24-mer used previously) that was labeled uniquely at its 3′ terminus with 32P-dAMP (Fig. 6). The time course of cleavage in the absence of ATP revealed the early appearance of products cleaved 5 and 6 nt from the 5′ end (at the same sites cleaved in the 5′-labeled substrate) and the subsequent accumulation of a predominant 16-mer product (Fig. 6). We surmise that the use of a 3′-labeled substrate allowed us to see AdnAB take two sequential “bites” from the 5′ direction; only the first bite was detectable with a 5′-labeled DNA. The electrophoretic mobility of the 3′ end-labeled 16-mer cleavage product was a half-nucleotide step faster than that of a 16-mer 5′-OH-terminated, 3′-[32P]dAMP-labeled oligonucleotide of identical 3′-terminal sequence as the 25-mer substrate (data not shown), signifying that cleavage by AdnAB at an internal phosphodiester generates a 5′-PO4 end. The addition of ATP stimulated the rate of consumption of the 3′-labeled 25-mer by a factor of 5–10 and triggered a switch in the product distribution to a predominant 32P-labeled 9-mer (Fig. 6), which arose by incision at the same site that gave rise to the 5′-labeled 16-mer (see above). ATP also promoted the late appearance of a 12-mer/13-mer doublet that we construe to represent a third incremental bite from the 5′ end, owing to the overall stimulation of nuclease activity. Just a trace amount of the 13-mer species was detected at late times in the reaction lacking ATP.
Figure 6.
Cleavage of 3′ end-labeled ssDNA. Reaction mixtures (70 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 3′ 32P-labeled 25-mer DNA substrate (depicted at the bottom), either no nucleotide or 1 mM ATP as indicated, and AdnAB (60 ng of AdnB) were incubated at 37°C. Aliquots (10 μL) were withdrawn at the times specified and quenched with formamide/EDTA. The reaction products were analyzed by Urea-PAGE and visualized by autoradiography. The principal sites of AdnAB incision of the 25-mer DNA in the absence of ATP are indicated by arrows above the sequence; the cleavage site induced by ATP is indicated below the sequence.
Evidence for dual nuclease activities
By varying the ssDNase reaction conditions, we accrued evidence that the ATP-independent and ATP-dependent DNA cleavage activities might reflect a dual nuclease organization of the AdnAB enzyme. Whereas both activities required a divalent cation cofactor, they had different magnesium optima. The ATP-dependent cleavages were evident in a narrow window between 1 and 2 mM MgCl2 and were suppressed at 5–10 mM MgCl2 (Fig. 5D). In contrast, ATP-independent cleavage was optimal over a broader range from 1 to 10 mM MgCl2 (Fig. 5D). A survey of various divalent cations at 2 mM concentration indicated that magnesium, manganese, and cobalt were able to activate the AdnAB nuclease, whereas cadmium, calcium, copper, zinc, and nickel were not (Supplemental Fig. S4A). Induction of ATP-dependent cleavages was limited to a narrow window of 0.5 mM MnCl2, yet ATP-independent cleavages occurred over a broad range from 0.5 to 10 mM MnCl2 (Supplemental Fig. S4C). A metal-mixing experiment showed that copper and zinc abolished the magnesium-dependent ssDNA activity (Supplemental Fig. S4B).
The two cleavage modes also had different pH optima, whereby the ATP-dependent cleavages were favored at pH 5.5–7.5 while the ATP-independent scissions were optimal at pH 8.0–9.5 (Fig. 5E). The disparate requirements for the ATP-independent and ATP-triggered cleavages hinted that they might be catalyzed by different active sites.
The AdnB nuclease domain catalyzes the ATP-triggered cleavage of ssDNA
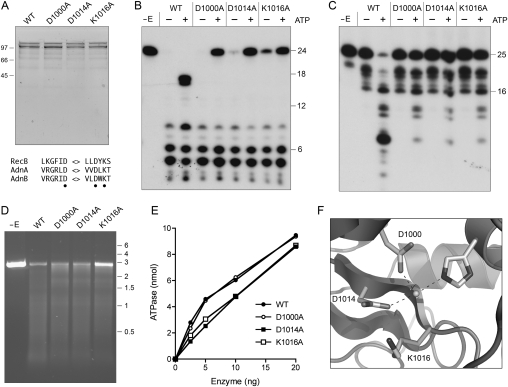
The C-terminal modules of AdnA and AdnB both have the signature active site motifs of the RecB nuclease domain (Fig. 7A; Supplemental Fig. S1). The RecB nuclease is demarcated in the RecBCD crystal structure by a metal-binding site composed of two aspartates and a histidine—corresponding to Asp1000, Asp1014, and His928 in AdnB and Asp920, Asp934, and His833 in AdnA—and a conserved lysine (Lys1016 in AdnB and Lys936 in AdnA) that is likely to coordinate the scissile phosphodiester (Fig. 7F; Singleton et al. 2004). To assess the contributions of the AdnB nuclease domain, we performed an alanine scan of three of the putative active site residues. AdnB mutants D1000A, D1014A, and K1016A were coexpressed in E. coli with wild-type AdnA. SDS-PAGE analysis of the peak glycerol gradient fractions corresponding to the AdnAB heterodimer revealed that the mutated and wild-type preparations were of similar purity and polypeptide composition (Fig. 7A). The D1000A, D1014A, and K1016A mutations did not adversely affect the DNA-dependent ATPase activity of AdnAB (Fig. 7E). The instructive findings were that the D1000A, D1014A, and K1016A mutations selectively ablated the ATP-dependent cleavages of the 5′ 32P-labeled 24-mer DNA, while having little or no impact on the ATP-independent cleavages at sites closer to the 5′ terminus (Fig. 7B). Indeed, the mutant proteins were actually less effective in DNA incision in the presence of ATP (as gauged by consumption of the 24-mer) than in the absence of nucleotide (Fig. 7B), suggesting that the AdnB subunit nuclease is responsible for the ATP-dependent cleavages and is the downstream “receptor” of a putative allosteric effect of ATP hydrolysis on overall nuclease activity. These results were seconded by analysis of mutational effects on cleavage of a 3′ 32P-labeled 25-mer DNA (Fig. 7C), which showed that the D1000A, D1014A, and K1016A mutations suppressed ATP-dependent formation of the 32P-labeled 9-mer product. Notwithstanding these effects on ssDNase activity, the mutant heterodimers retained activity in ATP-dependent exonucleolytic resection of linear plasmid DNA (Fig. 7D), with only the K1016A enzyme having a modest effect on the extent of consumption of the input substrate. Our inference is that the AdnA subunit can provide nuclease activity during end resection when the AdnB nuclease is disabled.
Figure 7.
Mutations of the AdnB nuclease domain. (A) Purification. Aliquots of the peak glycerol gradient fractions of heterodimeric wild-type AdnAB and the indicated AdnB nuclease domain mutants (containing 0.5 μg of AdnB polypeptide) were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the left. Conserved RecB nuclease motifs are shown at the bottom; the residues targeted for alanine scanning are indicated by ●. (B) ssDNase with 5′-labeled DNA. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA substrate, 1 mM ATP (where indicated by +), and wild-type or mutant AdnAB (10 ng of AdnB) were incubated for 5 min at 37°C. (C) ssDNase with 3′-labeled DNA. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 3′ 32P-labeled 25-mer DNA substrate, 1 mM ATP (where indicated by +), and wild-type or mutant AdnAB (5 ng of AdnB) were incubated for 5 min at 37°C. (D) dsDNA exonuclease. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng of linear SmaI-cut pUC19, and wild-type or mutant AdnAB as specified (2.5 ng of AdnB) were incubated for 5 min at 37°C. The reaction products were analyzed by native agarose gel electrophoresis in the presence of ethidium bromide. (E) ATPase. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 1 μg of salmon sperm DNA, 1 mM [γ32P]ATP, and wild-type or mutant AdnAB as specified were incubated for 5 min at 37°C. 32Pi release is plotted as a function of input enzyme (nanograms of AdnB polypeptide). (F) A view of the RecB nuclease active site (from PDB 1W36) depicting the catalytic metal ion coordinated by two conserved aspartates (Asp1000 and Asp1014 in AdnB) and a histidine. A conserved lysine (Lys1016 in AdnB) likely coordinates the scissile phosphodiester.
The AdnA nuclease domain is responsible for ATP-independent incision of ssDNA
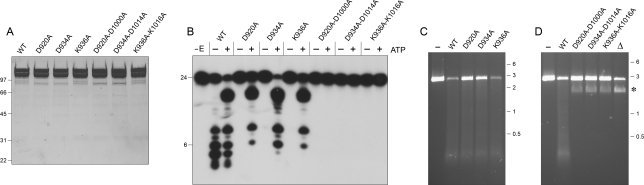
We extended the alanine scan to the three putative active site residues of the AdnA nuclease domain. AdnA mutants D920A, D934A, and K936A were coexpressed in E. coli with wild-type AdnB and purified in parallel with the wild-type heterodimer (Fig. 8A). The D920A, D934A, and K936A mutations selectively ablated the ATP-independent ssDNase activity, without affecting the ATP-dependent scission of the 5′ 32P-labeled 24-mer DNA substrate (Fig. 8B). These results, together with those in the preceding section, establish a division of labor whereby the AdnA nuclease domain cleaves close to the 5′ end in the absence of ATP and the AdnB nuclease domain responds to ATP and cleaves ssDNA at distal sites.
Figure 8.
Mutations of the AdnA nuclease domain. (A) Purification. Aliquots of the peak glycerol gradient fractions of heterodimeric wild-type AdnAB, the indicated AdnA nuclease domain mutants, and the indicated AdnA–AdnB double-nuclease mutants (containing 1.8 μg of AdnB polypeptide) were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the left. (B) ssDNase. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA substrate, 1 mM ATP (where indicated by +), and wild-type or mutant AdnAB (40 ng of AdnB) were incubated for 5 min at 37°C. The products were analyzed by denaturing PAGE and visualized by autoradiography. (C) dsDNA exonuclease. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 2 mM MgCl2, 1 mM ATP, 200 ng of linear BamHI-cut pUC19, and wild-type or mutant AdnAB as specified (100 ng of AdnB) were incubated for 5 min at 37°C. AdnAB was omitted from the control sample in lane −. The reaction products were analyzed by native agarose gel electrophoresis in the presence of ethidium bromide. (D) Helicase activity of nuclease-defective double mutants. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 2 mM MgCl2, 1 mM ATP, 200 ng of linear BamHI-cut pUC19, and wild-type or mutant AdnAB as specified (100 ng of AdnB) were incubated for 5 min at 37°C. AdnAB was omitted from the control sample in lane −. Lane △ contains 200 ng of linear pUC19 DNA that was heated for 5 min at 95°C and quenched on ice prior to electrophoresis. The species corresponding to single-stranded linear pUC19 DNA is indicated by the asterisk.
The AdnAB heterodimers with mutated AdnA nucleases had residual activity in ATP-dependent DSB end resection (Fig. 8C), as did the enzymes with AdnB nuclease mutations (vide supra), signifying that the dsDNA exonucleolytic processing can be performed by either the AdnA or the AdnB nuclease module. To consolidate these points, we purified AdnAB heterodimers in which the equivalent active site aspartates and lysines were substituted by alanine in both subunits (Fig. 8A). The AdnA(D920A)–AdnB(D1000A), AdnA(D934A)–AdnB(D1014A), and AdnA(K936A)–AdnB(K1016A) proteins were unreactive with 5′ 32P-labeled 24-mer ssDNA in the presence or absence of ATP (Fig. 8B) and were unreactive in ATP-dependent DSB resection (Fig. 8C). The nuclease-defective double mutants retained DNA-dependent ATPase activity (data not shown) and were capable of completely unwinding a fraction of the input linear duplex substrate to form a discrete faster migrating product that comigrated with heat-denatured single-stranded pUC19 DNA (Fig. 8C). Thus, ablation of the AdnAB nuclease allowed us to appreciate the intrinsic AdnAB helicase activity.
Disparate effects of mutations in the AdnA and AdnB motor domains on ATP hydrolysis
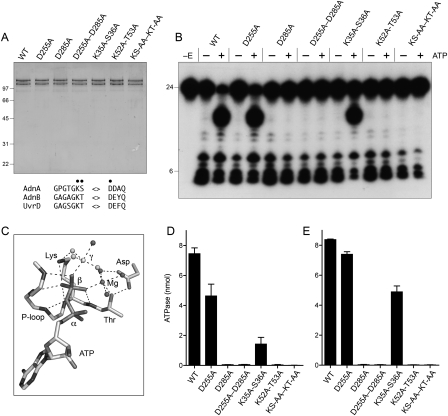
The N-terminal domains of AdnA and AdnB both have the signature active site motifs I and II of the superfamily I helicases, exemplified by E. coli UvrD (Fig. 9A; Supplemental Fig. S1). Motif I of AdnA (GPGTGKS) and AdnB (GAGAGKT) are the equivalent of the “P-loop” that binds the ATP β and γ phosphates in UvrD (Fig. 9C). The main chain amide nitrogens of the P-loop form an “oxyanion hole” for the ATP β phosphate. The conserved motif I lysine side chain contacts the nonbridging oxygens of the β and γ phosphates and stabilizes the pentacoordinate phosphorane transition state of the γ phosphate (Fig. 9B). An essential divalent cation contacts a different pair of nonbridging β and γ phosphate oxygens and stabilizes the transition state. The motif I threonine/serine side chain and the motif II aspartate (255 DDAH in AdnA; 285 DEYQ in AdnB) are the key enzymic components of the metal-binding site (Fig. 9C). To assess the contributions of the individual phosphohydrolase domains, we introduced an alanine in lieu of the metal-binding motif II aspartates of AdnA (D255A) or AdnB (D285A) and purified them as heterodimers with their respective wild-type partner subunits (Fig. 9A). We also purified an AdnA(D255A)–AdnB(D285A) heterodimer in which both motif II elements were mutated (Fig. 9A). The wild-type and mutated AdnAB proteins were assayed in parallel for ATP hydrolysis in the presence of salmon sperm DNA (Fig. 9D). The striking finding was that ATPase activity was abolished by a single motif II mutation of the AdnB subunit. In contrast, the equivalent change in the AdnA subunit caused only a 38% reduction in the extent of ATP hydrolysis compared with the wild-type enzyme (Fig. 9D).
Figure 9.
Mutations of the AdnAB phosphohydrolase motifs. (A) Purification. Aliquots of the peak glycerol gradient fractions of heterodimeric wild-type AdnAB, the indicated AdnA or AdnB phosphohydrolase motif mutants, and the indicated AdnA–AdnB double mutants (containing 0.53 μg of AdnB polypeptide) were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the left. Phosphohydrolase motifs I and II are shown at the bottom; the residues targeted for alanine scanning are indicated by ●. (B) ssDNase with 5′-labeled DNA. Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 5′ 32P-labeled 24-mer DNA substrate, 1 mM ATP (where indicated by +), and wild-type or mutant AdnAB (21 ng of AdnB polypeptide) were incubated for 15 min at 37°C. (C) Structure of the ATPase activity site of DNA-bound E. coli UvrD (PDB 2IS6) as a transition-state mimetic in complex with Mg2+, ADP, and a trigonal planar MgF3 with an apical water nucleophile (analogous to the pentacoordinate γ phosphate of the transition state). The atomic contacts of the P-loop (motif I) and the motif II aspartate are denoted by dashed lines. (D) ATPase reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 0.5 mM DTT, 1 μg of salmon sperm DNA, 1 mM [α32P]ATP, and wild-type or mutant AdnAB as specified (10 ng of AdnB polypeptide) were incubated for 15 min at 37°C. The extent of 32P-ADP formation was determined by TLC. Data are the average of three separate experiments ± SEM. (E) ATPase reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 0.5 mM DTT, 10 μM 24-mer ssDNA oligonucleotide, 1 mM [α32P]ATP, and wild-type or mutant AdnAB as specified (21 ng of AdnB polypeptide) were incubated for 15 min at 37°C. The extent of 32P-ADP formation was determined by TLC. Data are the average of four separate experiments ± SEM.
Similar results pertained when we introduced pairs of alanine mutations in motif I—either at the 35KS36 dipeptide of AdnA or the 52KT53 dipeptide of AdnB or simultaneously in both subunits. None of the mutations affected heterodimerization of the recombinant subunits, as gauged by the polypeptide compositions of the peak glycerol gradient fractions (Fig. 9A). Here again, we found that the motif I mutations in AdnB ablated ATP hydrolysis in the presence of salmon sperm DNA (Fig. 9D). The equivalent motif I mutations in AdnA reduced activity to one-fifth of the wild-type level (Fig. 9D). These results signify that the phosphohydrolase domain of AdnB is essential for all of the observed ATP hydrolysis of the AdnAB heterodimer. That AdnA mutations elicited modest reductions in activity could indicate that it makes a contribution to overall activity, but depends on AdnB to trigger its phosphohydrolase function.
We considered the prospect that AdnA requires AdnB to initiate unwinding and cleavage of the native salmon sperm DNA cofactor in order to convert it into a form that might trigger ATP hydrolysis by AdnA. Therefore, we performed the ATPase assays using a single-stranded 24-mer oligonucleotide as the nucleic acid cofactor (Fig. 9E). The results echoed the findings with salmon sperm DNA; i.e., the motif I and II mutations in AdnB ablated ssDNA-dependent ATP hydrolysis, whereas the motif I K35A–S36A mutation in AdnA merely reduced activity by 40% compared with wild-type AdnAB. These results show that the AdnB subunit is the principal DNA-dependent ATPase of the heterodimeric motor.
To address whether the AdnB phosphohydrolase activity requires the AdnA subunit, we produced AdnB singly in E. coli as a His10Smt3–AdnB fusion, isolated the recombinant protein from a soluble bacterial extract, removed the tag, and then analyzed the tag-free AdnB protein by zonal velocity sedimentation in the presence or absence of internal standards. AdnB sedimented as a single monomeric component on the “heavy” side of the BSA peak (Supplemental Fig. S5A). When analyzed by itself, AdnB sedimented identically and cosedimented with a comparatively feeble ATPase activity (Supplemental Fig. S5C). Side-by-side comparisons of the rates of ATP hydrolysis by equivalent amounts of AdnAB heterodimer and AdnB monomer under the standard reaction conditions employed to study AdnAB indicated that AdnB was 1800-fold less active than AdnAB (data not shown). We conclude that AdnB requires AdnA to manifest its motor activity. When the AdnA subunit was produced singly and purified in parallel, we observed that it sedimented diffusely (Supplemental Fig. S5B), signifying a tendency to aggregate or form multimers. (We did not detect an ATPase associated with the isolated AdnA subunit, and initial attempts to reconstitute the active heterodimer by simply mixing the isolated recombinant subunits have not been fruitful.)
Effects of mutations in the motor domains on AdnAB nuclease activity
We exploited the motor domain mutants to query whether the signal for the ATP-regulated single-strand nuclease activity of the AdnB subunit of AdnAB is propagated in cis by AdnB itself or in trans by the AdnA subunit. The AdnA motif I and motif II mutations did not affect the ATP-induced scission of the 24-mer ssDNA substrate at sites 16–17 nt from the 5′ end (Fig. 9B, cf. wild type [WT] and D225A and K35A–S36A). In contrast, the equivalent mutations in the ATPase active site of AdnB (D285A and K52A–T53A) abolished the ATP-triggered cleavages (Fig. 9B). We conclude that ATP hydrolysis by the AdnB motor domain activates the AdnB nuclease in cis. Similar results were obtained when we assayed mutational effects on GTP-dependent ssDNAase activity (data not shown), thereby vitiating the idea that different nucleotides might regulate the AdnB nuclease through different phosphohydrolase modules.
The effects of the motor domain mutations on the dsDNA end resection activity of AdnAB paralleled the effects on ATP hydrolysis. AdnAB digestion of linear plasmid DNA was ablated by the motif I and II mutations in the AdnB subunit, but was either unaffected or partially reduced by the corresponding mutations in the AdnA subunit (data not shown).
Mycobacterial Ku blocks DSB end resection by AdnAB
The majority of Ku-dependent NHEJ events at blunt and 5′ overhang DSBs in M. smegmatis occur without nucleolytic resection (Aniukwu et al. 2008). Genetic evidence implicates the DNA end-binding protein Ku and the dedicated NHEJ ligase LigD in protecting exposed DSB ends from deletion in vivo (Aniukwu et al. 2008). Here we found that purified Mycobacterium Ku inhibited AdnAB-catalyzed degradation of SmaI-cut pUC19 DNA (Fig. 3D), suggesting that Ku and AdnAB compete for access to the DSB. A plausible scenario is that when Ku wins this contest in vivo, LigD is recruited to the DSB and NHEJ is achieved, either faithfully or with LigD(POL)-catalyzed nucleotide additions at the repair junction (Aniukwu et al. 2008). However, if a motor–nuclease gains first access to the DSB, one or both ends will be trimmed and any subsequent NHEJ will score as a deletion (Aniukwu et al. 2008). The Ku protection of DSB ends from digestion by mycobacterial AdnAB echoes findings in eukarya, whereby yeast Ku is implicated in protecting telomere ends from digestion by the yeast nuclease Exo1 (Maringele and Lydall 2002; Bertuch and Lundblad 2004).
Discussion
Mycobacterial AdnAB is the founding member of a new family of DSB-resecting complexes composed of two motor-like domains (at least one of which has vigorous activity) and two nuclease modules. AdnAB is thereby distinguished from RecBCD, which has two motor domains and one nuclease, and AddAB, which has one motor domain and two nucleases. There are AdnAB homologs encoded by adjacent genes in multiple species of Mycobacterium, including M. smegmatis, M. tuberculosis, M. avium, M. bovis, M. abscessus, M. ulcerans, M. marinum, M. gilvum, and M. vanbaalenii (Supplemental Table 1). A remnant of the adnAB locus is present in the M. leprae genome, albeit peppered with numerous in-frame stop codons (GenBank locus AL583919). AdnAB homologs are also encoded by adjacent genes in species from many other bacterial genera: Streptomyces, Corynebacterium, Nocardia, Thermobifida, Acidothermus, Rhodococcus, Micrococcus, Frankia, Kineococcus, Janibacter, Athrobacter, Salinospora, Leifsonia, Saccharopolyspora, Brevibacterium, Kocuria, Clavibacter, Propionobacterium, and Tropheryma (Supplemental Table 1). All of the AdnAB-encoding bacteria belong to same taxon. Specifically, they are members of the order Actinomycetales of the Actinobacteridae subclass of the Actinobacteria class of the phylum Actinobacteria.
AdnAB has some properties that are typical of other NTP-driven motor–nucleases. For example, the AdnAB NTPase is quiescent in the absence of DNA and is triggered specifically by dsDNA with free ends. The energy of ATP hydrolysis is coupled to DSB end resection by the nuclease modules. However, ATP consumption triggered by free dsDNA ends is not affected by mutational inactivation of one or both of the nucleases. Indeed, the helicase activity of the AdnAB motor is revealed when the DNA cleavage function is ablated.
The ATP responsiveness and end-measuring capacity of the AdnAB ssDNase and the division of labor among the nuclease domains are what set it apart from the ssDNase activities of RecBCD and AddAB. The RecBCD ssDNase was recognized early (Wright et al. 1971; Goldmark and Linn 1972) as an endonuclease that degraded high-molecular-weight ssDNA to short oligonucleotide products with 5′-PO4 ends. When acting on a 32P end-labeled 26-mer oligonucleotide, RecBCD sequentially excises oligonucleotide fragments from the 3′ end of the ssDNA substrate in the presence of ATP, with the first “bite” liberating a 3′-terminal trinucleotide or tetranucleotide; digestion then continues until a 5′-terminal tetranucleotide is formed (Chen et al. 1998). Omission of ATP slows the rate of the RecBCD ssDNase by a factor of 3000 and shifts the initial sites of cleavage to near the 5′ terminus of the ssDNA olignucleotide (Chen et al. 1998). A rapid kinetic study of RecBCD digestion of a 40-mer ssDNA substrate verified that cleavage initiates from the 3′ end and proceeds inward in sequential oligonucleotide-releasing steps (Ghatak and Julin 2006). The 3′ directionality of the RecBCD ssDNase reaction is an intrinsic property of the 30-kDa C-terminal nuclease domain of the RecB subsunit (Sun et al. 2006).
This behavior is diametrically opposite that of AdnAB. First, AdnAB has a substantial ssDNase activity in the absence of ATP (apparently catalyzed by the AdnA subunit nuclease), and is stimulated only about fivefold in the presence of hydrolysable ATP. Second, AdnAB measures the distance from the free 5′ end to dictate the sites of incision of ssDNA, which are predominantly 5 or 6 nt from the 5′ end in the absence of ATP (and are unperturbed by lengthening the ssDNA substrate in the 3′ direction). Third, the molecular ruler of AdnAB is regulated by ATP, resulting in a displacement of the cleavage sites toward more distal positions, 16–17 nt from the 5′ terminus. ATP-dependent cleavage is apparently the province of the AdnB subunit nuclease and the AdnB motor domain. Fourth, a 12-mer is resistant to digestion by AdnAB, implying that it requires a longer segment of DNA 3′ of the scissile phosphodiester than does RecBCD.
Bacillus AddAB resembles mycobacterial AdnAB with respect to its heterodimeric quaternary structure and the presence of nuclease domains at the C termini of both subunits. The AddA subunit possesses the single motor activity of Bacillus AddAB and is, to our inspection, more similar in its organization to mycobacterial AdnB (the dominant motor in AdnAB) than to AdnA. Mycobacterial AdnA's nuclease domain is reminiscent of the Bacillus AddB nuclease module with respect to conservation of a cluster of three cysteine residues at the C terminus (CxxCxxxxxC) that comprises part of an iron–sulfur cluster (Yeeles et al. 2009). The overall ssDNase activity of Bacillus AddAB is stimulated by ATP (Haijema et al. 1996b). From the effects on mutations in the nuclease active sites, Yeeles and Dillingham (2007) concluded that AddA nuclease is partially active in ATP-dependent DSB resection when the AddB nuclease is disabled, and vice versa, but that the nuclease activity is ablated when both subunits are mutated. We noted similar effects of nuclease active site mutations in the individual AdnA and AdnB subunits, versus both subunits, on ATP-dependent DSB resection by mycobacterial AdnAB. Using tailed duplex substrates, Yeeles and Dillingham (2007) show that AddA nuclease is responsible for 3′-to-5′ cleavage of one of the strands displaced by the helicase motor, while AddB has a 5′-to-3′ nuclease that degrades the other displaced strand. Although the substrates used to study Bacillus AddAB nuclease are different than those used presently, we can say that there appears to be no 3′-to-5′ directionality to the nuclease of the mycobacterial AdnAB complex, at least with ssDNA substrates.
It is remarkable that AdnB alone provides the principal ATPase motor function of the AdnAB complex, which is abolished by mutations in the predicted phosphohydrolase active site of the AdnB subunit, but impacted only modestly by synonymous mutations in AdnA. It is conceivable that either (1) ATP hydrolysis by AdnB is required to trigger the AdnA ATPase, or (2) ATP binding by AdnA (with or without hydrolysis) serves to up-regulate the motor domain of AdnB. The latter model could account for the more significant impact of the motif I double mutation in AdnA on overall ATPase activity versus the motif II change, because K35A–T36A is expected to affect ATP binding plus metal coordination, whereas D255A is expected to affect metal coordination (and hence ATP hydrolysis), but not ATP binding. The apparent inability of AdnA to catalyze DNA-dependent ATP hydrolysis when AdnB is crippled was surprising to us, given that AdnA (like AdnB) seems to have most, if not all, of the helicase motifs identified in UvrD and other homologous superfamily I helicases. Indeed, AdnB has the full set of amino acids that comprise the phosphohydrolase active site of UvrD, as surmised from the crystal structure of the DNA-bound UvrD1–ADP–MgF3 transition state mimetic (Lee and Yang 2006). However, in hindsight, closer inspection and comparison of the aligned AdnA, AdnB, and UvrD sequences (Supplemental Fig. S1) hint that AdnA lacks several of the side chains that bind the DNA single-strand that activates the ATPase motor, including: (1) the motif Ia phenylalanine, which is glycine in AdnA; (2) the motif Id phenylalanine, replaced by alanine in AdnA; (3) the motif IVa asparagine, which is valine in AdnA; and (4) the motif VIa tyrosine/tryptophan, replaced by threonine in AdnA. Thus, AdnA might have a latent ATPase that responds to something other than generic ssDNA.
The identification and biochemical characterization of AdnAB will stimulate genetic studies of its role in recombination and repair in mycobacteria and other Actinomycetales species. The AdnAB complex is apparently not required for mycobacterial growth, insofar as a viable transposon insertion that disrupts the M. tuberculosis Rv3202c ORF (adnA) has been reported (Sassetti et al. 2003). Given that neither RecBCD nor AdnAB is essential in mycobacteria, and that ΔrecBCD has no overt DNA damage sensitivity or NHEJ phenotype (Stephanou et al. 2007), it appears that these two DSB end processing machines might be functionally redundant. Indeed, a significant fraction of the AdnAB-encoding species also encode a RecBCD homolog (Supplemental Table S1). These include members of the Mycobacterium, Rhodococcus, Nocardia, Kineococcus, and Propionibacterium genera. This phylogenetic distribution suggests that AdnAB might have evolved in an ancestral RecBCD+ actinobacterium; e.g., by duplications of a gene encoding a RecB-like helicase–nuclease. The RecBCD and AdnAB enzymes coexist in some present-day descendents, while other Actinomycetales have retained AdnAB and lost RecBCD. The coexistence of two distinct DSB-resecting apparati in mycobacteria echoes the recently elucidated scenario in eukarya, whereby the yeast Saccharomyces cerevisiae has multiple, functionally overlapping enzyme systems with helicase and nuclease components that can accomplish this key task (Mimitou and Symington 2008; Zhu et al. 2008). Such functional redundancy had been noted previously in B. subtilis, which relies on either the AddAB motor–nuclease complex or separate RecQ helicase and RecJ nuclease enzymes for end processing (Sanchez et al. 2006).
Materials and methods
AdnAB expression vectors
The ORF encoding M. smegmatis AdnA (MSMEG1941; http://www.tigr.org) was PCR-amplified from genomic DNA with primers that introduced a BglII site 5′ of the AUG start codon and a HindIII site 3′ of the stop codon, respectively. The amplified product, which contained an internal BglII site in the ORF, was digested with BglII. The 2122-base-pair (bp) BglII fragment encoding the AdnA N-terminal fragment was gel-purified and inserted into the BamHI site of pET28-HisSmt3. The resulting plasmid was digested with KpnI at nucleotide 873 of the AdnA ORF and HindIII (in the vector 3′ of the insert). Into this linearized plasmid DNA, we inserted a KpnI–HindIII fragment of the PCR-amplified adnA ORF that encodes the C-terminal portion of the AdnA polypeptide. The resulting pET-HisSmt3-AdnA expression plasmid encodes the full-length AdnA polypeptide fused to an N-terminal His10-Smt3 tag. The ORF encoding M. smegmatis AdnB (MSMEG1943; http://www.tigr.org) was PCR-amplified from genomic DNA with primers that introduced an NdeI site at the start codon (changed to AUG from GUG) and a BglII site 3′ of the stop codon. The PCR product was digested with NdeI and BglII and inserted between the NdeI and BamHI sites of pET3c. The resulting pET-AdnB plasmid encodes full-length AdnB without a tag. Alanine substitution mutations were introduced into the adnA and adnB genes by two-stage overlap extension PCR using mutagenic primers. The inserts of the wild-type and mutated AdnA and AdnB expression plasmids were sequenced completely to ensure that no unwanted changes were introduced during amplification and cloning.
Recombinant AdnAB
A mixture of pET-HisSmt3-AdnA and pET-AdnB DNA was electroporated into E. coli BL21(DE3) cells; cotranformants were selected on LB agar plates containing ampicillin and kanamycin. A 1-L culture amplified from a single colony was grown at 37°C in Luria-Bertani medium containing 0.1 mg/mL ampicillin and 50 μg/mL kanamycin until the A 600 reached 0.6. The culture was chilled on ice for 45 min and the expression of recombinant proteins was induced by adjusting the culture to 0.2 mM isopropyl-β-D-thiogalactopyranoside followed by incubation for 16 h at 17°C with constant shaking. Cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent steps were performed at 4°C. The thawed bacterial cell pellet was suspended in 50 mL of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 0.25 M NaCl, and 10% sucrose. Cell lysis was achieved by adding lysozyme and Triton X-100 to final concentrations of 1 mg/mL and 0.1% respectively. The suspensions were incubated for 60 min until lysis was complete and then sonicated to reduce the viscosity. Insoluble material was removed by centrifugation and the supernatant was applied to a 5-mL column of nickel-nitriloacetic acid-agarose (Qiagen) that had been pre-equilibrated with lysis buffer. The column was washed with buffer A (50 mM Tris-HCl at pH 8.0, 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol) and then eluted stepwise with buffer A containing 50, 100, 200, 500, and 1000 mM imidazole. The polypeptide compositions of the eluate fractions were monitored by SDS-PAGE. The recombinant Adn protein was recovered predominantly in the 100 and 200 mM imidazole eluates, which were pooled and dialyzed against buffer A. The dialysate was applied to a 3-mL DEAE-Sephacel column that had been equilibrated with buffer A. The Adn protein was recovered in the flowthrough fraction. The His10Smt3 tag was removed from AdnA by treatment with the Smt3-specific protease Ulp1 (at a Adn:Ulp1 ratio of ∼500:1) for ∼10 h (overnight) at 4°C. The Adn proteins were separated from the tag by passage over a second nickel-agarose column equilibrated in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.05% Triton X-100, and 10% glycerol. The AdnA and AdnB polypeptides were recovered in the flowthrough fraction. Protein concentrations were determined by using the Bio-Rad dye reagent with BSA as the standard. The yield of wild-type AdnAB at the second Ni-agarose step was 8 mg. The AdnAB nuclease and phosphohydrolase mutants were purified similarly.
Zonal velocity sedimentation
Aliquots (100 μg) of the second Ni-agarose flowthrough fraction of AdnAB—with or without marker proteins catalase (50 μg), ovalbumin (50 μg), and cytochrome c (100 μg)—were applied to 4.8-mL 15%–30% glycerol gradients containing 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 1 mM DTT, and 0.05% Triton X-100. The gradients were centrifuged for 13 h at 4°C in a Beckman SW55Ti rotor at 50,000 rpm. Fractions (∼200 μL) were collected from the bottom of the tubes and analyzed by SDS-PAGE. Fractions containing the AdnAB heterodimer (Fig. 2B) were used for functional characterization of AdnAB enzymatic activities. The protein concentrations of the peak AdnAB heterodimer fractions were determined by SDS-PAGE analysis in parallel with increasing known amounts of BSA. The Coomassie Blue-stained gel was scanned and the intensity of the AdnB and BSA polypeptides was quantified with Imagequant software. The concentration of AdnB was calculated by interpolation to the BSA standard curve. The figure legends specify the amounts of added enzyme (in nanograms) with respect to the AdnB subunit.
Nucleoside triphosphatase assay
Standard reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 1 mM [γ32P]ATP or [α32P]ATP (Perkin-Elmer Life Sciences), 1 μg of salmon sperm DNA, and AdnAB were incubated for 5 min at 37°C. The reactions were quenched with 2 μL of 5 M formic acid. Aliquots (2 μL) of each mixture were spotted on a PEI-cellulose TLC plate, which was developed with either 0.45 M ammonium sulfate or 0.5 M LiCl, 1 M formic acid. The radiolabeled material was visualized by autoradiography. The extents of 32Pi or 32P-ADP formation were quantified by scanning the TLC plate with a Fujix BAS2500 imager.
Assay of dsDNA nuclease activity
Standard reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng of linear pUC19 DNA, and AdnAB were incubated for 5 min at 37°C. The reactions were terminated by adding 5 μL of a solution containing 50% glycerol, 50 mM EDTA, and 0.6% SDS. The samples were analyzed by electrophoresis through a horizontal 0.7% agarose gel containing 45 mM Tris-borate, 1.2 mM EDTA, and 0.5 μg/mL ethidium bromide.
Assay of ssDNA nuclease activity
5′ 32P-labeled DNA substrates were prepared by phosphorylation of synthetic 5′-OH 30-mer, 24-mer, 18-mer, and 12-mer DNA oligonucleotides with T4 polynucleotide kinase and [γ32P]ATP. The radiolabeled DNAs were purified by electrophoresis through a native 18% polyacrylamide gel. The 3′ 32P-labeled 25-mer DNA substrate was prepared by annealing the 5′-OH 24-mer DNA oligonucleotide primer 5′-GCCCTGCTGCCGACCAACGAAGGT-3′ to a twofold excess of a complementary template oligonucleotide 3′-GGCTGGTTGCTTCCATAA-5′. The primer strand was extended by DNA polymerase I Klenow fragment in the presence of [α32P]dATP. The fill-in reaction was terminated with formamide/EDTA. The mixture was heat-denatured and the 3′ radiolabeled 25-mer strand was purified by electrophoresis through a native 18% polyacrylamide gel.
Nuclease reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.5 mM DTT, 0.1 μM 32P end-labeled ssDNA substrate, and ATP and AdnAB as specified were incubated for 5 min at 37°C. The reactions were terminated by adjustment to 30% formamide and 15 mM EDTA. The products were analyzed by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate and 1.2 mM EDTA and then visualized by autoradiography.
Acknowledgments
This research was supported by NIH grant AI64693.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1805709.
Supplemental material is available at http://www.genesdev.org.
References
- Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase–nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes & Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–1659. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases for Gram-positive bacteria: Insights into the initiation of DNA recombination. Mol Microbiol. 2002;43:823–834. doi: 10.1046/j.1365-2958.2002.02785.x. [DOI] [PubMed] [Google Scholar]
- Chen HW, Radle DE, Gabbidon M, Julin DA. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J Mol Biol. 1998;278:89–104. doi: 10.1006/jmbi.1998.1694. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- Ghatak A, Julin DA. Kinetics of ATP-stimulated nuclease activity of the Escherichia coli RecBCD enzyme. J Mol Biol. 2006;361:954–968. doi: 10.1016/j.jmb.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Goldmark PJ, Linn S. Purification and properties of the recBC DNase of Escherichia coli K12. J Biol Chem. 1972;247:1849–1860. [PubMed] [Google Scholar]
- Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J Biol Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: A low fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- Haijema BJ, Venema G, Kooistra J. The C terminus of the AddA subunit of the Bacillus subtilis ATP-dependent DNase is required for the ATP-dependent exonuclease activity but not for the helicase activity. J Bacteriol. 1996a;178:5086–5091. doi: 10.1128/jb.178.17.5086-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Meima R, Kooistra J, Venema G. Effects of lysine-to-glycine mutations in the ATP-binding consensus sequences in the AddA and AddB subunits on the Bacillus subtilis AddAB enzyme activities. J Bacteriol. 1996b;178:5130–5137. doi: 10.1128/jb.178.17.5130-5137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes & Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Tonkin LM, Daley JM, Palmbos PL, Green AJ, Velting TL, Brzostek A, Korycka-Machala M, Cresawn S, Dziadek J, et al. Mycobacteriophage exploit NHEJ to facilitate genome circularization. Mol Cell. 2006;23:743–748. doi: 10.1016/j.molcel.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Green AJ, Brzostek A, Korycka-Machala M, Dziadek J, Doherty AJ. NHEJ protects mycobacteria in stationary phase against the harmful effects of dessication. DNA Repair (Amst) 2007a;6:1271–1276. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Annu Rev Microbiol. 2007b;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:247–259. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Kidane D, Cozar MC, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis Re4cN to DNA double-strand breaks in the absence of DNA end processing. J Bacteriol. 2006;188:356–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Sinha KM, Stephanou NC, Gao F, Glickman MS, Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J Biol Chem. 2007;282:15114–15125. doi: 10.1074/jbc.M701167200. [DOI] [PubMed] [Google Scholar]
- Sinha KM, Stephanou NC, Unciuleac MC, Glickman MS, Shuman S. Domain requirements for DNA unwinding by mycobacterial UvrD2, an essential DNA helicase. Biochemistry. 2008;47:9355–9364. doi: 10.1021/bi800725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JZ, Julin DA, Hu JS. The nuclease domain of the Escherichia coli RecBCD enzyme catalyzes degradation of linear and circular single-stranded and double-stranded DNA. Biochemistry. 2006;45:131–140. doi: 10.1021/bi051150v. [DOI] [PubMed] [Google Scholar]
- Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structure of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Wright M, Buttin G, Hurwitz J. The isolation and characterization from Escherichia coli of an adenosine triphosphate-dependent deoxyribonuclease directed by recB,C genes. J Biol Chem. 1971;246:6543–6555. [PubMed] [Google Scholar]
- Yeeles JTP, Dillingham MS. A dual-nuclease mechanism for DNA processing by AddAB-type helicase nucleases. J Mol Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- Yeeles JTP, Cammack R, Dillingham MS. An iron–sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases DNA2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]