Abstract
Cells use strategic metabolites to sense the metabolome and accordingly modulate gene expression. Here, we show that the purine and phosphate pathways are positively regulated by the metabolic intermediate AICAR (5′-phosphoribosyl-5-amino-4-imidazole carboxamide). The transcription factor Pho2p is required for up-regulation of all AICAR-responsive genes. Accordingly, the binding of Pho2p to purine and phosphate pathway gene promoters is enhanced upon AICAR accumulation. In vitro, AICAR binds both Pho2p and Pho4p transcription factors and stimulates the interaction between Pho2p and either Bas1p or Pho4p in vivo. In contrast, SAICAR (succinyl-AICAR) only affects Pho2p–Bas1p interaction and specifically up-regulates purine regulon genes. Together, our data show that Bas1p and Pho4p compete for Pho2p binding, hence leading to the concerted regulation of cellular nucleotide synthesis and phosphate consumption.
Keywords: Phosphate utilization, purine pathway, small molecules, transcription factor, yeast
To maintain homeostasis, cells must permanently “estimate” the concentration of critical metabolites and modulate activity of highly interconnected metabolic pathways. Interestingly, recent evidence has pointed to specific metabolites as major regulators of transcription processes. Small molecules can affect chromatin remodeling complexes (Shen et al. 2003; Steger et al. 2003) as well as the ability of transcription factors to activate transcription (Sze et al. 1992; Flynn and Reece 1999). Several yeast transcription factors respond to critical metabolic intermediates, the abundance of which directly reflects the flux through the metabolic pathway (Sellick and Reece 2005). Thereby, the flow in the metabolic pathway can be modulated by regulating the synthesis of all the enzymes of the pathway at the transcriptional level. This role for small molecules in transcriptional control of their own synthesis pathway is now clearly established and in few instances has been reconstituted in vitro (Sellick and Reece 2005).
We found previously that two metabolic intermediates, AICAR (5′-phosphoribosyl-5-amino-4-imidazole carboxamide) and SAICAR (succinyl-AICAR), are involved in the transcriptional regulation of purine pathway genes (ADE genes) (Rebora et al. 2001, 2005). Activation of these purine regulon genes in response to purine limitation require the Bas1p and Pho2p transcription factors (Daignan-Fornier and Fink 1992). Based on genetic evidence, it was proposed that the increase of AICAR and/or SAICAR concentration was a molecular signal promoting Bas1p–Pho2p interaction, and consequently resulted in purine gene up-regulation (Rebora et al. 2001, 2005). Consistently, under purine limitation conditions, Pho2p is recruited to the ADE gene promoters in a Bas1p-dependent manner (Som et al. 2005). Importantly, Pho2p together with Pho4p, another transcription factor, activates the PHO regulon genes in response to phosphate limitation (Lenburg and O'Shea 1996), thus suggesting that regulation of purine and phosphate pathways could be interconnected.
Because small molecules such as AICAR and SAICAR play regulatory roles, their overabundance can be highly detrimental. Abnormal accumulation of AICAR and SAICAR is toxic for yeast cells (Rebora et al. 2005). Importantly, AICAR or SAICAR accumulation in human are associated with two monogenic disorders leading to severe mental retardation and autistic features (Stone et al. 1992; Marie et al. 2004). Recently, treatment of mice with AICAR has revealed a role for this molecule in improving endurance in the absence of training (Narkar et al. 2008). Therefore, AICAR accumulation has pleiotropic effects suggesting that its levels should be highly regulated. We took advantage of yeast genetics to investigate AICAR physiological roles in eukaryotes and identify underlying molecular mechanisms.
Here, we developed a set of yeast strains accumulating various amounts of AICAR. This well-controlled experimental design allowed us to uncover the effects of AICAR on transcription in a eukaryotic cellular model, and reveal that a small subset of yeast genes responds to AICAR. The vast majority of these genes require the transcription factor Pho2p and either Pho4p or Bas1p for their expression in response to AICAR. We show that both Pho2p and Pho4p directly bind AICAR in vitro. In vivo, AICAR promotes the interaction of Pho2p with both Pho4p and Bas1p, resulting in finely concerted activation of the purine biosynthesis and phosphate utilization pathways. Since purine nucleotide synthesis leads to significant phosphate consumption in yeast, coregulation of these pathways by both the transcription factor Pho2p and the small molecule AICAR provides an efficient mechanism to maintain purine–phosphate cellular homeostasis.
Results
AICAR accumulation stimulates the expression of phosphate and purine regulon genes in a concentration-dependent manner
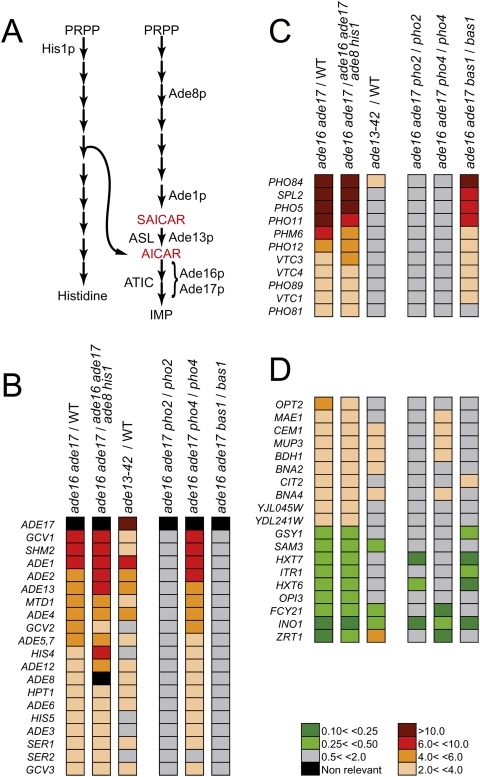
We first designed a set of isogenic yeast strains carrying mutations affecting AICAR metabolism (Fig. 1A) and used high-performance ion chromatography to precisely measure intracellular AICAR concentration in these strains (Supplemental Fig. 1A). In an ade16 ade17 double mutant lacking AICAR transformylase activity, AICAR accumulated (1.6 mM), while it was below detection level (<10 μM) in both an isogenic wild-type strain and an ade16 ade17 ade8 his1 mutant (referred to as “quadruple mutant” hereafter) that cannot synthesize AICAR due to the upstream blocks in both histidine and IMP (inosine 5′-monophosphate) biosynthesis pathways.
Figure 1.
Transcriptional effects of AICAR and SAICAR accumulation. (A) Schematic representation of purine and histidine de novo pathways in yeast. (ASL) Adenylosuccinate lyase; (ATIC) AICAR transformylase IMP cyclohydrolase; (PRPP) 5-phosphoribosyl-1-pyrophosphate. (B–D) Functional classes of genes differentially expressed (more than twofold) in strains accumulating or not AICAR and/or SAICAR. Data were obtained by DNA microarray analyses as described in Breton et al. (2008) and Lemoine et al. (2006), and AICAR-responsive genes were divided into functional classes: purine regulon (B), phosphate regulon (C), and other metabolisms (D). Raw data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13275.
By comparing transcript abundance in this set of strains, we found that AICAR accumulation resulted in differential expression of 50 genes, all encoding metabolic enzymes, and largely composed of induced (41) rather than repressed (nine) genes (Fig. 1B–D, rows 1 and 2). As expected, the purine regulon, which includes IMP biosynthesis genes and other genes (HIS4, SHM2, MTD1 …) known to be activated by the Bas1p and Pho2p transcription factors (Arndt et al. 1987; Daignan-Fornier and Fink 1992; Denis and Daignan-Fornier 1998), was strongly induced in the ade16 ade17 mutant (Fig. 1B; Rebora et al. 2005). More surprisingly, several genes involved in phosphate utilization and its regulation (Ogawa et al. 2000) were among the most up-regulated genes in response to AICAR accumulation (Fig. 1C). The other regulated genes did not belong to a clear unique metabolic pathway (Fig. 1D).
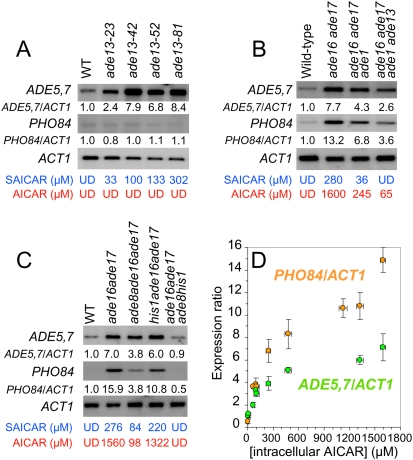
Importantly, in the ade16 ade17 double mutant, not only AICAR but also the metabolite immediately upstream in the pathway (Fig. 1A), SAICAR, accumulated (0.3 mM) far above wild-type level (<10 μM) (Supplemental Fig. 1A). To differentiate between the effects of SAICAR and AICAR on transcription, we used an ade13 mutant, which accumulated SAICAR to a level comparable with the ade16 ade17 mutant (0.1 mM range) but did not contain any detectable AICAR (Supplemental Fig. 1B). In this mutant, the purine regulon, but not the phosphate regulon genes, were up-regulated (Fig. 1B,C, row 3). A set of ade13 mutants accumulating various amounts of SAICAR further confirmed that these two regulons respond differently to SAICAR (Fig. 2A). Importantly, no other blocks in the IMP biosynthesis pathway resulted in up-regulation of PHO5 or PHO84 expression (Supplemental Fig. 2A,B). Therefore, expression of the PHO regulon cannot be induced by accumulation of either SAICAR alone or any upstream intermediates. Finally, we used an ade16 ade17 ade1 ade13 mutant that is totally unable to synthesize SAICAR (Supplemental Fig. 1C) due to the ade1 block in the de novo pathway combined to the ade13 mutation that prevents reverse synthesis of SAICAR from AICAR (Fig. 1A). Using this strain, we established that AICAR alone was sufficient for transcriptional up-regulation of the phosphate and purine regulon genes (Fig. 2B). Results shown in Figure 2B suggest that the transcriptional response was affected by intracellular AICAR concentration. Using several combinations of mutants in the purine and histidine pathways, we determined the AICAR dose-dependent response of ADE5,7 and PHO84 transcription (Fig. 2B–D). These results reveal that expression of these two genes respond to a wide range of intracellular AICAR concentrations.
Figure 2.
Expression of PHO84 and ADE5,7 is stimulated by AICAR in a concentration-dependent manner. (A) SAICAR accumulation stimulates ADE5,7 but not PHO84 transcription. ADE5,7 and PHO84 expression was monitored by Northern blotting on exponentially growing cells. ADE5,7/ACT1 and PHO84/ACT1 ratios were determined using a phosphorimager and were set at 1 for wild-type cells. Ratio values correspond to the average of at least three independent Northern blot analyses. Variations to the mean were <10%. AICAR and SAICAR concentrations determined by chromatography, correspond to an average of two independent injections done with at least three independent metabolite extractions. Variations to the mean were below 10%. (UD) Undetectable, the minimum detection level in our conditions corresponding to concentrations below 10–20 μM for both AICAR and SAICAR. (B) ADE5,7 and PHO84 transcription is stimulated in response to AICAR alone. (C) PHO84 and ADE5,7 expression is tightly correlated to intracellular AICAR concentration. (D) Correlation between intracellular AICAR concentration and expression of PHO84 or ADE5,7. Data correspond to a compilation of transcript and AICAR measurements presented in the figure and data not shown. Error bars correspond to variation to the mean on each axis. The PHO84/ACT1 ratio was set at 1 for wild-type cells.
Pho2p occupancy at ADE and PHO promoters is strongly enhanced in the ade16 ade17 mutant
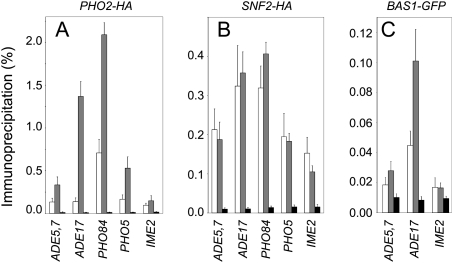
Activation of both purine and phosphate pathways involves Pho2p respectively combined with Bas1p or Pho4p, (Shao et al. 1996; Zhang et al. 1997). The requirement of these three transcription factors for AICAR-induced transcriptional response was assayed by introducing separately bas1, pho2, and pho4 mutations in the ade16 ade17 double mutant. The PHO regulon genes were totally dependent on Pho4p but not on Bas1p, while the purine regulon genes were fully dependent on Bas1p but not on Pho4p (Fig. 1B,C, rows 5 and 6; Supplemental Fig. 3A). Importantly, all but three AICAR-responsive transcripts were strongly dependent on Pho2p (Fig. 1B–D, row 4). Of note, all three Pho2p-independent genes are down-regulated in response to AICAR. This central role of Pho2p in this transcriptional response prompted us to investigate whether AICAR accumulation could modulate the presence of Pho2p at the endogenous ADE and PHO promoters. Chromatin immunoprecipitation (ChIP) revealed a much higher occupancy of the ADE and PHO promoters by Pho2p in the ade16 ade17 mutant compared with the isogenic wild-type strain (Fig. 3A). In a similar experiment, AICAR accumulation had no effect on occupancy of the same promoters by Snf2p (Fig. 3B), which is required for ADE5,7, ADE17, PHO84, and PHO5 expression (Sudarsanam et al. 2000; Neef and Kladde 2003; Koehler et al. 2007). We then investigated the occupancy of the ADE gene promoters by Bas1p and found that Bas1p was slightly more abundant on the ADE gene promoters when yeast cells accumulate AICAR (Fig. 3C). Thus, Pho2p is clearly required for the AICAR transcriptional response, and its presence at up-regulated gene promoters is strongly enhanced under conditions where AICAR accumulates. Importantly, we found that Pho2p protein level was not significantly different in the ade16 ade17 mutant compared with the isogenic wild-type (Supplemental Fig. 3B).
Figure 3.
Pho2p occupancy at ADE and PHO promoters is enhanced in the ade16 ade17 mutant. ChIP assays were performed on the indicated promoters using antibodies against the HA tag on Pho2p (A) and Snf2p (B) and the GFP tag on Bas1p (C). In all cases, results are presented as percent of immunoprecipitation (immunoprecipitation signal/input signal) in strain either accumulating AICAR (gray bars) or not (white bars), or in the untagged control strain (black bars). In this experiment, IME2, which is not AICAR-responsive, was used as a control. Error bars correspond to variation to the mean.
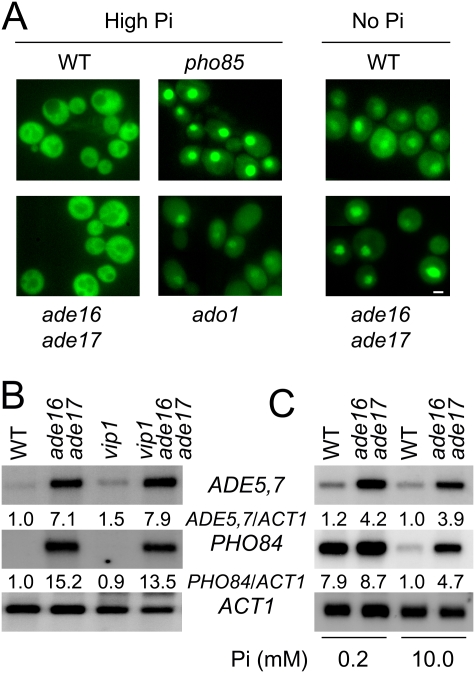
AICAR activation of the PHO regulon is not associated with constitutive nuclear localization of Pho4p
Transcriptional regulation of the PHO regulon in response to extracellular phosphate availability occurs through a well-characterized regulatory cascade involving synthesis of inositol heptakisphosphate (IP7) by Vip1p (Lee et al. 2007) and subsequent inhibition of nuclear relocalization of Pho4p (Komeili and O'Shea 1999). Wild-type strain and the ade16 ade17 double mutant were indiscernible for Pho4p localization. Indeed, under phosphate-replete conditions, Pho4-GFP was found both in the cytoplasm and the nucleus of the two strains (Fig. 4A; Supplemental Fig. 4A). The existence of a nuclear pool of Pho4p under these conditions is further attested by the significant expression of PHO84 compared with that measured in a pho4 deletion mutant (Supplemental Fig. 3A). In contrast, other mutations, such as pho85 or ado1, leading to constitutive expression of the PHO regulon, caused a constitutive nuclear localization of Pho4p (Fig. 4A), as reported previously by others (O'Neill et al. 1996; Huang and O'Shea 2005). Finally, in both wild-type and ade16 ade17 strains, Pho4p relocalized to the nucleus when cells were shifted to phosphate-free medium (Fig. 4A). Together, these results establish that AICAR effect on the PHO regulon is not associated with a constitutive relocalization of Pho4p to the nucleus and thus does not involve the low-phosphate—IP7-sensing—pathway (Lee et al. 2007). Consistently, activation of the PHO pathway in the ade16 ade17 mutant was not dependent on synthesis of IP7 by Vip1p (Fig. 4B), indicating that AICAR and IP7 act through separated signaling pathways. However, under low-phosphate conditions where Pho4p is constitutively nuclear, PHO84 expression was not further up-regulated by the ade16 ade17 mutation (Fig. 4C) nor by purine limitation (Supplemental Fig. 4B). Thus, although AICAR and IP7 signaling pathways are separated, the resulting transcriptional outcome is not strictly additive.
Figure 4.
AICAR activation of the PHO regulon is not associated with constitutive nuclear localization of Pho4p. (A) Pho4p localization was monitored by microscopy using an integrative Pho4p-3xGFP fusion. Cells were grown in High Pi medium, diluted in the same medium containing orphophosphate (10 mM) or not, and observed 16 h later. Bar, 2 μm. (B) Vip1p is not required for expression of the PHO pathway in response to AICAR accumulation. ADE5,7 and PHO84 expression levels were determined by Northern blotting as in Figure 2A. (C) PHO84 expression is not further up-regulated by the ade16 ade17 mutation under low-phosphate conditions. Expression of ADE5,7 and PHO84 in wild-type and ade16 ade17 mutants grown under low- or high-phosphate conditions was monitored by Northern blotting as in Figure 2A.
AICAR accumulation stimulates Pho2p–Pho4p interaction in vivo
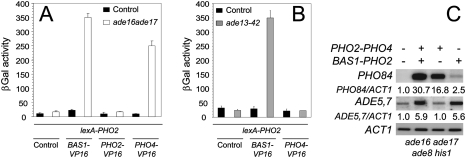
The fact that Pho4p is not constitutively localized in the nucleus of AICAR-accumulating cells suggests that the mechanism leading to up-regulation of the PHO regulon operates somewhere downstream from Pho4p nuclear localization. We therefore developed an in vivo assay to establish whether AICAR accumulation could stimulate interactions between Pho4p and Pho2p. This assay, based on the two-hybrid method performed in parallel in yeast strains accumulating, or not, AICAR, revealed that Pho2p–Pho4p interaction was very low in the control strain but strongly stimulated in the ade16 ade17 mutant accumulating AICAR (Fig. 5A). In the same experiment, we found that interaction between Pho2p and Bas1p was also enhanced by AICAR, while no Pho2p–Pho2p interactions could be detected (Fig. 5A). Importantly, AICAR accumulation had no effect on the individual transcription factors fused to lexA (Supplemental Fig. 5). Consistently with our transcriptome analysis (Fig. 1), Bas1p–Pho2p interaction was stimulated in the ade13-42 mutant accumulating only SAICAR, as previously reported (Rebora et al. 2001), but this accumulation had no effect on Pho2p–Pho4p interaction (Fig. 5B).
Figure 5.
AICAR, but not SAICAR, stimulates Pho2p–Pho4p interaction. (A) Two-hybrid assay reveals that AICAR stimulates Pho2p interaction with its partners Bas1p and Pho4p. βGAL activities were measured in cells transformed with a lexAop-LacZ plasmid and the indicated plasmids. Error bars correspond to variations to the mean. (B) SAICAR stimulates Pho2p interaction with Bas1p but not with Pho4p. βGAL activities were measured in cells transformed with a lexAop-LacZ plasmid and the indicated plasmids. (C) BAS1–PHO2 and PHO2–PHO4 chimeras, respectively, restore ADE5,7 and PHO84 transcription in ade16 ade17 ade8 his1 quadruple mutant cells.
In view of these results, a simple assumption is that forced interactions between transcription factors, via fusion chimeras, should bypass AICAR requirement for transcriptional activation in vivo. PHO2–PHO4 and BAS1–PHO2 fusions should, respectively, make PHO and ADE gene expression AICAR-independent. Indeed, a PHO2–PHO4 fusion expressed under the control of the PHO2 promoter on a centromeric plasmid in the quadruple mutant, which cannot synthesize AICAR, led to strong expression of PHO84 but had no effect on ADE5,7 expression (Fig. 5C). Conversely, the BAS1–PHO2 fusion up-regulated ADE5,7 expression but had little effect on PHO84 (Fig. 5C).
AICAR binds to Pho2p and Pho4p in vitro
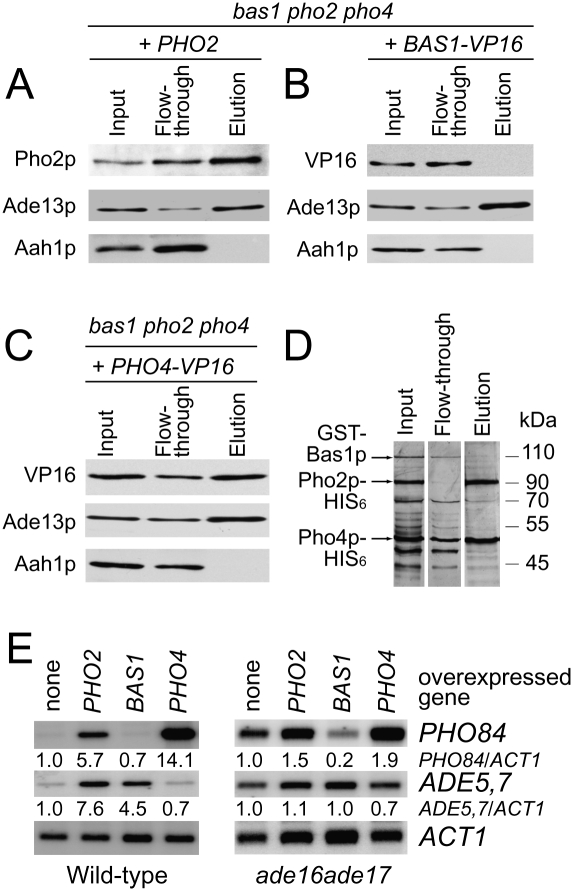
To address the question as to how AICAR affects Pho2p–Bas1p and Pho2p–Pho4p interactions, we expressed each of the three transcription factors individually and, once purified from yeast, assayed their respective ability to bind an AICAR affinity column. Pho2p (Fig. 6A) and Pho4p (Fig. 6C) were effectively retained on the AICAR column, whereas Bas1p was clearly not retained (Fig. 6B). Moreover, adenylosuccinate lyase (Ade13p), the enzyme responsible for AICAR synthesis (positive control), selectively bound the AICAR column (Fig. 6A–C), while, as expected, the adenine deaminase (Aah1p) used as a negative control was not retained on the column (Fig. 6A–C). At this point, we could not rule out that binding of Pho2p and Pho4p to AICAR required accessory factors or post-translational modification. To address this issue, we further performed the AICAR-binding experiment with the three transcription factors independently purified from Escherichia coli. Clearly, Pho2p and Pho4p were still efficiently retained while Bas1p was not (Fig. 6D), demonstrating that binding of Pho2p and Pho4p to AICAR does not require additional yeast proteins.
Figure 6.
Pho2p and Pho4p but not Bas1p can bind AICAR in vitro. (A–C) Pho2p (A), Bas1p (B), and Pho4p (C) binding on an AICAR affinity resin. A triple bas1 pho2 pho4 mutant strain was transformed with plasmids allowing expression of PHO2, BAS1–VP16, or PHO4–VP16. Protein extraction and binding to an AICAR affinity resin were performed as described in the experimental procedures section. Proteins from input, flowthrough and eluted fractions were separated on SDS-PAGE and revealed by Western blotting using anti-Pho2p, anti-VP16, anti-Ade13p, and anti-Aah1p. (D) Purified His6-Pho2p and His6-Pho4p bind the AICAR affinity resin. His6-Pho2p, His6-Pho4p, and GST-Bas1p proteins individually purified from E. coli were mixed and tested for their ability to bind an AICAR affinity resin. Proteins from input, flowthrough and eluted fractions were separated by SDS-PAGE and revealed by silver staining. Additional bands correspond to His6-Pho2p and GST-Bas1p degradation products as revealed by anti-Pho2p and anti-GST antibodies (data not shown). (E) Effect of BAS1, PHO2, or PHO4 overexpression on PHO84 and ADE5,7 transcription in wild-type and ade16 ade17 double mutant strains. Northern blot analyses were done as in Figure 2A on cells transformed with BAS1, PHO2, or PHO4 (each expressed under its own promoter on a 2μ plasmid).
The transcriptional response to AICAR is strongly dependent on Pho2p combined with either Pho4p or Bas1p. This implies that if Pho2p is limiting, these two transcription factors should compete for Pho2p binding in vivo. To assess this assumption, we independently overexpressed each of the transcription factors in a wild-type strain and monitored the expression of genes of the purine and PHO pathways. Clearly, PHO84 expression was enhanced by overexpression of Pho2p and even more efficiently by overexpression of Pho4p (Fig. 6E, left panel). Similarly, ADE5,7 expression was strongly activated by overexpression of Pho2p and Bas1p to a lesser extent (Fig. 6E, left panel). Thus, all three transcription factors appear limiting under these conditions. Importantly, in conditions of AICAR accumulation, in an ade16 ade17 mutant, overexpression of Bas1p strongly down-regulated PHO84 expression (Fig. 6E, right panel) further revealing an effective competition between Pho4p and Bas1p transcription factors for Pho2p binding.
Discussion
Here, we found that yeast cells take profit of an intermediate metabolite to modulate the expression of different metabolic pathways. Our data reveal a new role for the purine metabolic intermediate AICAR, which not only regulates its own pathway but also modulates expression of the phosphate utilization pathway. Importantly, while we found that a wide range of AICAR concentrations could be sensed (Fig. 2D), the highest transcriptional variations were found at low AICAR concentrations that are closer to physiological conditions. Interestingly, the AICAR dose response curves of both ADE and PHO genes had similar shapes suggesting a common mechanism. We propose that the transcription factor Pho2p is the major player in the AICAR response because it is required for up-regulation of all the AICAR-responsive genes, it binds AICAR in vitro, and its presence is enhanced at the target gene promoters under AICAR accumulation conditions. Importantly, the presence of Bas1p was also enhanced (about twofold) on the ADE gene promoters when yeast cells accumulate AICAR. However, this enrichment was much lower than the one observed for Pho2p. These results are very similar to those reported by Rolfes and coworkers (Som et al. 2005), who found a twofold increase in the binding of Bas1p when yeast cells are shifted from adenine-replete to adenine-limited medium. Som et al. (2005) also reported a Bas1p-dependent recruitment of Pho2p under derepressing (low adenine) conditions. A simple explanation for enhanced binding of both Bas1p and Pho2p to the ADE promoters in the ade16 ade17 mutant is that AICAR favors Bas1p–Pho2p interaction and promotes cooperative binding of Bas1p and Pho2p. While such a cooperative effect has not been observed in vitro (Rebora et al. 2001), cooperative binding of Bas1p and Pho2p was proposed to occur in vivo based on ChIP analysis (Som et al. 2005). Together, our results and those published by the Rolfes laboratory (Som et al. 2005) suggest that AICAR promotes both cooperative binding of Bas1p and Pho2p and further recruitment (or retainment) of Pho2p on the ADE gene promoters.
We found that AICAR accumulation stimulates interaction of Pho2p with both Bas1p and Pho4p. Utilization of a common transcription factor together with a common activating metabolite results in competition for activation of the purine and phosphate pathways and thereby probably contributes to fine tuning of both pathways. Intriguingly, accumulation of the upstream metabolic intermediate SAICAR only stimulates Pho2p–Bas1p interaction and specifically up-regulates the Bas1p-responsive genes. The physiological reasons for such a differential response to AICAR and SAICAR are unclear, but could reflect the dual role of adenylosuccinate lyase, the SAICAR metabolizing enzyme, in both the IMP and AMP biosynthesis pathways (Guetsova et al. 1997). This enzyme is feedback inhibited by its products, and therefore SAICAR levels could reflect both the flux in the de novo pathway and the AMP/GMP balance, while AICAR concentration would mostly reflect the flux in the de novo pathway. Importantly, we found that AICAR accumulation mimics purine starvation in multiple respects, leading to induction of the PHO genes only under phosphate replete conditions (Fig. 4C; Supplemental Fig. 4B), resulting in increased Pho2p at the ADE gene promoters (Fig. 3A; Som et al. 2005) and stimulating Bas1p–Pho2p interaction (Fig. 5A; Zhang et al. 1997). Thus, our results strongly support AICAR as a secondary messenger of cellular purine availability in yeast. Whether this role could be extended to other eucaryotes remains to be established. In mammalian cells, AICAR is widely used as an “artificial” activator of the AMP-dependent protein kinase (AMPK) (Towler and Hardie 2007) and as such was shown to have anti-diabetic properties in rodents (Buhl et al. 2002) and mimic some effects of exercise (Narkar et al. 2008). The yeast homolog of AMPK is encoded by the SNF1 gene. To our surprise, none of the genes described as strongly regulated by Snf1p (Young et al. 2003) were found in the 50 AICAR-responsive genes. While it has not been established whether AICAR can stimulate Snf1p in vitro or in vivo, the direct interaction of AICAR with both Pho2p and Pho4p in vitro indicates that the regulation of their target genes in response to AICAR is most probably not dependent on AICAR activation of Snf1p.
Because purine nucleotides are abundant molecules, their synthesis leads to significant phosphate consumption. There is growing evidence that yeast cells have evolved complex regulatory mechanisms to coordinate phosphate utilization and nucleotide synthesis. Recently, two purine metabolism mutants affecting the adenosine kinase (ADO1) and the adenylate kinase (ADK1) genes were shown to strongly stimulate expression of Pho5p (Auesukaree et al. 2005; Huang and O'Shea 2005). In both mutants, this effect was associated to the relocalization of Pho4p to the nucleus under high-phosphate conditions (Huang and O'Shea 2005) and in the case of adk1, the up-regulation was inversely correlated to ATP levels independently of AICAR (Gauthier et al. 2008). Together, our data indicate that the PHO pathway regulation is highly responsive to the cellular purine level through sensing of both the final product (ATP) and an intermediate of the pathway (AICAR). The regulation by AICAR is clearly independent of the previously described phosphate-sensing response, since it does neither affect nuclear localization of the transcription factor Pho4p nor require the Vip1p-dependent synthesis of IP7. Instead, we found that AICAR accumulation stimulates Pho2p–Pho4p and Pho2p–Bas1p interactions, thereby unraveling a novel mechanism for a small molecule to regulate transcription.
Overall our data support the idea that cells use a subset of critical metabolites to permanently sense their metabolome and maintain homeostasis. It becomes clear that various mechanisms have evolved for metabolite sensing from “ancestral” riboswitches to protein–protein signaling. Identification of such key cellular metabolites and their effectors will significantly contribute to our understanding of cellular homeostasis.
Materials and methods
Strains and media
Strains used in this study are described in the Supplemental Material. Phosphate-free SD medium was prepared with yeast nitrogen base without phosphate (Bio101).
Two-hybrid assay
For the two-hybrid assay, bas1 pho2 pho4 ade16 ade17, ade13-42, and isogenic control cells were transformed with a lexAop-LacZ plasmid, and with either a lexA-BAS1 (Zhang et al. 1997), or a lexA-PHO2 (Zhang et al. 1997) plasmid and with a plasmid allowing expression of either BAS1–VP16 (Pinson et al. 2000), PHO2–VP16 (Barbaric et al. 1998), or PHO4–VP16 (Barbaric et al. 1998). βGal activities were measured in transformants as previously described (Daignan-Fornier and Bolotin-Fukuhara 1988).
GST-Bas1p, Pho2p-His6, and Pho4p-His6 cloning, expression, and purification
GST-Bas1p, Pho2p-His6, and Pho4p-His6 plasmids construction details are described in the Supplemental Material. Protein expression and purification were done as described (Brazas and Stillman 1993; Pinson et al. 1998).
AICAR affinity column
An AICAR resin was obtained by coupling AICAR to an epoxy-activated sepharose 6B resin (GE Healthcare). For affinity binding with yeast extracts, a bas1 pho2 pho4 strain was transformed with plasmids allowing expression of either Pho2p, Bas1p–VP16, or Pho4p–VP16. Yeast protein extracts were prepared by vortex grinding with glass beads (5 min in Na2HPO4/HCl at pH 7.2, 100 mM NaCl) followed by 2-min sonication at maximum intensity (FB15050; Thermo-Fischer Scientific). Cellular debris were eliminated by successive centrifugations (5000g, 5 min, 4°C, and 25,000g, 30 min, 4°C). The supernatant was dialyzed at 4°C against buffer A (Na2HPO4/HCl at pH 7.2, 10 mM NaCl). The extract was then incubated with the AICAR affinity resin for 1 h and the suspension was loaded onto a poly-prep chromatography column (Biorad) and rinsed with buffer A. Proteins were eluted with a NaCl gradient in buffer A as described by the manufacturer (GE Healthcare). Ade13p, Aah1p, Bas1p–VP16, Pho4p–VP16, and Pho2p were revealed by Western blotting with anti-Ade13p (1/50,000) (Escusa et al. 2006), anti-Aah1p (1/10,000) (Escusa et al. 2006), anti-VP16 (1/2000; Abcam) and anti-Pho2p antibodies (1/5000; O.S. Gabrielsen). For binding of E. coli purified proteins to AICAR affinity resin, GST-Bas1p, Pho2p-His6 and Pho4p-His6 were mixed (2 nmol each) in buffer A and then treated as for yeast protein extracts.
Microscopy
Three tandem copies of GFP were fused in frame at the C terminus of PHO4 in wild-type, pho85, ado1, ade16, ade17 strains. Details of construction are described in the Supplemental Material. For imaging, strains were grown in liquid free Pi SD casa WAU medium, either supplemented (High Pi condition) or not (No Pi) with KPi 10 mM, and imaged as described in Laporte et al. (2008)
ChIP assays
ChIP experiments were performed as described previously (Bernard et al. 2008). Yeast strains (Y4790, Y4757, Y4751, Y4836, and Y4838) were grown in SDcasaWAU medium OD600 ∼1.0. Cells were fixed for 30 min with formaldehyde 1%, NaCl 10 mM, EDTA 0.1 mM; EGTA 0.05 mM, and Tris/HCl 5 mM (pH 8). Cross-linking was stopped by the addition of Glycine (125 mM). Fifty-microliter cultures were used for each ChIP. Cells were lysed in a Fastprep-24 (four runs of 20 sec at 6.5 m/sec). Extracts were sonicated to shear chromatin to an average DNA fragment size of 0.5–1 kb using a bioruptor sonicator (Diagenode), twice (seven cycles 30 sec on/30 sec off) at maximum power. GFP- and HA-tagged proteins were immunoprecipitated by incubating chromatin extracts with 2 μL of polyclonal anti-GFP antibody (A-11122, Invitrogen) or 15 μL of monoclonal anti-HA antibody (Sc-7392, Santa Cruz Biotechnologies), respectively. Immunoprecipitations were carried out overnight at 4°C in 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, Tris/HCl (pH 8). Immune complexes were collected using ChIP Adembeads protein-A (Ademtech, #04240), washed, and recovered according to manufacturer instructions. ChIP quantifications were performed by real time PCR using a MX3000P cycler (Stratagene) and Absolute qPCR Sybr Green Mix (ABGene). Primers used are described in Supplemental Table 3. A 10-fold dilution series of genomic DNA was used to calibrate the quantification. DNA was quantified in the input and immunoprecipitated samples and the ratio was calculated (percent of immunoprecipitation). Immunoprecipitations were performed in duplicate.
Northern blots, microarray analyses, and metabolites quantification
Northern Blot, DNA microarray analyses, and intracellular metabolites determination by high-performance ionic chromatography were performed as described in Denis et al. (1998), Breton et al. (2008), and Diaz-Ruiz et al. (2008), respectively. The microarray data were normalized without background substraction by the global Lowess method performed with the Goulphar software (Lemoine et al. 2006).
Acknowledgments
We thank Drs. D. Appling, O. Gabrielsen, C. Goding, L. Kuras, R. Rolfes, and A. Vershon for sharing biological materials; C. Saint-Marc, N. Durand, and J. Ceschin for technical assistance; and Drs. A. Devin and M. Escobar-Henriques for comments on the manuscript. This work was supported by Conseil Régional d'Aquitaine and ANR grant #BLAN06-1_136912 to B.D.-F.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.521809.
Supplemental material is available at http://www.genesdev.org.
References
- Arndt KT, Styles C, Fink GR. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25127–25133. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- Barbaric S, Munsterkotter M, Goding C, Horz W. Cooperative Pho2–Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol Cell Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JP. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2008;27:111–121. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazas RM, Stillman DJ. Identification and purification of a protein that binds DNA cooperatively with the yeast SWI5 protein. Mol Cell Biol. 1993;13:5524–5537. doi: 10.1128/mcb.13.9.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A, Pinson B, Coulpier F, Giraud MF, Dautant A, Daignan-Fornier B. Lethal accumulation of guanylic nucleotides in Saccharomyces cerevisiae HPT1-deregulated mutants. Genetics. 2008;178:815–824. doi: 10.1534/genetics.107.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl ES, Jessen N, Pold R, Ledet T, Flyvbjerg A, Pedersen SB, Pedersen O, Schmitz O, Lund S. Long-term AICAR administration reduces metabolic disturbances and lowers blood pressure in rats displaying features of the insulin resistance syndrome. Diabetes. 2002;51:2199–2206. doi: 10.2337/diabetes.51.7.2199. [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B, Fink GR. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B, Bolotin-Fukuhara M. In vivo functional characterization of a yeast nucleotide sequence: Construction of a mini-Mu derivative adapted to yeast. Gene. 1988;62:45–54. doi: 10.1016/0378-1119(88)90578-1. [DOI] [PubMed] [Google Scholar]
- Denis V, Daignan-Fornier B. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1998;259:246–255. doi: 10.1007/s004380050810. [DOI] [PubMed] [Google Scholar]
- Denis V, Boucherie H, Monribot C, Daignan-Fornier B. Role of the myb-like protein bas1p in Saccharomyces cerevisiae: A proteome analysis. Mol Microbiol. 1998;30:557–566. doi: 10.1046/j.1365-2958.1998.01087.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R, Averet N, Araiza D, Pinson B, Uribe-Carvajal S, Devin A, Rigoulet M. Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J Biol Chem. 2008;283:26948–26955. doi: 10.1074/jbc.M800408200. [DOI] [PubMed] [Google Scholar]
- Escusa S, Camblong J, Galan JM, Pinson B, Daignan-Fornier B. Proteasome- and SCF-dependent degradation of yeast adenine deaminase upon transition from proliferation to quiescence requires a new F-box protein named Saf1p. Mol Microbiol. 2006;60:1014–1025. doi: 10.1111/j.1365-2958.2006.05153.x. [DOI] [PubMed] [Google Scholar]
- Flynn PJ, Reece RJ. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol. 1999;19:882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier S, Coulpier F, Jourdren L, Merle M, Beck S, Konrad M, Daignan-Fornier B, Pinson B. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol Microbiol. 2008;68:1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- Guetsova ML, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, O'Shea EK. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics. 2005;169:1859–1871. doi: 10.1534/genetics.104.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler RN, Rachfall N, Rolfes RJ. Activation of the ADE genes requires the chromatin remodeling complexes SAGA and SWI/SNF. Eukaryot Cell. 2007;6:1474–1485. doi: 10.1128/EC.00068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O'Shea EK. Regulation of a cyclin–CDK–CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: Rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Marie S, Heron B, Bitoun P, Timmerman T, Van Den Berghe G, Vincent MF. AICA-ribosiduria: A novel, neurologically devastating inborn error of purine biosynthesis caused by mutation of ATIC. Am J Hum Genet. 2004;74:1276–1281. doi: 10.1086/421475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARδ agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Kladde MP. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol Cell Biol. 2003;23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin–CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Pinson B, Sagot I, Borne F, Gabrielsen OS, Daignan-Fornier B. Mutations in the yeast Myb-like protein Bas1p resulting in discrimination between promoters in vivo but notin vitro. Nucleic Acids Res. 1998;26:3977–3985. doi: 10.1093/nar/26.17.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson B, Kongsrud TL, Ording E, Johansen L, Daignan-Fornier B, Gabrielsen OS. Signaling through regulated transcription factor interaction: Mapping of a regulatory interaction domain in the Myb-related Bas1p. Nucleic Acids Res. 2000;28:4665–4673. doi: 10.1093/nar/28.23.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora K, Desmoucelles C, Borne F, Pinson B, Daignan-Fornier B. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol Cell Biol. 2001;21:7901–7912. doi: 10.1128/MCB.21.23.7901-7912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora K, Laloo B, Daignan-Fornier B. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: A central role for a small molecule. Genetics. 2005;170:61–70. doi: 10.1534/genetics.104.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Reece RJ. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem Sci. 2005;30:405–412. doi: 10.1016/j.tibs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Shao D, Creasy CL, Bergman LW. Interaction of Saccharomyces cerevisiae Pho2 with Pho4 increases the accessibility of the activation domain of Pho4. Mol Gen Genet. 1996;251:358–364. doi: 10.1007/BF02172527. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Som I, Mitsch RN, Urbanowski JL, Rolfes RJ. DNA-bound Bas1 recruits Pho2 to activate ADE genes in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1725–1735. doi: 10.1128/EC.4.10.1725-1735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RL, Aimi J, Barshop BA, Jaeken J, Van den Berghe G, Zalkin H, Dixon JE. A mutation in adenylosuccinate lyase associated with mental retardation and autistic features. Nat Genet. 1992;1:59–63. doi: 10.1038/ng0492-59. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Woontner M, Jaehning JA, Kohlhaw GB. In vitro transcriptional activation by a metabolic intermediate: Activation by Leu3 depends on α-isopropylmalate. Science. 1992;258:1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- Zhang F, Kirouac M, Zhu N, Hinnebusch AG, Rolfes RJ. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol Cell Biol. 1997;17:3272–3283. doi: 10.1128/mcb.17.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]