Abstract
We reported previously that when cells are arrested in S phase, a subset of p53 target genes fails to be strongly induced despite the presence of high levels of p53. When DNA replication is inhibited, reduced p21 mRNA accumulation is correlated with a marked reduction in transcription elongation. Here we show that ablation of the protein kinase Chk1 rescues the p21 transcription elongation defect when cells are blocked in S phase, as measured by increases in both p21 mRNA levels and the presence of the elongating form of RNA polymerase II (RNAPII) toward the 3′ end of the p21 gene. Recruitment of specific elongation and 3′ processing factors (DSIF, CstF-64, and CPSF-100) is also restored. While additional components of the RNAPII transcriptional machinery, such as TFIIB and CDK7, are recruited more extensively to the p21 locus after DNA damage than after replication stress, their recruitment is not enhanced by ablation of Chk1. Significantly, ablating Chk2, a kinase closely related in substrate specificity to Chk1, does not rescue p21 mRNA levels during S-phase arrest. Thus, Chk1 has a direct and selective role in the elongation block to p21 observed during S-phase arrest. These findings demonstrate for the first time a link between the replication checkpoint mediated by ATR/Chk1 and the transcription elongation/3′ processing machinery.
Keywords: Chk1, Chk2, S-phase checkpoint, p53, transcription elongation
The initial stage of the eukaryotic transcription cycle is the recruitment of the RNA polymerase II (RNAPII) enzyme, complexed with general transcription factors (GTFs), to the region of the transcription start site to form a preinitiation complex (PIC) (Orphanides et al. 1996). Recent genome wide analysis of RNAPII occupancy has revealed that the PIC is preformed on a large number of inactive genes, suggesting that the rate-limiting step for transcription activation at these loci is the conversion of a preloaded, paused RNAPII into an elongation-competent form (Kumar et al. 1998; Muse et al. 2007). This mode of regulation is prevalent among inducible genes activated by developmental cues, various cell signaling pathways, and stress stimuli. Some, but not all, p53 target genes are regulated in this fashion (Espinosa et al. 2003; Gomes et al. 2006). Phosphorylation of the RNAPII C-terminal domain (CTD) heptad (YSPTSPS) repeats seems to play an important role in this process. For example, phosphorylation of Ser5 within the CTD heptad occurs at the 5′ end of genes, increases upon transcriptional activation, and is necessary for the 5′ mRNA capping enzymes to bind RNAPII (Komarnitsky et al. 2000). Protein kinases known to mediate Ser5 phosphorylation include CDK7 (associated with the TFIIH general transcription factor), CDK8 (associated with the Mediator complex) and CDK9 (subunit of the positive transcription elongation factor, P-TEFb) (Oelgeschlager 2002). The precise contribution of these and other kinases to Ser5 phosphorylation and regulation of early elongation steps in mammalian cells remains to be elucidated. Chemical genetic inhibition of CDK7, pharmacological inhibition of CDK9 with DRB, and knockdown of CDK8 do not significantly reduce levels of Ser5 phosphorylation in vivo, revealing the action of multiple, possibly redundant Ser5 kinases in the cell nucleus (Gomes et al. 2006; Larochelle et al. 2007; Knuesel et al. 2009). This regulatory abundance may facilitate exquisite promoter- and stimulus-specific control of gene expression after PIC assembly (Luecke and Yamamoto 2005; Gomes et al. 2006).
Many non-GTF proteins exist that regulate mammalian transcription at both the initiation and elongation phases. Yamaguchi et al. (1999) have characterized the negative elongation factors that are present at the promoters of RNAPII-transcribed genes. DRB sensitivity-inducing factor (DSIF), a dimer of SPT4–SPT5, is required for the effects of DRB inhibition on CDK9 in vitro (Yamaguchi et al. 1999). DSIF and negative elongation factor (NELF) function synergistically to pause RNAPII at the promoter, and their inhibition is relieved upon phosphorylation of DSIF by CDK9 (Yamada et al. 2006). NELF and DSIF are also likely to play gene-specific effects in cells (Gomes et al. 2006; Gilchrist et al. 2008). As it progresses toward the 3′ end of the gene, RNAPII becomes phosphorylated at Ser2 of its CTD heptad repeat. CDK9 is the likely kinase responsible for this modification (Phatnani and Greenleaf 2006). Intimate links also exist between transcription and RNA processing (Komarnitsky et al. 2000; Pandit et al. 2008). Notably, the phosphorylated CTD of RNAPII serves to recruit the CstF and CPSF complexes that are necessary for mRNA cleavage and polyadenylation (Hirose and Manley 2000; SH Ahn et al. 2004; Proudfoot 2004).
Arguably, the p53 protein's most important function is to act as a transcription factor that regulates a specific subset of the cell's RNAPII-transcribed genes. These p53 target genes produce multiple outcomes such as cell cycle arrest (e.g., p21, GADD45, and 14–3–3), apoptosis (e.g., pig3, FAS, PUMA, and NOXA), senescence (e.g., PAI-1), autophagy (e.g., DRAM 1), and others, or, they can regulate the p53 pathway itself (e.g., MDM2) (Murray-Zmijewski et al. 2008). P53 binds as a tetramer to its response element where it can recruit a plethora of transcriptional coregulators such as histone modifying enzymes, chromatin remodeling factors, and/or subunits of the Mediator complex to modulate RNAPII activity at target loci (Laptenko and Prives 2006).
The p53 transcriptional program is regulated in a stimulus-specific fashion (Zhao et al. 2000; Donner et al. 2007a; Murray-Zmijewski et al. 2008). Distinct subsets of p53 target genes are induced in response to different p53-activating agents, which may allow cells to tailor their response to different types of stress. We reported previously that p53 is transcriptionally impaired in inducing certain target genes such as p21, mdm2, and cyclin G when cells are arrested in S phase after treatment with hydroxyurea (HU), an inhibitor of the ribonucleotide reductase enzyme, or aphidicolin, that blocks DNA polymerase (Gottifredi et al. 2001). In contrast, these target genes are efficiently activated in response to daunorubicin-mediated DNA damage. At the p21 locus, this phenomenon correlates with, and likely results from, a block in transcription elongation (Mattia et al. 2007), but the precise mechanism driving differential p21 expression remains to be elucidated.
DNA damaging agents frequently activate the PI3K family of sensor kinases, which include ATM and ATR (Traven and Heierhorst 2005). In response to DNA damage, ATM/ATR activate the kinases Chk1 and Chk2 (Niida and Nakanishi 2006). Both ATM/ATR and Chk1/Chk2 phosphorylate p53, on Ser15 and Ser20, respectively (Bartek and Lukas 2003), although p53 can still accumulate after HU or other treatments if Chk1 and Chk2 have been down-regulated (Ahn et al. 2003; Jallepalli et al. 2003; Ho et al. 2006). In mammalian cells, it is predominantly Chk1 that is activated by ATR in response to S-phase arrest, although Chk2 has been shown to be activated by ATR under certain conditions (Feijoo et al. 2001; Ho et al. 2006). When activated during S phase, Chk1 activity leads to hyperphosphorylation of Cdc25A and the subsequent inhibition of the CDK2/cyclin E complex (Sorensen et al. 2003; Xiao et al. 2003). This results in decreased DNA replication origin firing, activation of DNA repair pathways, the stabilization of stalled replication forks, and the arrest of cells in S phase (Chen and Sanchez 2004). Chk1-null mice die early in development (Takai et al. 2000), indicating that the proper regulation of these processes by Chk1 is not only necessary in response to exogenous DNA damage, but is also required during unperturbed S phase (Petermann and Caldecott 2006). In addition to its well-established roles in checkpoint control, Chk1 has been shown to have noncanonical roles in various cellular pathways including alternative splicing, phospho-histone H3-dependent transcriptional repression, and the suppression of a caspase-2-dependent apoptotic response to IR (Katzenberger et al. 2006; Shimada et al. 2008; Sidi et al. 2008).
In this study, we establish a novel link between S-phase arrest signaling and transcriptional elongation control at the p21 gene. Specifically, we demonstrate that Chk1, but not Chk2, mediates the transcriptional elongation block at the p21 locus in response to HU treatment. Our results demonstrate that signaling cascades triggered by different genotoxic insults regulate the p53 transcriptional program at post-RNAPII recruitment steps.
Results
Ablation of ATR rescues p21 protein and mRNA levels when DNA replication is blocked
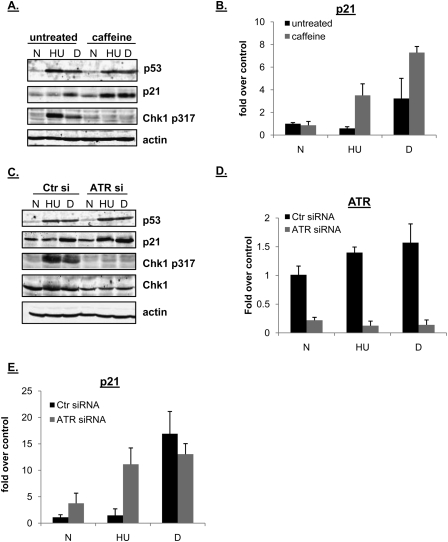
We showed previously that treatment of RKO or HCT116 cells with HU or daunorubicin (dauno) induces the accumulation of p53 protein to roughly the same level, but p21 mRNA and protein fail to accumulate significantly after HU (Gottifredi et al. 2001) and the small amount of p21 mRNA that does accumulate after HU is p53-independent (Mattia et al. 2007). Since ATR is the main sensor kinase responsible for detecting stalled replication forks (Paulsen and Cimprich 2007), we investigated its role in our system. RKO cells were treated with caffeine, a compound that inhibits ATR (as well as ATM, although less specifically) followed by treatment with either HU or dauno (Fig. 1A,B). As expected, treatment with HU alone led to markedly less p21 protein or mRNA than seen with dauno treatment, as well as increased levels of Chk1 phosphorylated at Ser317. On the other hand, significantly increased levels of p21 protein and mRNA accumulated in cells that were treated with both HU and caffeine. P21 mRNA was also increased when cells were treated in combination with caffeine and dauno. We tested whether this increase was due to the ability of caffeine to also inhibit ATM that is activated by treatment with dauno. ATR was down-regulated by siRNA and cells were then treated with HU or dauno as above. The efficiency of ATR knockdown was measured both by the reduction in Chk1 phosphorylation at Ser317 (Fig. 1C) and the decrease in steady-state ATR mRNA levels (Fig. 1D). While levels of Chk1 were consistently reduced after ATR knockdown, p53 accumulation after HU or dauno was unaffected by ATR ablation, and p21 protein (Fig. 1C) and mRNA (Fig. 1E) levels increased significantly after the combination of HU treatment and ATR ablation. Similar results were obtained in HCT116 cells (data not shown). Note that although there was more p21 mRNA in untreated cells containing ATR siRNA compared with the control, the relative increase was considerably greater when cells were also treated with HU. Markedly, the combination of ATR ablation and dauno treatment did not cause an increase in p21 mRNA over and above treatment with dauno alone.
Figure 1.
ATR ablation increases p21 protein and mRNA levels when DNA replication is stalled. (A) RKO cells were pretreated or not with 4 mM caffeine for 1 h, and then left untreated (N), treated with HU (HU, 1.7 mM, 24 h) or dauno (D, 0.22 μM, 8 h), lysed, and subjected to immunoblot analysis with indicated antibodies. (B) RKO cells were treated as in A; RNA was extracted and subjected to qRT–PCR using primers for the p21 mRNA transcript. Values were normalized to those of the hprt1 gene. Graphs are representative of at least six independent PCRs from three independent experiments. In all experiments, error bars included in the graphs represent one standard deviation from the mean. (C) RKO cells were transfected with control (Ctr) or ATR siRNA duplexes for 36 h and then drug-treated with HU or dauno as in A. The cells were lysed and subjected to immunoblot analysis with antibodies as indicated in the figure. (D,E) RKO cells were treated as in C, and RNA was extracted and subjected to qRT–PCR as in B using primers for the p21 or ATR mRNA transcript.
These results imply a specific role for ATR, but not ATM, in negatively regulating p21 transcription.
Chk1 ablation rescues p21 protein and mRNA levels during the S-phase checkpoint
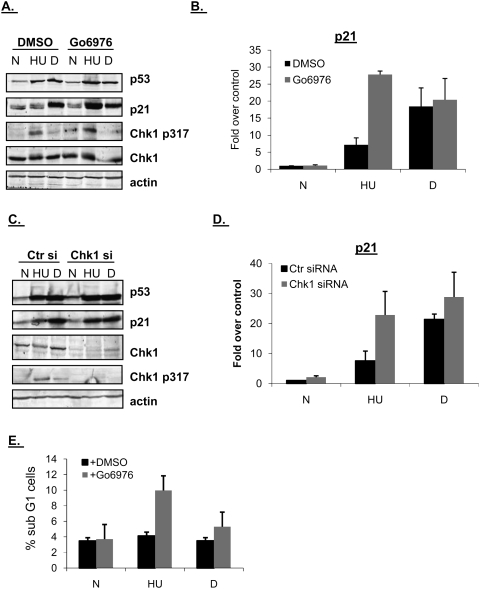
When DNA replication is blocked, ATR activates Chk1 by phosphorylating it on Ser317 and Ser345 leading to S-phase arrest (Liu et al. 2000; Zhao and Piwnica-Worms 2001). To determine whether ATR's role in negatively regulating p21 transcription was dependent on Chk1, HCT116 cells were pretreated with the Chk1 inhibitor Gö6976 (Kohn et al. 2003), followed by treatment with HU or dauno. Gö6976 did inhibit Chk1 activity as assayed by the phosphorylation status of Cdc25C at Ser216 (Supplemental Fig. S1), a substrate of Chk1 even in the absence of DNA damage (Kaneko et al. 1999). P53 accumulation after HU or dauno was virtually unaffected by pretreatment with Gö6976. Treatment of cells with Gö6976 along with HU, however, resulted in dramatic increases in the levels of p21 protein (Fig. 2A) and mRNA (Fig. 2B). Note that while HCT116 cells show a slightly stronger p21 response to HU than RKO cells (Fig. 1A–D), at equivalent levels of p53, the p21 response in HCT116 cells is significantly impaired when compared with the response to daunorubicin treatment (Fig. 2A,B). Note that the level of p21 mRNA was unchanged when pretreatment with Gö6976 was combined with dauno treatment (Fig. 2B), presumably because dauno leads to the activation of the ATM/Chk2 signaling cascade, and thus the inhibition of Chk1 would have little effect in this case. RKO cells provided essentially similar results (Supplemental Fig. S2A,B). Chk1 inhibition by Gö6976 did not cause significant p21 mRNA or protein accumulation in p53-null HCT116 cells, indicating that this rescue was p53-dependent (Supplemental Fig. S3A,B). In HCT116 cells, HU but not dauno treatment induced phosphorylation of Chk1 on Ser317, which was further increased after Gö6976 treatment (Fig. 2A), in line with other reports that Chk1 inhibition leads to decreased activity of the Chk1-activated PP2A phosphatase (Leung-Pineda et al. 2006).
Figure 2.
Chk1 ablation rescues p21 protein and mRNA levels during S-phase arrest in HCT116 cells. (A) HCT116 cells were pretreated with DMSO or 1 μM Gö6976 for 1 h, and then left untreated (N), treated with HU (HU, 1.7 mM, 24 h) or dauno (D, 0.22 μM, 8 h), lysed, and subjected to immunoblot analysis with antibodies as indicated in the figure. (B) HCT116 cells were treated as in A; RNA was extracted and subjected to qRT–PCR using primers for the p21. Values were normalized to those of the hprt1 gene. (C) HCT116 cells were transfected with control (Ctr) or Chk1 siRNA duplexes for 36 h and then drug-treated with HU or dauno as in A. Cells were lysed and subjected to immunoblot analysis with antibodies as indicated in the figure. (D) HCT116 cells were treated as in C, RNA was extracted and subjected to qRT–PCR as in B. (E) FACS analysis was performed on HCT116 cells that were subjected to different combinations of drug treatments as in A and the data was analyzed using the ModFit program.
We next sought to support these results with Chk1 pharmacological inhibition by using RNAi. Down-regulation of Chk1 by siRNA led to a ∼90% decrease in protein levels (Fig. 2C). Chk1 ablation did not affect p53 accumulation after treatment with HU or dauno, but, as with Chk1 pharmacological inhibition, p21 protein (Fig. 2C) and mRNA (Fig. 2D) levels were increased when Chk1 ablation was combined with HU treatment. Similar results were obtained in RKO cells (Supplemental Fig. S2C,D).
Notably, our results with Chk1 inhibition were supported by cell cycle profiles of the various treated cells. HU-treated HCT116 cells arrest at the G1/S transition while dauno arrests cells in G2 (Gottifredi et al. 2001). Here we found that pretreatment of HCT116 cells with Gö6976 and subsequent treatment with HU led to a significant increase in apoptosis as measured by the subG1 population of cells. Apoptosis was much less pronounced when Chk1 inhibition was combined with dauno treatment (Fig. 2E). Upon treatment with HU and subsequent release by removal of this inhibitor, cells with low levels of p21 can proceed normally through the cell cycle, whereas cells with high levels of p21 in S phase are unable to do so (Gottifredi et al. 2004). Since Chk1 inhibition in combination with HU treatment induced high levels of p21 protein in S phase (Fig. 2A), we wanted to determine whether cells with chemically inactivated Chk1 could recover from S-phase arrest. Others have reported that Chk1 is necessary for cells to recover from double thymidine block (Rodriguez and Meuth 2006). In line with this, RKO cells pretreated with the Chk1 inhibitor Gö6976 could not recover from an HU-induced S-phase block once HU had been removed, as measured by the population of cells able to proceed to G2 phase (Supplemental Fig. S5).
These results strongly implicate Chk1 as being critical for the blunting of the p53 response that occurs when DNA replication is stalled.
Unlike Chk1, Chk2 exerts a positive effect on p21 transcription in stressed cells
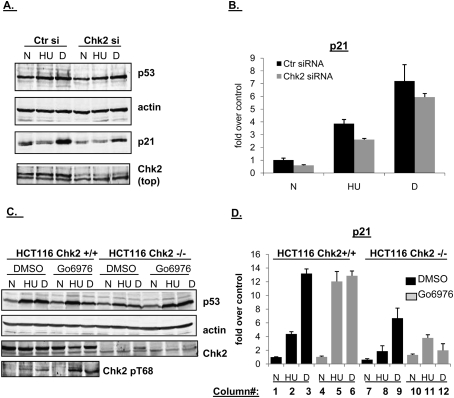
Chk1 and Chk2 have similar (but not identical) phosphorylation consensus sequences (O'Neill et al. 2002) and in certain scenarios, replication stress has been shown to activate the Chk2 pathway in addition to the ATR–Chk1 pathway (Feijoo et al. 2001; Ho et al. 2006). Therefore, we examined Chk2's contribution to the block to p21 transcription during S-phase arrest by ablating Chk2 by siRNA or using HCT116 cells in which both alleles were deleted. Chk2 siRNA resulted in significant reduction of Chk2 protein levels and did not significantly affect p53 levels when cells were treated with HU or dauno (Fig. 3A). Yet, in marked contrast to our results with Chk1 inhibition, Chk2 siRNA did not rescue p21 protein (Fig. 3A) or mRNA (Fig. 3B) accumulation after HU treatment. Even more compellingly, in HCT116 cells in which both Chk2 alleles have been disrupted by homologous recombination (Jallepalli et al. 2003), there was again no increase in p21 mRNA accumulation after replication stress (Fig. 3D, cf. lanes 2 and 8). Indeed, Chk2 was apparently required for full activation of the p21 gene since in the HCT116 Chk2-null cells, accumulation of p21 mRNA was impaired after both HU and dauno, regardless of Chk1 inhibition status (Fig. 3D). Importantly, when both Chk1 and Chk2 were inhibited (by treating Chk2-null cells with Gö6976), the p21 transcriptional response was muted after treatment with HU (with respect to the HCT116 Chk2 +/+ cells) (Fig. 3D, cf. lanes 5 and 11), but levels of p21 mRNA were still increased about twofold when compared with Chk2-null cells treated with HU alone (Fig. 3D, cf. lanes 8 and 11). Thus, Chk1 ablation rescued the transcriptional defect seen with HU even in the background of Chk2 deprivation.
Figure 3.
Unlike Chk1, Chk2 exerts a positive effect on p21 transcription. (A) HCT116 cells were transfected with control or Chk2 siRNA duplexes for 36 h, and then left untreated (N), treated with HU (HU, 1.7 mM, 24 h) or dauno (D, 0.22 μM, 8 h), lysed, and subjected to immunoblot analysis with antibodies as indicated in the figure. (B) HCT116 cells were treated as in A, and RNA was extracted and subjected to qRT–PCR using primers for p21. Values are normalized to those of hprt1. (C) HCT116 Chk2+/+ or Chk2-null cells were treated as in Figure 1A, lysed, and subjected to immunoblot analysis with the indicated antibodies. Note that the extra band in the dauno-treated Chk2-null cells is nonspecific. (D) HCT116 Chk2+/+ or Chk2-null cells were treated as in C and RNA was extracted and subjected to qRT–PCR using primers for p21 as in B.
It is acknowledged that when Chk2 siRNA was used with HCT116 cells there was only a modest decrease in p21 mRNA accumulation (Fig. 3B), and there was no reduction at all in Chk2 siRNA-treated RKO cells (data not shown). This could be due to the inability to fully ablate Chk2 using siRNA and may reflect cell type-specific differences as well.
Double-strand breaks caused by various agents activate ATM, leading to phosphorylation of Chk2 at Thr68, after which Chk2 can oligomerize and phosphorylate itself in cis or in trans (Schwarz et al. 2003; J Ahn et al. 2004). As previously shown (Ho et al. 2006), phosphorylation of Chk2 on Thr68 was similar after replication stress and DNA damage (Fig. 3C; Supplemental Fig. S4). Chk2 Thr68 phosphorylation was increased when either treatment was combined with Chk1 inhibition (Fig. 3C; Supplemental Fig. S4), or when cells were treated with HU and dauno in combination (Supplemental Fig. S4). This is perhaps reflective of the increased levels of DNA damage in these cells. Note also that treating cells with HU and dauno in combination did not increase levels of p21 protein (Supplemental Fig. S4) or RNA (Mattia et al. 2007) above the levels seen with treatment with HU alone. Thus, activation of Chk2 alone is not sufficient to induce strong transcription of p21.
We conclude that Chk2 activity is not responsible for the block to p21 expression when cells are arrested in S phase, rather our data indicate that, at least in some cells, Chk2 activation is necessary, but not sufficient, for full transcriptional induction of the p21 gene.
Association of p53 with the p21 promoter is not affected by Chk1 inhibition
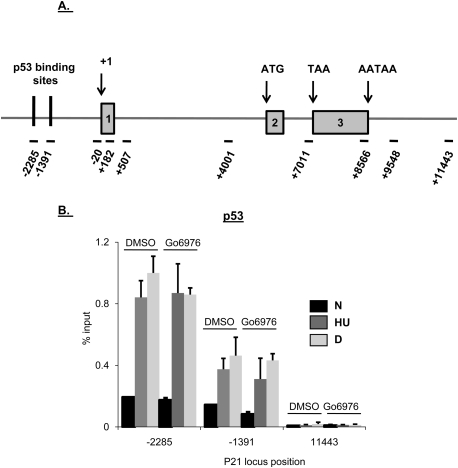
A possible explanation for the increased levels of p21 transcription after Chk1 inhibition in S-phase-arrested cells could be increased p53 recruitment to the p21 promoter. We showed previously that in RKO cells, p53 recruitment to p21, and also to other p53 target genes, is equivalent after treatment with HU or dauno (Mattia et al. 2007). Here, HCT116 cells treated or not with Gö6976 and either HU or dauno were analyzed by quantitative chromatin immunoprecipitation (q-ChIP) using p53-specific antibodies. DNA was amplified for both p53-binding sites on p21 (−2285 and −1391) and a region 3′ to the p21 polyadenylation signal (+11,443) was used as a negative control. Figure 4A depicts the location of the primers used on the p21 locus in this and subsequent experiments. Consistent with other reports (Espinosa et al. 2003; Mattia et al. 2007), the distal p21-binding site recruited ∼2.5-fold more p53 than the proximal binding site (Fig. 4B). In either case p53 recruitment was unaffected by Chk1 inhibition. Thus, enhanced p53 recruitment to the p21 promoter is not responsible for the increase in p21 transcription seen after the combination of Chk1 inhibition and HU treatment.
Figure 4.
Chk1 inhibition does not affect p53 recruitment to the p21 promoter. (A) Diagram depicting the primers used to amplify the different regions of p21 for ChIP and nascent RNA transcript analyses. (B) ChIP was performed in HCT116 cells following drug treatment as in Figure 2A. In all ChIP experiments, cells were cross-linked in 1% formaldehyde, lysed in RIPA buffer, and sonicated to produce chromatin fragments of ∼500 base pairs. Samples were then immunoprecipitated with anti-p53 monoclonal antibodies (1801 and DO-1). Five percent of the input protein was subjected to immunoblot analysis. ChIP-enriched DNA was quantified by qRT–PCR using the indicated amplicons on p21. Values are expressed as percentage of input DNA immunoprecipitated, normalized to the highest immunoprecipitation signal. For all qChIPs, graphs represent at least six independent PCRs from three separate immunoprecipitations and cell cultures.
Chk1 inhibition stimulates distal transcription elongation on the p21 promoter after HU treatment
After certain types of stress, such as UV, global transcription initiation is blocked due to the depletion of the pool of unphosphorylated RNAPII molecules that are competent for PIC formation (Heine et al. 2008). We showed in both a semiquantitative fashion (Mattia et al. 2007) and by q-ChIP (Supplemental Fig. S6A) that RNAPII is roughly equivalently recruited to the p21 promoter after HU or dauno treatment, indicating that if this global block to transcription initiation occurs after HU treatment, it is not affecting p21. Furthermore, Chk1 inhibition did not significantly affect RNAPII recruitment to p21 after HU or dauno, although it did increase the basal levels of RNAPII recruited to this promoter, possibly because Chk1 depletion itself may be a form of stress to cells (Supplemental Fig. S6A).
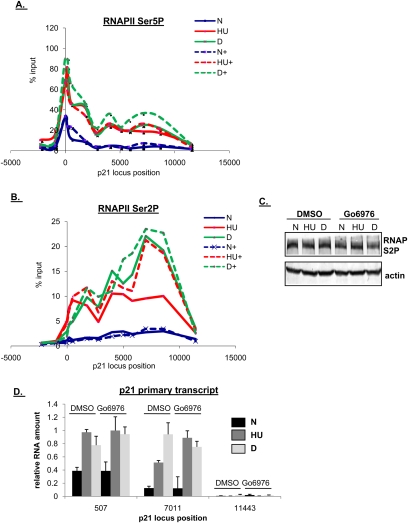
We next determined whether impaired promoter clearance could account for the weak p21 response seen when cells are blocked in S phase. Phosphorylation of the CTD at Ser5 is a hallmark of promoter clearance (Kumar et al. 1998). Using a high-resolution q-ChIP tiling array of the p21 locus with an antibody against this modification, we found that levels of RNAPII Ser5P at the p21 promoter were equivalent after both drug treatments and were unaffected by Chk1 inhibition in either case (Fig. 5A).
Figure 5.
Chk1 inihibition stimulates distal transcription elongation on the p21 locus after HU treatment. (A–C) ChIP was performed in HCT116 cells following treatment as in Figure 2A. Cells were cross-linked in 1% formaldehyde, lysed in RIPA buffer, and sonicated. Lysates were precleared and then immunoprecipitated with antibodies recognizing RNAPII phosphorylated at Ser5 (A) or Ser2 (B) of its heptad repeat, as indicated in the figure. Solid lines refer to samples pretreated with DMSO; dashed lines refer to samples pretreated with Gö6976. Quantification of ChIP-enriched DNA and replicates were performed as in Figure 4. (C) Five percent of the input protein was subjected to immunoblot analysis. (D) HCT116 cells were treated as above; RNA was extracted and subjected to qRT–PCR using primers recognizing different regions of the nascent p21 transcript. Values were normalized to those of hprt1.
As RNAPII transits a gene locus, it becomes increasingly phosphorylated on Ser2 of its CTD heptad repeats. Using a similar tiling array as for examination of RNAPII Ser5P, we found that after dauno treatment, the amount of RNAPII phosphorylated at Ser2 progressively increased along the p21 locus (Fig. 5B). In marked contrast, after HU treatment, levels of Ser2-phosphorylated RNAPII failed to increase along the length of the gene (Fig. 5B). As a result, the elongating, Ser2-phosphorylated form of RNAPII was significantly less abundant at the very distal p21 region (3′ to approximately +6000 kb) after HU treatment when compared with cells treated with dauno (Fig. 5B, cf. solid red line and solid green line). This was not due to a change in the overall levels of Ser2-phosphorylated RNAPII (Fig. 5C). Most importantly, the progressive increase in Ser2-phosphorylated RNAPII observed in HU-treated cells was completely restored when Chk1 inhibition was combined with replication stress (Fig. 5B, cf. solid and dashed red lines). Similar results were observed using Chk1 siRNA, although the effect was not as dramatic, most likely due to the residual levels of Chk1 protein that remained after knockdown by siRNA (Supplemental Fig. S6B).
To confirm that the defect in p21 mRNA accumulation was well correlated with a deficiency in RNA elongation, we assessed the presence of specific regions of the nascent p21 RNA transcript after the combinations of drug treatments described above. Total RNA from HCT116 cells was amplified for regions that are exclusively present in the nascent p21 transcript (i.e., introns that are spliced out of mature p21 mRNA). Short p21 nascent transcripts, as assayed for by an amplicon close to the transcription start site but within the first intron of the gene (+507), were equally present after HU or daunorucibin treatment and little affected by Go6976 treatment (Fig. 5D). Note that we do not believe the +507 amplicon is significantly present in the longer nascent transcripts due to cotranscriptional splicing. However, longer p21 RNA transcripts, as assayed for by an amplicon in p21 located just before the third exon (+7011), weakly accumulate after HU treatment, but are strongly induced after the combination of Gö6976 and HU treatment (Fig. 5D). A region not expected to be present in either the nascent or mature mRNA transcript of p21 (+11,443, after the polyadenylation site) was amplified as a negative control. As a positive control, an amplicon was assayed that is present in the mature p21 mRNA transcript (+8566). As expected, the signal from the positive control was ∼100-fold stronger than the signals from the primary transcript amplifications (data not shown).
Taken together, these data imply that Chk1 functions to block p21 transcription elongation specifically at the distal end of the gene.
Chk1 inhibition does not affect assembly of general transcription initiation factors at the p21 promoter when cells are blocked in S phase
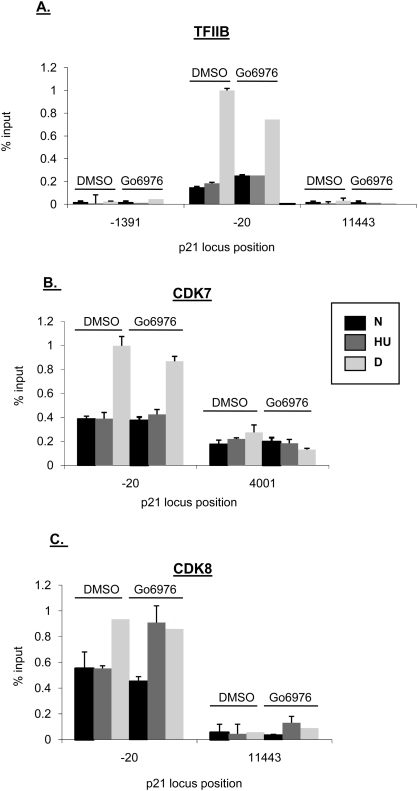
It is conceivable that the increased transcription resulting from the combination of Chk1 inhibition and HU treatment is caused by augmented PIC assembly at the p21 promoter, which could indicate a greater ability for RNAPII to reinitiate transcription. Indeed, two components of the PIC, TFIIB and TFIIH/CDK7, were strongly recruited to the p21 promoter after dauno but not HU treatment (Fig. 6A,B, respectively), although their levels in cells were unchanged under the two conditions (data not shown). Nevertheless, in neither case was their differential recruitment rescued by Chk1 inhibition. Other components of the PIC, such as TBP, showed little difference in recruitment after either HU or dauno (Mattia et al. 2007). Since neither TFIIB nor TFIIH recruitment was affected after the combination of HU treatment and Chk1 inhibition, it is unlikely that other components of the basal transcription machinery are responsible for these increased levels of p21 mRNA.
Figure 6.
Chk1 inhibition does not affect PIC assembly at the p21 promoter when cells are blocked in S phase. (A–C) ChIP was performed in HCT116 cells following treatment as in Figure 2A. Cells were cross-linked in 1% formaldehyde, lysed in RIPA buffer, and sonicated. Lysates were precleared and then immunoprecipitated with antibodies as indicated in the figure. Five percent of the input protein was subjected to immunoblot analysis. Quantification of ChIP-enriched DNA and replicates were performed as in Figrue 4.
CDK7 is the canonical kinase that phosphorylates the CTD of RNAPII at Ser5. RNAPII Ser5P levels were equivalent at the p21 promoter region after HU and dauno, and were unaffected by Chk1 inhibition (Fig. 5A). Yet TFIIH/CDK7 was not significantly recruited to p21 after HU treatment, in contrast to its increased presence after dauno treatment (Fig. 6B). This implies that another kinase substitutes for CDK7 at p21 after HU treatment. One such kinase reported to phosphorylate RNAPII at Ser5, CDK8, was also strongly recruited to the 5′ region of the p21 locus after dauno but not HU treatment; however, unlike CDK7, CDK8 recruitment was increased after inhibiting Chk1 and treating cells with HU (Fig. 6C). This is consistent with another report that recruitment of CDK8 correlates positively with strong p21 transcription (Donner et al. 2007b). Nevertheless, since there was no difference in Ser5 phosphorylation under the different conditions tested, it is unlikely that differential recruitment of CDK8 affects p21 transcription via modification of this residue. It is unclear at this point how CDK8 may contribute to Ser2 phosphorylation and late elongation steps at this gene locus.
These data together indicate that on the p21 gene, S-phase arrest affects PIC formation in a Chk1-independent fashion, and that a kinase other than CDK7 or CDK8 phosphorylates RNAPII at Ser5 after HU treatment.
Chk1 inhibition selectively alters the recruitment of key factors to the 3′ end of the p21 gene during S-phase arrest
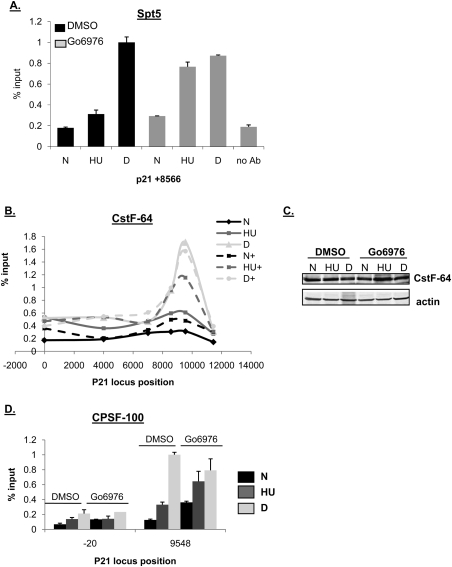
We asked whether increased RNAPII pausing at the transcription initiation region of the p21 promoter was responsible for the decreased transcription from this locus after HU treatment. In unstressed cells, low amounts of RNAPII are poised in the p21 TATA region, which facilitates rapid transcription of this gene upon DNA damage (Espinosa et al. 2003). In the absence of stress, Spt5/DSIF functions, in concert with NELF, to pause RNAPII at certain promoters such as hsp70, jun-B, and p21 (Wu et al. 2003; Aida et al. 2006; Gomes et al. 2006). Recent work has also demonstrated a positive role for DSIF, but not NELF, in the regulation of transcription elongation where, upon phosphorylation by P-TEFb, Spt5 travels into the body of the gene with RNAPII (Cheng and Price 2007). In our experiments, neither Spt5 (Supplemental Fig. S6C) nor NELF (data not shown) were differentially recruited to the p21 TATA region under any combination of stresses examined. Thus, after HU treatment increased transcriptional pausing of RNAPII in the TATA region of p21 is not responsible for the block to p21 transcription.
Despite the equivalent recruitment of Spt5 to the p21 promoter after HU and dauno, at the distal end (+8566) of the p21 gene its presence was significantly reduced after HU treatment, and this reduction was attenuated in the presence of Chk1 inhibition (Fig. 7A). As Spt5 tracks with RNAPII, this is perhaps reflective of the increased transiting of the polymerase through this region under these conditions.
Figure 7.
Chk1 inhibition selectively alters the recruitment of certain factors to the 3′ end of the p21 gene after HU treatment. (A–D) ChIP was performed in HCT116 cells following treatment as in Figure 2A. Cells were cross-linked in 1% formaldehyde, lysed in RIPA buffer, and sonicated. Lysates were precleared and then immunoprecipitated with antibodies as indicated in the figure. Five percent of the input protein was subjected to immunoblot analysis. Quantification of ChIP-enriched DNA and replicates were performed as in Figure 4.
Phosphorylation of RNAPII at Ser2 of its CTD heptad repeat has been shown to provide a docking point for several proteins that regulate transcription elongation (Hampsey and Reinberg 2003). One such example is the Set2 methyltransferase, which trimethylates histone H3 on Lys36. In metazoans high levels of this modification are a mark of efficiently transcribed genes (Bannister et al. 2005). Consistently, this histone modification was increased in cells ablated for Chk1 that were treated with HU (Supplemental Fig. S6D).
Because the block to p21 transcription elongation after HU was limited to the far distal region of the gene within 1–2 kb of the polyadenylation site, we determined whether Chk1 had any effect on the localization of 3′ mRNA processing factors to this region. Certain subunits of the metazoan CPSF and CstF components of polyadenylation complexes—for example, CPSF-100 and CstF-64—can be recruited to active promoters via interaction with general transcription factors (such as TFIID) and then remain associated with RNAPII as it traverses through the gene (Dantonel et al. 1997; Calvo and Manley 2003; Glover-Cutter et al. 2008). When we examined CstF-64 association with p21, we found that it was recruited equivalently to the TATA region after HU and dauno treatment, and that its recruitment to this region was unaffected by Chk1 inhibition (Fig. 7B). In contrast, at the distal region of p21 (i.e., beginning at approximately +7 kb from the transcription start site and extending to 1 kb downstream from the polyadenylation site), there was a significant increase in association of CstF-64 in dauno-treated cells, while there was no such increase in CstF-64 detected after HU treatment (Fig. 7B). This could not be attributed to differing cellular levels of CstF-64 (Fig. 7C). Furthermore, CstF-64 localization to this distal region on p21 was significantly enhanced in the HU-treated cells after Chk1 inhibition (Fig. 7B). Similar results were seen with two components of the CPSF complex, CPSF-100 (Fig. 7D) and CPSF-73 (data not shown). Thus, our results indicate that when cells are blocked in S phase, Chk1 has a role in regulating the localization of polyadenylation factors at the p21 distal region.
Discussion
Here we report a crucial link between the regulation of p21 transcriptional elongation and Chk1 kinase signaling. When cells are arrested in S phase and Chk1 is ablated, the levels of p21 mRNA are increased, and this up-regulation is strongly linked to increased elongation through the distal region of the gene. Our findings are summarized in Figure 8.
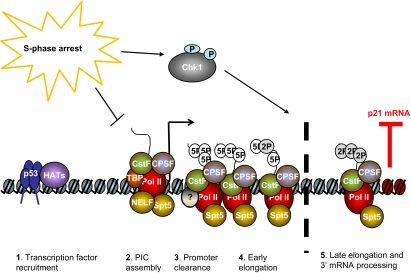
Figure 8.
A model for Chk1 function in the transcription elongation block to p21 during S-phase arrest. In cells treated with replication stress, p53 and HAT assembly at the upstream region of the p21 promoter is unaffected. When cells are blocked in S phase, a Chk1-independent differential PIC assembly at p21 is observed, which excludes TFIIB, CDK7, and CDK8 and possibly other components. RNAPII is efficiently recruited to the TATA region of p21 under these conditions, and upon phosphorylation of its Ser5 residue by an unknown kinase (circle with question mark, which is neither CDK7 nor CDK8), it is able to clear the promoter. RNAPII then transits the p21 gene, becoming minimally phosphorylated on Ser2 of its CTD heptad repeats, until it reaches the distal end of the gene (approximately +5 kb to +7 kb), whereupon it encounters a specific, Chk1-regulated, block to elongation. This block correlates with the decreased recruitment of DSIF, CPSF-100, CstF-64 to the distal region on p21. H3K36 trimethylation in this region is also blocked. These conditions lead to greatly reduced p21 mRNA production.
Two explanations can be proposed for why down-regulating Chk1 increases p21 transcription elongation: Chk1 itself may be performing an active role in repressing p21 transcription elongation after HU treatment, or the ablation of Chk1 may lead to the activation of a secondary pathway that is not normally functioning in cells arrested in S phase. As an example of the latter possibility, Chk1 is known to stabilize stalled replication forks, and its absence may lead to the processing of these forks into DNA double-stranded breaks leading to activation of the ATM/Chk2 pathway (Paulsen and Cimprich 2007). Two lines of evidence exclude this latter scenario. First, when HU treatment and dauno treatment are combined, p21 mRNA is significantly decreased when compared with treatment with dauno alone, indicating that HU treatment activates a dominant pathway that actively represses p21 transcription (Mattia et al. 2007). The fact that Chk1 is significantly more phosphorylated after HU than after dauno supports an active role for this kinase after replication stress (Figs. 1A, 2A). Second, after HU treatment, Chk2 is activated to a comparable level as seen after dauno treatment (as measured by levels of Chk2 phosphorylated at Thr68), indicating that Chk2 activation is not in itself sufficient to mediate full transcriptional induction of p21 after replication stress. Indeed, using Chk2-null HCT116 cells our data suggest that Chk2 activity is necessary for full activation of p21 transcription after HU or dauno. This is in agreement with findings that p53-mediated transcription of multiple target genes is compromised in Chk2-null mice (Takai et al. 2002). It is interesting that Chk2 activity may be required for full p21 transactivation while Chk1 activity appears to negatively regulate p21 transcriptional elongation. Although these two kinases have generally similar consensus sequences for phosphorylation (O'Neill et al. 2002), they can differentially phosphorylate sites within the C terminus of p53 (Shieh et al. 2000). This may not only result in a selective impact on p53; perhaps key components of the transcription machinery are also differentially recruited or phosphorylated by these two checkpoint kinases.
While our results implicate the ATR/Chk1 signaling cascade in regulating p21 transcription elongation during S-phase arrest, the participation of parallel signaling pathways cannot be ruled out. For example, treating mouse embryonic stem cells with Chk1 inhibitors such as UCN-01 activates the p38 MAP kinase pathway (Rodriguez-Bravo et al. 2007; Vitale et al. 2008). Moreover, ATR itself has been shown to activate the p38–MAPK pathway under certain conditions (Im and Lee 2008), and p53-deficient cells rely on the p38–MAPK pathway for survival after DNA damage (Reinhardt et al. 2007). A DNA replication checkpoint response that is dependent on Chk1 and claspin, but not on ATR, ATM, p38, or Rad17, has been described in HeLa cells but not HCT116 cells (Rodriguez-Bravo et al. 2006). These findings together mandate further investigation of the role of p38 signaling in the block to p21 transcription during S-phase arrest.
The signaling activity downstream from Chk1 should also be considered. Upon activation of the S-phase checkpoint, Chk1 inhibits CDK2/cyclin E activity by triggering the degradation of the cdc25A protein (Xiao et al. 2003; Sorensen et al. 2004). Ablating Chk1 thus restores CDK2 activity. Since many proteins are substrates of CDK2/cyclin E, further work is necessary to define the role, if any, of the downstream effectors of Chk1 in the transcription elongation block to p21 during S phase.
Although the initiation phase of transcription was originally thought to be the stage at which most regulatory events occur, more recent findings show transcription elongation can also be an important point of control (Sims et al. 2004). In line with this, we showed that elongation is specifically regulated by ATR and Chk1. We also found that the recruitment of certain PIC components, namely, TFIIB and CDK7, to the p21 promoter is reduced after HU but not dauno treatment, although their recruitment is not rescued after Chk1 inhibition. This indicates that while HU treatment certainly impacts PIC assembly, this occurs in a Chk1-independent fashion. Further, this result supports our conclusion that transcription initiation is not the sole determinant of the extent of p21 transcription (see model; Fig. 8). Although it is possible that sustained levels of TFIIB and CDK7 contribute to increased reinitiation from the p21 promoter after dauno treatment (Espinosa et al. 2003), high levels of TFIIB at the promoter do not directly correlate with strong p21 mRNA induction (JM Espinosa, unpubl.). Moreover, the amount of RNAPII loaded at the p21 promoter does not reflect the differential PIC assembly that we observe.
Intriguingly, as mentioned before, given the occupancy profiles of the cyclin-dependent kinases at p21 after HU treatment, our data imply that neither CDK7 nor CDK8 is responsible for phosphorylating Ser5 of RNAPII at this locus after replication stress. In the past, CDK7 was thought to be the main RNAPII Ser5 kinase. However, recent chemical genetic experiments discourage this notion (Larochelle et al. 2007). Other kinases have been implicated in Ser5 phosphorylation, including CDK8 and CDK9, as well as DNA-PK, ERK1/2 (Oelgeschlager 2002). Of these, CDK1 is unlikely to be required to phosphorylate Ser5 when cells are arrested in S phase because it is only active in the G2/M phase of the cell cycle (Clute and Pines 1999). More likely, either ERK or DNA-PK modifies RNAPII Ser5 after HU, as both kinases have been shown to be activated by S-phase block (Wu et al. 2006). Further work is required to determine whether either kinase is phosphorylating RNAPII Ser5 when DNA replication is blocked.
The data presented here link the failure to induce p21 transcription after HU treatment with a defect in transcription elongation, as assayed for by ChIPs that detect RNAPII phosphorylated at Ser2 (Fig. 5B; Supplemental Fig. S5C). How is this elongation block accomplished? Two possibilities exist: In the first case, the polymerase would fail to efficiently escape the promoter, leading to decreased density of RNAPII throughout the entire p21 gene. Alternatively, the polymerase would escape the p21 promoter region normally but would stall upon encountering one or more distal blocks to transcription. ChIPs for RNAPII phosphorylated at Ser5 (which assays promoter escape) strongly support the latter mechanism (see model in Fig. 8). It should be mentioned, however, that phosphorylation of RNAPII at Ser2 is not required for p21 mRNA accumulation, based on its resistance to DRB treatment (Gomes et al. 2006). Nevertheless, while phosphorylation of RNAPII at Ser2 may not be essential for p21 transcription or RNA processing, the absence of this modification is certainly correlated with the decrease in p21 mRNA levels when DNA replication is blocked.
In this study, we establish a previously unknown link between the polyadenylation machinery and a DNA damage effector kinase. What is perhaps most striking about our results is that several proteins (DSIF, CPSF-100, and CstF-64) show no difference in recruitment to the TATA region of the p21 promoter after HU, but show a progressive loss in occupancy (relative to the sharp increase brought about by dauno treatment) as the p21 gene is traversed by RNAPII. This is most pronounced at or downstream from the poly(A) site (see model in Fig. 8). DSIF, CPSF-100, and CstF-64 have all been shown to associate with RNAPII along the gene, and treatment of cells with DRB, which inhibits RNAPII phosphorylation at Ser2, decreases the association of CstF and CPSF subunits with distal regions of p21 (Glover-Cutter et al. 2008). Therefore, we cannot at this point rule out that the decreased recruitment of these processing factors at the 3′ end of p21, and their subsequent rescue by Chk1 inhibition, does not simply reflect the decrease in Ser2-phosphorylated RNAPII under these conditions. However, a more interesting hypothesis is that these polyadenylation factors are regulated by Chk1 in such a way as to affect the efficiency of transcription elongation. Several reports have shown that polyadenylation factors impact transcription, especially elongation and termination (Calvo and Manley 2003; Buratowski 2005; Rosonina et al. 2006), and several SR proteins have been shown to stimulate transcription elongation (Dermody et al. 2008; Lin et al. 2008). Intriguingly, our preliminary data indicate that Chk1 is recruited to the TATA region of p21 after HU treatment (Supplemental Fig S7). This provides further support for a direct role for Chk1 in regulating transcription elongation at this locus. In future experiments we hope to determine whether Chk1 directly phosphorylates CstF and CPSF subunits and if so, whether this affects transcription elongation.
Others have shown that the role of Chk1 differs in tumor and nontumor cell lines. While Chk1 is essential in maintaining genomic integrity during S-phase block in tumor cell lines such as HCT116, this is not the case in nontransformed cell lines such as NRK, NIH3T3, and MDCK cells (Rodriguez-Bravo et al. 2007). This observation has obvious implications for cancer therapy. It will be of great interest to determine whether the Chk1-dependent block to p21 transcription elongation is also conserved in nontransformed cell lines, and if not, whether this is the mechanism that accounts for the differential responses of tumor and nontumor cell lines to Chk1 inhibition.
Materials and methods
Cell culture, FACS analysis, and drugs
RKO cells and HCT116 colon carcinoma cells (wild type, p53−/−, and Chk2−/−) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 37°C. Drugs used in this study are as follows: 1.7 mM HU (Sigma), 0.22 μM dauno (Sigma), 1 μM Gö6976 (Sigma), and 4 mM caffeine (Sigma). For FACS analysis, cells were treated as described in the figure legends, trypsinized, and centrifuged at 1500 rpm. The pellet was resuspended in 200 μL of PBS and then fixed with 200 μL of ice-cold ethanol for at least 30 min at 4°C. The fixed cells were then centrifuged and washed in 400 μL of cold PBS. The cells were resuspended in 250–400 μL of PBS solution containing RNase A (50 μg/mL) and propdium iodide (60 μg/mL; Sigma). The cells were analyzed in a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson) for DNA content, and cell cycle stages were analyzed by using the ModFit LT version 3.0 program.
siRNA
siRNA duplexes were synthesized by Qiagen Inc. The negative control siRNA was the Qiagen All-stars duplex, and the ATR siRNA oligos were designed by Qiagen. Chk1 and Chk2 siRNA oligos have been described previously (Urist et al. 2004; Rodriguez and Meuth 2006). For RNAi experiments, HCT116 or RKO cells were plated at 30% confluency and transfected with 50 nM siRNA duplex using Dharmafect I (Dharmacon). Fresh media was added to the cells 6 h after transfection. The cells were then left untreated or exposed to 0.22 μM dauno or 1.7 mM hydroxyurea as indicated in the figure legends. Cells were lysed in TEGN buffer buffer (10 mM Tris at pH 8; 1 mM EDTA; 10% glycerol; 400 mM NaCl; 1% NP-40; 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitors [1 μM benzamidine, 3 μg/mL leupeptin, 0.1 μg/mL bacitracin, 1 μg/mL macroglobulin]) for immunoblotting, pelleted and frozen in liquid N2 for RNA extraction, or cross-linked and lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris at pH 8, 5 mM EDTA, protease inhibitors as described for TEGN buffer) for ChIP analysis.
Protein extraction and immunoblotting
Cells were lysed using TEGN buffer. Lysates were centrifuged to remove cellular debris, and the whole-cell extract was subjected to SDS–polyacrylamide gel electrophoresis followed by transfer to nitrocellulose. Membranes were blocked in 5% milk and then probed using the antibodies described in Supplemental Table S1. Anti-mouse or anti-rabbit secondary antibodies conjugated to IRDye 800 or 680 (Licor) were incubated with membranes and detected by the Odyssey fluorescence system (Licor).
RNA extraction and quantitative RT–PCR (qRT–PCR) analysis
RNA was harvested using a Qiagen RNeasy Mini kit, and first-strand cDNA synthesis was performed with the SuperScript III Supermix for qRT–PCR kit (Invitrogen) according to the manufacturers' specifications. A “No-RT” reaction, in which RNA was subjected to the conditions of cDNA synthesis without reverse transcriptase enzyme, was included as a negative control in all RT–PCR experiments to confirm the purity of RNA samples. Samples were analyzed by quantitative real-time PCR on an ABI 7300 real-time PCR instrument using the ΔΔCT method. PCR reaction mixtures contained 2 ng of cDNA, 1× SYBR Green Mix (Applied Biosystems), and 100 nM primers. Values were normalized to those of the hprt1 housekeeping gene. All primer sequences are available upon request.
ChIP
The antibodies used for ChIP are described in Supplemental Table 1. In all ChIP experiments, conditions that allow at least 90% immunodepletion of the epitope were used, and a no-antibody control confirmed the specificity of the immunoprecipitation. In cases where immunoprecipitations were performed using antibodies against the phosphorylated form of RNAPII (anti-Pol II-CTD Ser 2P or Ser 5P), a mixture of phosphatase inhibitors (Calbiochem) was added to the lysis buffer to protect against phosphatase activity. ChIP assays were performed essentially as described (Gomes et al. 2006) with the following modification: ChIP DNA was purified using the Qiaquik PCR Purification Kit (Qiagen). PCR reaction mixtures containing 1× SYBR Green Mix (Applied Biosystems), 1/50th fraction of ChIP-enriched DNA, and 100 nM primers in a total volume of 20 μL were set up for analysis by real-time PCR. Standard curves from 0.1–100 ng of sonicated genomic DNA were run along with the ChIP samples. All primer sequences are available upon request.
Acknowledgments
We thank Ella Freulich for expert technical assistance. This work was supported by NIH grants CA77742 and CA87497 (to C.P.), CA117907 (to J.M.E.), and GM 28983 (to J.L.M.); NSF grant MCB 0842974 (to J.M.E); the Cancer League of Colorado (to J.M.E.); and a Canadian NSERC post-graduate scholarship (to R.B).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1795709.
Supplemental material is available at http://www.genesdev.org.
References
- Ahn J, Urist M, Prives C. Questioning the role of checkpoint kinase 2 in the p53 DNA damage response. J Biol Chem. 2003;278:20480–20489. doi: 10.1074/jbc.M213185200. [DOI] [PubMed] [Google Scholar]
- Ahn J, Urist M, Prives The Chk2 protein kinase. DNA Repair (Amst) 2004;3:1039–1047. doi: 10.1016/j.dnarep.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Calvo O, Manley JL. Strange bedfellows: Polyadenylation factors at the promoter. Genes & Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sanchez Y. Chk1 in the DNA damage response: Conserved roles from yeasts to mammals. DNA Repair (Amst) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle. 2007a;6:2594–2598. doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007b;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: A role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi V, McKinney K, Poyurovsky MV, Prives C. Decreased p21 levels are required for efficient restart of DNA synthesis after S phase block. J Biol Chem. 2004;279:5802–5810. doi: 10.1074/jbc.M310373200. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Heine GF, Horwitz AA, Parvin JD. Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J Biol Chem. 2008;283:9555–9561. doi: 10.1074/jbc.M707700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Ho CC, Siu WY, Lau A, Chan WM, Arooz T, Poon RY. Stalled replication induces p53 accumulation through distinct mechanisms from DNA damage checkpoint pathways. Cancer Res. 2006;66:2233–2241. doi: 10.1158/0008-5472.CAN-05-1790. [DOI] [PubMed] [Google Scholar]
- Im JS, Lee JK. ATR-dependent activation of p38 MAP kinase is responsible for apoptotic cell death in cells depleted of Cdc7. J Biol Chem. 2008;283:25171–25177. doi: 10.1074/jbc.M802851200. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C, Vogelstein B, Bunz F. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem. 2003;278:20475–20479. doi: 10.1074/jbc.M213159200. [DOI] [PubMed] [Google Scholar]
- Kaneko YS, Watanabe N, Morisaki H, Akita H, Fujimoto A, Tominaga K, Terasawa M, Tachibana A, Ikeda K, Nakanishi M. Cell-cycle-dependent and ATM-independent expression of human Chk1 kinase. Oncogene. 1999;18:3673–3681. doi: 10.1038/sj.onc.1202706. [DOI] [PubMed] [Google Scholar]
- Katzenberger RJ, Marengo MS, Wassarman DA. ATM and ATR pathways signal alternative splicing of Drosophila TAF1 pre-mRNA in response to DNA damage. Mol Cell Biol. 2006;26:9256–9267. doi: 10.1128/MCB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn EA, Yoo CJ, Eastman A. The protein kinase C inhibitor Go6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res. 2003;63:31–35. [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KP, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, Shokat KM, Jallepalli PV, Fisher RP. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25:839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol. 2006;26:7529–7538. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes & Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C. p53-Dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol. 2007;27:1309–1320. doi: 10.1128/MCB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nature reviews. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager T. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J Cell Physiol. 2002;190:160–169. doi: 10.1002/jcp.10058. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA. The ATR pathway: Fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell. 2006;17:402–412. doi: 10.1091/mbc.E05-07-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Guaita-Esteruelas S, Florensa R, Bachs O, Agell N. Chk1- and claspin-dependent but ATR/ATM- and Rad17-independent DNA replication checkpoint response in HeLa cells. Cancer Res. 2006;66:8672–8679. doi: 10.1158/0008-5472.CAN-05-4443. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Guaita-Esteruelas S, Salvador N, Bachs O, Agell N. Different S/M checkpoint responses of tumor and non tumor cell lines to DNA replication inhibition. Cancer Res. 2007;67:11648–11656. doi: 10.1158/0008-5472.CAN-07-3100. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Kaneko S, Manley JL. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Schwarz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes & Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9–Rad1–Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes & Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, Anderson CW, Appella E, Nakanishi M, Suzuki H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Heierhorst J. SQ/TQ cluster domains: Concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes & Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Galluzzi L, Criollo A, Vivet S, Castedo M, Kroemer G. Chk1 inhibition activates p53 through p38 MAPK in tetraploid cancer cells. Cell Cycle. 2008;7:1956–1961. doi: 10.4161/cc.7.13.6073. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Chen B, Parihar K, He L, Fan C, Zhang J, Liu L, Gillis A, Bruce A, Kapoor A, et al. ERK activity facilitates activation of the S-phase DNA damage checkpoint by modulating ATR function. Oncogene. 2006;25:1153–1164. doi: 10.1038/sj.onc.1209148. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Chen Z, Gunasekera AH, Sowin TJ, Rosenberg SH, Fesik S, Zhang H. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem. 2003;278:21767–21773. doi: 10.1074/jbc.M300229200. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes & Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]