Abstract
Xenopus Cadherin-11 (Xcad-11) is expressed when cranial neural crest cells (CNC) acquire motility. However, its function in stimulating cell migration is poorly understood. Here, we demonstrate that Xcad-11 initiates filopodia and lamellipodia formation, which is essential for CNC to populate pharyngeal pouches. We identified the cytoplasmic tail of Xcad-11 as both necessary and sufficient for proper CNC migration as long as it was linked to the plasma membrane. Our results showing that guanine nucleotide exchange factor (GEF)-Trio binds to Xcad-11 and can functionally substitute for it like constitutively active forms of RhoA, Rac, and cdc42 unravel a novel cadherin function.
Keywords: Trio, cadherin-11, cell migration, filopodia, neural crest, small GTPases
The cranial neural crest (CNC) is a multipotent and highly migratory cell population that shapes the vertebrate face contributing to the craniofacial bones, cartilage, and ganglia (Le Lievre and Le Douarin 1975; Le Lievre 1978; Santagati and Rijli 2003). Balanced levels of Wnt, FGF, and BMP signaling induce the CNC at the neural plate border, giving rise to the neural fold from which CNC cells emigrate (Sauka-Spengler and Bronner-Fraser 2008). Before emigration, the CNC segregates into the mandibular, hyoid, and branchial segments, which follow distinct migratory routes to their final destinations (Kontges and Lumsden 1996).
Although it is extremely motile, the CNC continually expresses the cell adhesion molecule cadherin-11 (Kimura et al. 1995; Hadeball et al. 1998; Vallin et al. 1998). In adult tissues, up-regulation of cadherin-11 coincides with tumor progression (Pishvaian et al. 1999; Tomita et al. 2000; Chu et al. 2008) and cartilage invasion by synoviocytes in inflammatory arthritis (Lee et al. 2007). These observations point to a migration-stimulating property of cadherin-11 distinct from its cell-adhesive function. The latter was studied in detail by cell adhesion assays (Valencia et al. 2004), gain-of-function experiments in embryos (Borchers et al. 2001), and crystal structure analysis of the EC1 ectodomain (Patel et al. 2006), as well as by single-molecule-binding activity measurements (Heupel et al. 2008). Cadherin-11 shares both the p120-binding and β-catenin (β-cat)-binding sites (Kimura et al. 1995; Hadeball et al. 1998; Vallin et al. 1998) with all classical cadherins and the extended adhesive interface of the EC1 domain with all type II cadherins (Patel et al. 2006). Because of these similarities, the question arises of what makes cadherin-11 unique in stimulating cell migration.
Here we report on a novel function of cadherin-11 in regulating filopodia and lamellipodia formation in Xenopus CNC, which relies on its cytoplasmic tail and includes the activity of guanine nucleotide exchange factor (GEF)-Trio and the small Rho GTPases. This migration-stimulating function of the cytoplasmic tail is required for Xenopus CNC to enter the pharyngeal pouches, a prerequisite for proper formation of the craniofacial cartilage.
Results and Discussion
Xenopus cadherin-11 (Xcad-11) is required for CNC to populate the pharyngeal pouches and to form craniofacial cartilage structures
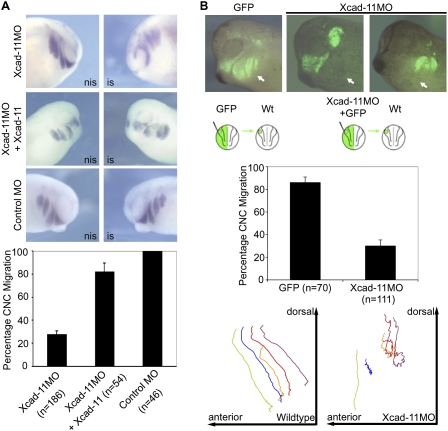
To identify the role of Xcad-11 in CNC migration, we performed a knockdown approach by single-sided antisense morpholino (Xcad-11MO or Xcad-11MO2) injection into two-cell stage embryos. The specificity of the MOs was demonstrated by suppressed translation and by injection of a control MO (CoMO) that showed no effect (Supplemental Fig. S1A–C). In situ hybridizations (ISHs) for twist, a marker for CNC, revealed that in the absence of Xcad-11, the hyoid and branchial CNC populations started to migrate but did not enter the pharyngeal pouches. The mandibular CNC population remained unaffected (Fig. 1A). Coinjection of full-length Xcad-11 (FLXcad-11) (Fig. 1A) and human Cadherin-11 but not XB-cadherin (type I) and mCadherin-6 (type II) were able to rescue CNC migration (Supplemental Fig. S1D). Transplantations of Dextran-labeled CNC from embryos injected with the Xcad-11MO confirmed that the hyoid and branchial CNC cells accumulated halfway on their route (Fig. 1B; Supplemental Movies S2, S3). Single-cell tracking revealed that these cells were unable to progress ventrally (Fig. 1B). At the tadpole stage, morphant embryos showed strong defects in cartilage formation together with an increase in melanocytes, which was confirmed by RT–PCR. In addition, Xcad-11MO-injected CNC transplants with impaired migration survived and differentiated into melanocytes or ganglia instead of cartilage (Supplemental Fig. S4A–E). Taken together, these observations point to a specific Xcad-11 function in a subpopulation of CNC. Xcad-11 and its human ortholog but none of the other cadherins tested allowed migration of CNC into the pharyngeal pouches, which is essential for craniofacial cartilage formation.
Figure 1.
Depletion of Xcad-11 blocks CNC migration. (A) Whole-mount ISH. Lateral view of Xenopus CNC at stage 26, analyzed by expression of twist. (Left column) Noninjected side (nis). (Right column) Injected side (is). Embryos injected with Xcad-11MO (2 pmol) showed twist signal adjacent to the brain but not in pharyngeal pouches (72%), indicating impaired CNC migration. The mandibular population remained unaffected. Coinjection of Xcad-11 mRNA (75 pg) restored the migration phenotype in 82%. CoMO-injected (2 pmol) embryos showed no effect. (B) Transplantation to follow migration in vivo in CNC. Wild-type CNC grafts exhibited normal migration (86%). Grafts taken from Xcad-11-depleted embryos were unable to invade hyoidal and branchial arches (70%; marked by arrows). Mandibular CNC remained unaffected. Single cell tracking demonstrated wild-type CNC migrating into the pharyngeal arches while Xcad-11 depletion stopped migration.
Xcad-11 is localized to cell protrusions and induces filopodia and lamellipodia formation
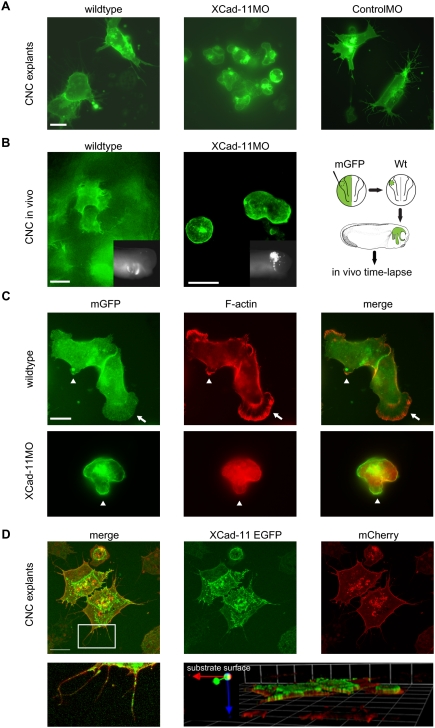
Isolated Xenopus CNC plated on fibronectin exhibits extensive protrusive activity and a leading edge to trail polarity (Kuriyama and Mayor 2008), as also observed for chicken NC in vivo (Kasemeier-Kulesa et al. 2005). In an effort to identify a putative cellular function of Xcad-11, we isolated CNC labeled by mGFP-RNA injection (termed wild type) or coinjected with either Xcad-11MO or CoMO and monitored their protrusion formation capacity by time-lapse recording. Wild-type or CoMO-injected CNC exhibited leading edge to trail polarity and intensive protrusive activity. However, CNC from morphants completely lost the ability to form filopodia and lamellipodia and instead produced membrane blebbing (Fig. 2A; Supplemental Movies S5, S6). This change in cell motility behavior was confirmed in vivo by time-lapse studies of transplants (Fig. 2B; Supplemental Movies S7, S8). When the stability of filopodia and lamellipodia was evaluated, no significant difference was observed between CNC from wild-type or CoMO-injected embryos as well as Xcad-11MO-injected embryos rescued with FLXcad-11 mRNA (Supplemental Fig. S9C). The blebbing was not due to cell death, as we measured an increase in proliferation rather than in apoptosis (Supplemental Fig. S4F). Instead, the nonapoptotic membrane blebbing points to a switch in cell motility from a fibroblast-like to an amoeboid-like mode of locomotion. This was supported by the lack of F-actin in blebs (Fig. 2C) characteristic for nonapoptopic membrane blebbing (Fackler and Grosse 2008).
Figure 2.
Cell shape changes and localization of Xcad-11. (A) Analysis of CNC migration in vitro. Control cells injected with mGFP (200 pg) showed extensive protrusion activity. CNC from Xcad-11-depleted embryos exhibited membrane blebbing. CoMO-injected cells showed the same cell behavior as wild-type cells. Bar, 10 μm. (B) Protrusion formation of CNC in vivo. Wild-type cells showed extensive protrusive activity while Xcad-11MO-injected cells were unable to form filopodia or lamellipodia. The right-hand scheme displays the experimental procedure. The inserts show an outside view of transplanted embryos at stage 26. Anterior is to the right side. Bar, 20 μm. (C) F-actin staining of wild-type and Xcad-11MO-injected CNC explants. In wild type, F-actin was enriched in small (arrowhead) and large (arrows) lamellipodia. In Xcad-11MO-treated explants, no F-actin was seen in blebs (arrowheads). Bar, 10 μm. (D) Localization of Xcad-11 in CNC explants. Embryos were injected with 1 ng of Xcad-11-EGFP and mCherry (200 pg). (Bottom left) Xcad-11-EGFP was visible in filopodia and lamellipodia, while mCherry labeled cell membranes. The boxed area is enlarged in the bottom left panel. Z-stack revealed localization of Cad11-EGFP at the substrate-facing cell surface. Bar, 10 μm.
The dramatic alteration in cell shape and cell behavior of morphant CNC raises the question of where Xcad-11 is localized in these cells. Since the antibodies against Xcad-11 do not work for immunostaining, we expressed an Xcad-11-EGFP fusion protein. We found Xcad-11-EGFP enriched in the membrane facing the substrate and in the cell protrusions (Fig. 2D). As Xcad-11 binds β-cat (Borchers et al. 2001), we stained the Xcad-11-EGFP-positive cells for β-cat. In CNC clusters, both proteins colocalized at cell–cell contacts but also in filopodia and at the leading edges of cells attached to the substrate (Supplemental Fig. S11A,B). Taken together, the subcellular localization of Xcad-11 coincides with its requirement for filopodia and lamellipodia formation.
CNC migration and protrusion formation depends on the membrane-anchored cytoplasmic tail of Xcad-11
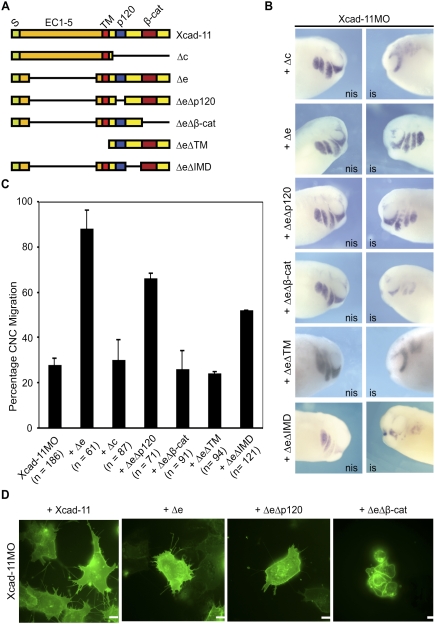
Classical cadherins can be subdivided in different functional domains. Their extracellular part mediates cell–cell adhesion through homophilic binding, while the cytoplasmic tail contains binding sites for different catenins. Catenin p120ctn binds to the juxtamembrane region and regulates cadherin–adhesion complex assembly (Anastasiadis and Reynolds 2000), lateral clustering of E-cadherin (Yap et al. 1998), and filopodia dynamics (Boguslavsky et al. 2007). The binding site for β-cat instead is located close to the C terminus of cadherins. β-cat is required to anchor the cadherin to the actin cytoskeleton via α-catenin (Rimm et al. 1995). To identify the domain in Xcad-11 that is essential for cell protrusion formation, we tested the rescue ability of different deletion mutants (Fig. 3A; Supplemental Fig. S9A). To our surprise, Xcad-11 lacking the extracellular domain (Δe) exhibited the best rescue ability (Fig. 3B,C). The rescue effect was even stronger than with FLXcad-11 (Fig. 1A), confirming previous results that showed that Δe behaves as a hypermorph in regard to CNC migration (Borchers et al. 2001). In vivo time-lapse imaging of CNC grafts coinjected with Xcad-11MO and Δe transplanted into wild-type hosts revealed that the Δe construct re-established dorsoventral pathfinding and migration into the pharyngeal pouches (data not shown). In contrast, Xcad-11 lacking the cytoplasmic tail (Δc) failed to rescue migration in Xcad-11MO-injected embryos (Fig. 3B,C). This suggests that Xcad-11-mediated cell–cell adhesion is not essential for CNC cells when they populate the pharyngeal pouches. The latter is in agreement with most recent results of McCusker et al. (2009), who found that Xcad-11 is cleaved by the metalloproteinase ADAM13, which is expressed in CNC and is required for their migration (Alfandari et al. 2001). The metalloproteinase produces an extracellular soluble fragment and a membrane-bound cytoplasmic tail of Xcad-11. While McCusker et al. (2009) emphasized the role of the extracellular EC1-3 fragment in blocking Xcad-11-mediated adhesion, our rescue data point to an independent role of the cytoplasmic tail in stimulating cell migration.
Figure 3.
The β-cat-binding site and the TM are essential for Xcad-11 function. (A) Xcad-11 constructs for reconstitution. (S) Signal peptide (green); (EC) extracellular domains (orange); TM (red); (p120/β-cat) p120-binding site (blue) or β-cat-binding site (dark red); intracellular domain (yellow). (B) Whole-mount ISH. Lateral view of CNC at stage 26, analyzed by expression of twist. (Left column) Noninjected side (nis). (Right column) Injected side (is). Embryos injected with Xcad-11MO (2 pmol) together with 50 pg of Δe or ΔeΔp120 showed twist signal in pharyngeal pouches, while coinjection of 75 pg of Δc, 50 pg ΔeΔβ-cat, ΔeΔTM, or ΔeΔIMD did not. (C) Statistics for reconstitution. In the case of Xcad-11MO injection, only 28% of embryos showed normal migration. Coinjection of 75 pg of FLXcad-11 resulted in 82%, 75 pg of Δe in 88%, and 75 pg of ΔeΔp120 in 82% of embryos with CNC present in pharyngeal pouches; in the case of Δc, ΔeΔβ-cat, ΔeΔTM, and ΔeΔIMD coinjection, only 30%, 27%, 28%, and 52%, respectively. (D) CNC explants. Coinjection of FLXcad-11, Δe, or ΔeΔp120 with Xcad-11MO restored formation of lamellipodia and filopodia in CNC; ΔeΔβ-cat coinjection showed blebbing. Bar, 10 μm.
To determine the required functional domain within the cytoplasmic tail, we deleted the p120ctn- or the β-cat-binding site in the Δe mutant. Twist-ISH showed that ΔeΔp120 injections in morphant embryos resulted in a rescue of ∼70%, whereas ΔeΔβ-cat did not rescue (Fig. 3B,C). We also investigated the rescue capacities of deletion mutants and FLXcad-11 to initiate cell protrusions. FLXcad-11, Δe, or ΔeΔp120 expression resulted in the rescue of both filopodia and lamellipodia formation (Fig. 3D; see also Supplemental Fig. S9C). Expression of the ΔeΔβ-cat mutant, however, led to membrane blebbing without cell protrusions, as seen in the morphant CNC (Fig. 3D). The results indicate that the β-cat-binding site but not the p120ctn-binding site in Xcad-11 is essential for proper migration of CNC into the pharyngeal pouches in vivo, as well as the formation of filopodia and lamellipodia in vitro.
Recent studies of chicken trunk NC have reported that a soluble N-cadherin fragment promotes β-cat/Wnt-dependent target gene expression (Shoval et al. 2007). To distinguish between a nuclear or membrane function, we produced a new mutant corresponding to the cytoplasmic domain of Xcad-11. This ΔeΔTM construct does not encode the transmembrane domain (TM) and the signal peptide and could not rescue Xcad-11MO-injected embryos (Fig. 3A–C). Therefore, we conclude that both the TM and the β-cat-binding site in Xcad-11 are essential for proper CNC migration. Together with the subcellular location of Xcad-11 (Fig. 2), the latter results suggest that the Xcad-11 cytoplasmic domain is critical at the plasma membrane.
The GEF-Trio and the small GTPases cdc42, Rac1, and RhoA act downstream from Xcad-11
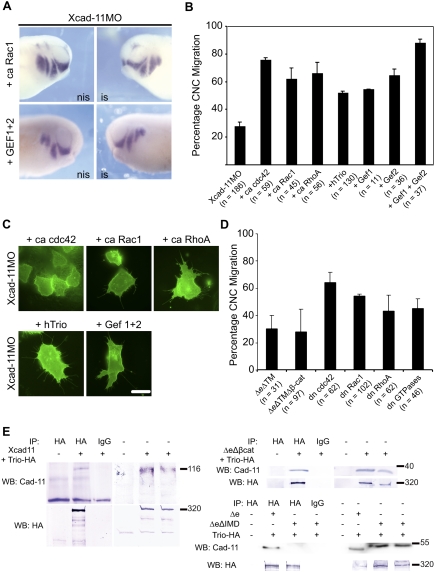
Cell protrusion formation depends on the activity of the small GTPases cdc42, Rac1, and RhoA (Jaffe and Hall 2005). To investigate whether these molecules act downstream from Xcad-11, we performed rescue experiments with constitutively active (ca) forms of cdc42, Rac1, and RhoA. Each of them restored CNC migration in embryos injected with Xcad-11MO (Fig. 4A,B). In vivo imaging of isolated CNC cells further confirmed that ca cdc42, ca Rac1, and ca RhoA rescued cell protrusion formation (Fig. 4C; Supplemental Movie S10, RhoA as an example). The involvement of the GTPases was further substantiated by dominant-negative (dn) expression of cdc42, Rac1, and RhoA, which resulted in 30%–50% inhibition of CNC migration (Fig. 4D) and by application of Toxin B (Supplemental Fig. S11B). In the search for a molecule that is able to activate small GTPases but interact specifically with Cadherin-11, we identified the GEF-Trio. This protein contains two GEF domains and was reported previously to interact with Cadherin-11 but not with N-Cadherin (Backer et al. 2007). GEF 1 in Trio activates Rac1 and RhoG/cdc42 while GEF 2 activates RhoA/ROK, thereby influencing cytoskeletal dynamics (Bateman and Van Vactor 2001). Reconstitution experiments with human Trio as well as the single Trio-GEF domains abolished the block in CNC migration (Fig. 4A,B). The best rescue was observed when both GEF 1 and GEF 2 were coexpressed. The slightly weaker rescue efficiency of the full-length form compared with GEF 1 and GEF 2 is not surprising since the C terminus of Trio varies between different species in containing additional regulatory elements; e.g., a SH3 domain or a serine/threonine kinase domain (Bateman and Van Vactor 2001).
Figure 4.
Xcad-11 function is mediated by small GTPases and GEF-Trio. (A) Whole-mount ISH. Injection of ca Rac1 and two GEF subunits of hTrio together with Xcad-11MO rescued CNC migration, demonstrated by twist ISH. (B) Statistics of reconstitution experiments with 10 pg of ca cdc42, Rac1, or RhoA. Coinjection with Xcad-11MO rescued CNC migration into pharyngeal pouches in 76%, 62%, or 66%, respectively. Twenty picograms of Trio or single GEF domains restored disturbed invasion in pharyngeal pouches in 52% (Trio), 55% (Gef1), 65% (Gef2), or 88% (Gef 1 + 2). (C) In Xcad-11-depleted CNC explants, coinjection of ca cdc42, Rac1, or RhoA, as well as with Trio or the two GEF subunits, restored formation of lamellipodia and filopodia. Bar, 10 μm. (D) Statistics of gain-of-function analysis with soluble Xcad-11 constructs; dn GTPases cdc42, Rac1, and RhoA; as well as a combination of all three GTPases together. Overexpression of the cytoplasmic domain of Xcad-11 with or without the β-cat-binding site resulted in a strong reduction of CNC migration (30% and 28% migrating CNC). The three dn GTPases cdc42, Rac1, and RhoA showed migration in 65%, 55%, and 44% of the embryos, respectively. A combination of all three dn GTPases together resulted in 46% migrating CNC. (E) Coimmunoprecipitation of Trio with different Xcad-11 constructs. (Bottom left panel) Precipitation of Trio with HA antibody from transfected COS cells. Trio was detected in Western blot. Western blot for Xcad-11 showed successful coprecipitation of Xcad-11 (top left) as well as of Xcad-11ΔeΔβ-cat (top right). In the case of Xcad-11ΔeΔIMD, no coprecipitation was detected. (Right half of panels) Input.
We also tested whether Xcad-11 forms a complex with Trio, as has been reported for the mCadherin-11 (Backer et al. 2007). Since the Xenopus Trio homolog has not been cloned yet, we performed coprecipitations with Xcad-11 or its mutants and hTrio after transfection in COS7 cells. FLXcad-11, Δe, and ΔeΔβ-cat (Fig. 4E) were found in complex with hTrio, excluding an interaction mediated by β-cat. Therefore, we produced another mutant lacking 85 amino acids between the p120-binding site and the β-cat-binding site (ΔeΔIMD) (Fig. 3A; Supplemental Fig. S9B). This mutant did not coprecipitate with hTrio (Fig. 4E), indicating that Trio binds to the intermediate domain (IMD) in Xcad-11. However, it remains to be clarified whether this interaction is direct or indirect. Functional studies revealed that ΔeΔIMD possesses only partial rescue ability (Fig. 3B,C), which points to a requirement of this region for proper CNC migration. The latter was further supported by expression of ΔeΔTM and ΔeΔTMΔβ-cat to see if they could compete for the binding of Trio and affect CNC migration. Both mutants blocked CNC migration with nearly the same efficiency as the MOs independent of the presence of the β-cat-binding site (Fig. 4D). Taken together, we identified three domains in the cytoplasmic tail of Xcad-11 that collaborate in promoting CNC migration: the TM-binding site, the Trio-binding site, and the β-cat-binding site. This indicates that at the plasma membrane, a Cadherin-11/β-cat/Trio complex is assembled.
Next, we asked whether a Xenopus Trio homolog is expressed in the CNC. We isolated a partial sequence of XTrio that covers the region of base pairs 2281–2900. It shares an amino acid identity of 71% with the murine or human ortholog in total, but 90% within the GEF 1 domain (Supplemental Fig. S12). Based on this partial sequence, we examined the temporal and spatial XTrio expression in the early embryo. By ISH, we found XTrio in migratory CNC cells, the neural plate, and the neural tube from late neurula stage 19 onward. RT–PCR revealed that expression starts in stage 16 (mid-neural stage) (Supplemental Fig. S12).
Our results offer a new insight toward understanding how the mesenchymal cadherin-11 promotes cell migration. Apart from its presence in cell–cell contacts, Xcad-11 localizes in both filopodia and lamellipodia (Fig. 2). Up to date, cadherin localization in filopodia has been reported as an early step in adherence junction formation depending on homophilic binding of the extracellular domains (Kobielak and Fuchs 2004). Our data, however, demonstrate that the membrane-anchored cytoplasmic domain by itself is sufficient to initiate cell protrusive activity (Fig. 3) and to drive cell migration. Xcad-11-mediated protrusive activity needs Trio and the active small GTPases RhoA, Rac1, and cdc42 in combination with the Xcad-11/β-cat membrane complex. Because Trio binds adjacent to β-cat in Xcad-11, we suggest that both work together in recruiting and dynamically reorganizing F-actin at the membrane. Our current work provides, to our knowledge, the first evidence for a cadherin-regulated cell protrusion activity in migrating cells. Since the human cadherin-11 rescues depletion of the Xenopus ortholog, our findings will have strong implications on understanding cell invasion in a variety of different diseases.
Materials and methods
Constructs
pcDNA3.1 Xcad-11 was described previously (Borchers et al. 2001). Deletion constructs were generated by the following procedures: For Δe, Xcad-11 was incubated with BstXI and BamHI to remove the extracellular domain. For Δc, Xcad-11 was incubated with EcoRV to remove the intracellular domain. For ΔeΔp120, the Δe construct was incubated with EcoRV and the fragment with the β-cat-binding site was ligated to the remaining Δe fragment. For the ΔeΔβ-cat construct, the Δe construct was amplified by PCR with a reverse primer removing the β-cat site (Fw, CCAAGCTGGCTAGCGTTTA; Rv, CATTTGGTACCCGGAGCTGCAGGGTCATTATC). ΔeΔIMD was constructed synthetically (MWG). For further details, see Supplemental Figure S9. For the Xcad-11-EGFP construct, the Xcad-11 ORF was amplified by PCR using StuI–Xcad11-Fw, 5′-TAGGCCTATGAAGAAAGACTTTTGCTTACA-3′; and AgeI–Xcad11-Rv, 5′-CGACCGGTGCAGAATCATCTTCACAAGTGT-3′. The GFP vector was generated from pCSH2B-EGFP, and cDNA of Histone H2B was replaced with Xcad11-ORF. The Xcad-11MO and CoMO oligonucleotides were purchased from GeneTools, LLC. For mCherrry, cherry was subcloned in pCR2.1 and inserted in pCS2+ mGFP replacing GFP ORF, but retaining membrane anchor. pSP64 XB-cadherin was published previously (Dufour et al. 1994).
Embryo manipulations, staining methods, and RT–PCR
Handling of Xenopus laevis embryos and CNC transplantations were performed as described (Borchers et al. 2000). For explants, the method by Alfandari et al. (2003) was used. Toxin B (20 ng/mL; Sigma) was added in culture medium. Live cell images were taken with a Leica-DMRE2 microscope with 63× plan apochromat NA 1.32 Ph3 oil objective. Live cell images in vivo were taken with a Perkin Elmer Ultra View ERS spinning disc confocal microscope with 60× plan apo VC NA 1.2 multi-immersion objective.
Cartilage staining and whole-mount 3A10 immunostaining was as described by Schuff et al. (2007). ISH was mentioned earlier (Harland 1991). Digoxigenin-labeled antisense RNA probes were generated according to the manufacturer's (Roche) description. TUNEL was described in Hensey and Gautier (1998). For immunostaining, NC explants were fixed in PFA before permeabilization with 0.5% Triton in PBS, and incubation with primary β-cat antibody or Phalloidin Alexa 568 (1:100; Invitrogen). As a secondary antibody, goat α-rat-Cy3 (Dianova) was used (1:200). Whole-mount pH3 immunostaining was described earlier (Kunz et al. 2004). Confocal images were taken with a Zeiss LSM 5 using the 63× LCI-Neofluar Imm Corr DIC objective (NA 1.3). For RT–PCR, RNA was extracted from embryos of the recommended stage (Macherey-Nagel). Reverse transcription was followed by standard PCR. The primer sequences used were ODC fwd, GGAGCTGCAAGTTGGAGA, and rev, CTCAGTTGCCAGTGTGGTC; Trio fwd, GGGTGTTGCATTTCTGGACT, and rev, CCGCCATGATGAATTCTTTT; H4 fwd, CGGGATAACATTCAGGGTATCACT, and rev, ATCCATGGCGGTAACTGTCTTCTT; and Trp2 fwd, TGTTACTTGGGTTACC, and rev, GCACTTATCACGGGTC.
Immunoprecipitation
Cos7 cells were transfected using TransPass (New England Biolabs). Cells were harvested after 48 h and lysed with NOP buffer (10 mM HEPES at pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40) supplemented with Complete protease inhibitor (Roche). Trio was pulled down with rabbit anti-HA (Abcam). For Western blotting, proteins were transferred to PVDF membrane and incubated with primary antibody αXcad-11 polyclonal antibody generated against the cytoplasmic domain of Xcad-11 protein, anti-XB-cadherin (6D5), anti-myc (9E10), and anti-HA, followed by incubation with alkaline phosphatase-labeled goat anti-rabbit or mouse IgG (Dianova) and were developed using NBT/BCIP.
Acknowledgments
We thank the following persons for providing material for this study: A. Debant (Montpellier, France) for human Trio, T. Inoue (Kansas City, USA) for mouse cadherin-6, E. DeRobertis (Los Angeles, USA) for pCS2 + mGFP, J.-K. Han (Kyoung-Buk, South Korea) for ca Cdc42, K. Giehl (Ulm, Germany) for RhoA and Rac1, J. Small (Vienna, Austria) for cherry, R.W. Köster (Pasadena, USA) for pCSH2B-EGFP, and R. Rupp (München, Germany) for β-cat antibody from rat. Special thanks to U. Engel and the Nikon Center University Heidelberg for help with spinning disc microscopy. We thank C. Van Lishout and C. Winter for technical assistance. J.K. was funded by the Virtual Institute for Bio-Imaging of the Helmholtz-Gemeinschaft. D.A. was supported by grants from NIH (DE016289). J.K., A.K., R.M., and D.W. designed research and wrote the manuscript. J.K. and A.K. conducted the research including embryonic experiments and explants. J.K. prepared the immunoprecipitation, immunostaining, live-cell images, and cell tracking. A.K. performed the cloning of constructs, alignments, and TNT experiments. S.K. constructed the Xcad-11-EGFP clone. D.A. developed Xcad-11 antibody.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.519409.
Supplemental material is available at http://www.genesdev.org.
References
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Hoffstrom BG, DeSimone DW. Integrin α5β1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev Biol. 2003;260:449–464. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Reynolds AB. The p120 catenin family: Complex roles in adhesion, signaling and cancer. J Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- Backer S, Hidalgo-Sanchez M, Offner N, Portales-Casamar E, Debant A, Fort P, Gauthier-Rouviere C, Bloch-Gallego E. Trio controls the mature organization of neuronal clusters in the hindbrain. J Neurosci. 2007;27:10323–10332. doi: 10.1523/JNEUROSCI.1102-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J, Van Vactor D. The Trio family of guanine-nucleotide-exchange factors: Regulators of axon guidance. J Cell Sci. 2001;114:1973–1980. doi: 10.1242/jcs.114.11.1973. [DOI] [PubMed] [Google Scholar]
- Boguslavsky S, Grosheva I, Landau E, Shtutman M, Cohen M, Arnold K, Feinstein E, Geiger B, Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci. 2007;104:10882–10887. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers A, Epperlein HH, Wedlich D. An assay system to study migratory behavior of cranial neural crest cells in Xenopus . Dev Genes Evol. 2000;210:217–222. doi: 10.1007/s004270050307. [DOI] [PubMed] [Google Scholar]
- Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, Yu-Lee LY, Zhang S, Yeh ET, Hu MC, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6:1259–1267. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour S, Saint-Jeannet JP, Broders F, Wedlich D, Thiery JP. Differential perturbations in the morphogenesis of anterior structures induced by overexpression of truncated XB- and N-cadherins in Xenopus embryos. J Cell Biol. 1994;127:521–535. doi: 10.1083/jcb.127.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadeball B, Borchers A, Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wg/Wnt signal. Mech Dev. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. Programmed cell death during Xenopus development: A spatio-temporal analysis. Dev Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Heupel WM, Baumgartner W, Laymann B, Drenckhahn D, Golenhofen N. Different Ca2+ affinities and functional implications of the two synaptic adhesion molecules cadherin-11 and N-cadherin. Mol Cell Neurosci. 2008;37:548–558. doi: 10.1016/j.mcn.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: Biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Kulesa PM, Lefcort F. Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development. 2005;132:235–245. doi: 10.1242/dev.01553. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Matsunami H, Inoue T, Shimamura K, Uchida N, Ueno T, Miyazaki T, Takeichi M. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev Biol. 1995;169:347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. α-Catenin: At the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kunz M, Herrmann M, Wedlich D, Gradl D. Autoregulation of canonical Wnt signaling controls midbrain development. Dev Biol. 2004;273:390–401. doi: 10.1016/j.ydbio.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Kuriyama S, Mayor R. Molecular analysis of neural crest migration. Philos Trans R Soc Lond B Biol Sci. 2008;363:1349–1362. doi: 10.1098/rstb.2007.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lievre CS. Participation of neural crest-derived cells in the genesis of the skull in birds. J Embryol Exp Morphol. 1978;47:17–37. [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: Analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lee DM, Kiener HP, Agarwal SK, Noss EH, Watts GF, Chisaka O, Takeichi M, Brenner MB. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007;315:1006–1010. doi: 10.1126/science.1137306. [DOI] [PubMed] [Google Scholar]
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Mol Biol Cell. 2009;20:78–89. doi: 10.1091/mbc.E08-05-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, et al. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Pishvaian MJ, Feltes CM, Thompson P, Bussemakers MJ, Schalken JA, Byers SW. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–952. [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schuff M, Rossner A, Wacker SA, Donow C, Gessert S, Knöchel W. FoxN3 is required for craniofacial and eye development of Xenopus laevis . Dev Dyn. 2007;236:226–239. doi: 10.1002/dvdy.21007. [DOI] [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Tomita K, van Bokhoven A, van Leenders GJ, Ruijter ET, Jansen CF, Bussemakers MJ, Schalken JA. Cadherin switching in human prostate cancer progression. Cancer Res. 2000;60:3650–3654. [PubMed] [Google Scholar]
- Valencia X, Higgins JM, Kiener HP, Lee DM, Podrebarac TA, Dascher CC, Watts GF, Mizoguchi E, Simmons B, Patel DD, et al. Cadherin-11 provides specific cellular adhesion between fibroblast-like synoviocytes. J Exp Med. 2004;200:1673–1679. doi: 10.1084/jem.20041545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin J, Girault JM, Thiery JP, Broders F. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mech Dev. 1998;75:171–174. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]