Abstract
Insulators are protein-bound DNA elements that are thought to play a role in chromatin organization and the regulation of gene expression by mediating intra- and interchromosomal interactions. Suppressor of Hair-wing [Su(Hw)] and Drosophila CTCF (dCTCF) insulators are found at distinct loci throughout the Drosophila melanogaster genome and function by recruiting an additional protein, Centrosomal Protein 190 (CP190). We performed chromatin immunoprecipitation (ChIP) and microarray analysis (ChIP–chip) experiments with whole-genome tiling arrays to compare Su(Hw), dCTCF, boundary element-associated factor (BEAF), and CP190 localization on DNA in two different cell lines and found evidence that BEAF is a third subclass of CP190-containing insulators. The DNA-binding proteins Su(Hw), dCTCF, and BEAF show unique distribution patterns with respect to the location and expression level of genes, suggesting diverse roles for these three subclasses of insulators in genome organization. Notably, cell line-specific localization sites for all three DNA-binding proteins as well as CP190 indicate multiple levels at which insulators can be regulated to affect gene expression. These findings suggest a model in which insulator subclasses may have distinct functions that together organize the genome in a cell type-specific manner, resulting in differential regulation of gene expression.
Keywords: Insulators, chromatin organization, BEAF, CTCF, gypsy
Insulators have been implicated in the regulation of gene expression through the formation of chromatin loops. Insulators are classically described by two experimentally defined properties, barrier activity and enhancer-blocking activity (Gaszner and Felsenfeld 2006). Barrier activity stops the spread of a chromatin state into neighboring regions, whereas enhancer-blocking activity interferes with the communication between a regulatory element and a promoter in a directional manner so that neither the regulatory element nor the promoter is inactivated. Drosophila melanogaster has five known insulators characterized by their DNA-binding proteins, Suppressor of Hair-wing [Su(Hw)], Drosophila CTCF (dCTCF), boundary element-associated factor (BEAF), Zeste-white 5 (Zw5), and GAGA factor (GAF) (Maeda and Karch 2007). In contrast, the only known insulator protein in vertebrates is CTCF (Wallace and Felsenfeld 2007). Although it has been shown that some of the D. melanogaster insulator complexes can interact with one another to form chromatin loops (Blanton et al. 2003; Gerasimova et al. 2007; Mohan et al. 2007), it is unclear whether the various complexes serve redundant or distinct functions in genome regulation.
Two of the Drosophila insulator complexes that are thought to interact through the formation of chromatin loops are Su(Hw) and dCTCF. Each of these two proteins can be visualized at hundreds of sites on highly replicated polytene chromosomes without substantial overlap; however, in diploid nuclei they colocalize at five to 25 distinct loci, termed insulator bodies (Gerasimova and Corces 1998; Gerasimova et al. 2007; Mohan et al. 2007). The interaction among individual Su(Hw) and dCTCF insulator sites in insulator bodies is facilitated by the common use of a protein known as Centrosomal Protein 190 (CP190). CP190 is necessary for both insulator body formation and the enhancer-blocking activity of the Su(Hw) and dCTCF insulators (Pai et al. 2004; Gerasimova et al. 2007; Mohan et al. 2007). Although CP190 is not thought to bind to specific DNA sequences on its own, it is found on polytene chromosomes at loci that do not bind Su(Hw) or dCTCF, suggesting that there are other unknown DNA-binding proteins that can recruit CP190. These observations have led to a model in which Su(Hw), dCTCF, and potentially other proteins bind to insulator elements found throughout the genome. They then come together through protein–protein interactions mediated by CP190 and association with the nuclear lamina to form insulator bodies, resulting in the organization of the intervening chromatin into loops (Capelson and Corces 2005).
Interaction between insulator sites to form chromatin loops is characteristic of many different insulators, and in some cases has been shown to affect gene expression. The scs and scs' insulator elements of Drosophila have been shown to interact through chromatin looping in a manner dependent on their DNA-binding proteins, Zw5 and BEAF (Blanton et al. 2003). Similarly, the vertebrate insulator protein CTCF is thought to form loops through its interaction with the nucleolus and itself (Pant et al. 2004; Yusufzai et al. 2004). More specifically, CTCF-dependent loop formation that is necessary for allele-specific transcription has been observed within the mouse β-globin locus and at the imprinting control region (ICR) in the H19/Igf2 locus (Murrell et al. 2004; Kurukuti et al. 2006; Splinter et al. 2006; Yoon et al. 2007). Additionally, the insertion of an ectopic insulator into the β-globin locus results in the formation of a new CTCF-mediated loop that disrupts normal loop formation as well as normal transcription (Hou et al. 2008).
Since insulators appear to be involved in the regulation of gene expression through the formation of chromatin loops, it is possible that regulation of insulator elements is required during development to create specific transcription profiles during cell specification. Evidence for the regulation of insulators has been shown at the ICR in the H19/Igf2 region of vertebrates, where differential DNA methylation modulates CTCF binding, which in turn regulates gene expression (Bell and Felsenfeld 2000; Hark et al. 2000). This suggests that a possible mechanism for regulating insulators might be to modulate the affinity of the interaction between insulator proteins and their target DNA. If this is the case, and if insulators are involved in establishing distinct patterns of gene expression during cell fate specification, one would expect to find different configurations of insulator protein occupancy in the genome of different cell types. In fact, when human CTCF-binding sites were mapped globally in specific cell lines, though many sites were constant between cell lines, a subset of the identified sites were found to be cell type-specific (Kim et al. 2007; Cuddapah et al. 2009). In order to evaluate the involvement of the various Drosophila insulator proteins in cell type-specific chromatin organization, we must first determine their DNA localization patterns in various cell types.
Here, we perform a chromatin immunoprecipitation (ChIP) and microarray analysis (ChIP–chip) of Drosophila insulator proteins using whole-genome tiling arrays to investigate the in vivo binding profiles of Su(Hw), dCTCF, CP190, and BEAF in two different cell lines. This analysis revealed a new subclass of CP190 insulators defined by the presence of BEAF. Su(Hw), dCTCF, and BEAF insulator subclasses show distinct genomic localization patterns and cell type-specific distributions that correlate with changes in gene expression. These results suggest that these insulator proteins may play different roles in chromatin organization and may be important to establish cell type-specific gene expression profiles. We suggest that the three subclasses of CP190 insulators have distinct functions that together organize the genome in a cell type-specific manner.
Results
Genome-wide localization of insulator proteins
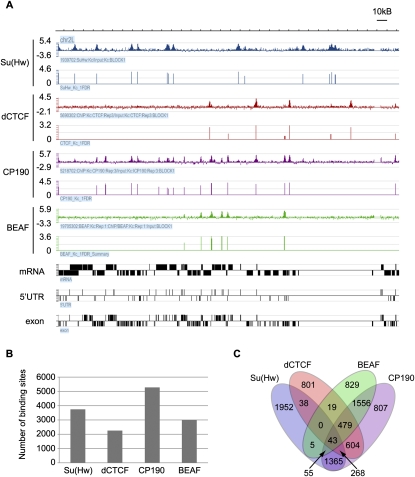
In order to identify sites of insulator protein occupancy on a genome-wide scale, we performed ChIP–chip with antibodies against Su(Hw), dCTCF, and CP190. This analysis was first carried out in Kc cells, a cell line originating from a heterogeneous embryonic culture and suggested to be of neural or glial origin (Cherbas et al. 1977). Microarray analysis was performed on Nimblegen whole-genome tiling arrays, and ChIP–chip experiments were done in triplicate for each protein (Fig. 1A). Additionally, we used Nimblegen peak analysis and applied a cutoff of a <5% false discovery rate (FDR). We also required that a peak be found in at least two out of the three biological replicates to be considered. With this cutoff of a 5% FDR, we found 4715 Su(Hw) peaks, 3001 dCTCF peaks, and 7845 CP190 peaks. With a more stringent cutoff of <1% FDR, we found 3747 Su(Hw) peaks, 2266 dCTCF peaks, and 5272 CP190 peaks (Fig. 1B). The remainder of the analysis was done with the more stringent data sets unless stated otherwise.
Figure 1.
Genome-wide localization of insulator proteins. (A) Representative ChIP–chip data for Su(Hw), dCTCF, CP190, and BEAF over a 250-kb region of Chromosome 2L. The Y-axis contains the log2 value of the ratio of ChIP/input signal. For each antibody, the top row represents the normalized data from one biological replicate and the bottom row represents the analyzed peak data that summarizes three biological replicates. Gene data that depict mRNA, 5′UTR, and exon information are shown in black, such that boxes above the line represent genes on the plus strand and genes below the line represent genes on the minus strand. (B) A summary of the whole-genome peak analysis for Su(Hw), dCTCF, CP190, and BEAF using the more stringent cutoff of <1% FDR. (C) A Venn diagram indicating the number of binding sites that overlap between Su(Hw), dCTCF, BEAF, and CP190.
To determine the specificity of the microarray analysis, we randomly chose 25 peaks for each of the three proteins and performed conventional ChIP with real-time PCR analysis (Supplemental Tables S1–S3). With this method we verified 100% of both the dCTCF and CP190 peaks and 91% of the Su(Hw) peaks. Results from these experiments suggest the absence of a large number of false positives in the data.
In order to verify the sensitivity of the data sets, we tested whether data from the ChIP–chip experiments contained the known binding sites for the proteins analyzed. Nine previously characterized Su(Hw)-binding sites throughout the genome all show a strong Su(Hw) signal in the ChIP–chip data, as do the 10 known dCTCF-binding sites in the bithorax region (Supplemental Tables S4, S5; Golovnin et al. 2003; Parnell et al. 2003, 2006; Ramos et al. 2006; Holohan et al. 2007). Of these 19 known Su(Hw)- and dCTCF-binding sites, all but one Su(Hw) site and two dCTCF sites show CP190 signal. These three sites might represent false negatives due to the method. Alternatively, they could indicate true, biologically significant CP190-independent sites since the nine Su(Hw) sites were not previously tested for the presence of CP190 and the CP190 analysis at the dCTCF sites was performed with third instar larval brains and therefore could be cell type-specific (Mohan et al. 2007).
Analysis of the relationship between insulator proteins reveals BEAF as a third subclass of CP190 insulators
Since insulator proteins are known to work in complexes, we wanted to understand how the localization patterns of these various insulator proteins relate to one another. Therefore, we compared the genome-wide distribution patterns of Su(Hw), dCTCF, and CP190. Previous immunofluorescence microscopy studies using polytene chromosomes have suggested that Su(Hw) and dCTCF do not themselves overlap, but colocalize with CP190 at distinct loci (Gerasimova et al. 2007; Mohan et al. 2007). In addition, ChIP data at the 3-Mb Adh region for dCTCF and Su(Hw) indicate that these two proteins do not show substantial overlap in their localization patterns (Holohan et al. 2007). Results from the whole-genome ChIP–chip analyses show that 47% of Su(Hw) sites and 62% of dCTCF sites colocalize with CP190 sites (Fig. 1C; Supplemental Table S6). In addition, only 349 sites are shared by Su(Hw) and dCTCF (i.e., 9% of Su(Hw) sites and 16% of dCTCF sites). Although these trends are what was predicted, the large number of Su(Hw) sites and dCTCF sites that do not recruit CP190 was unexpected. Given the requirement of CP190 for proper function of both Su(Hw) and dCTCF insulators, the absence of this protein from a subset of Su(Hw) and dCTCF sites suggests the possibility that a protein other than CP190 may interact with these sites or that they represent primed insulators waiting for CP190 recruitment to establish insulator function.
CP190 itself cannot directly bind DNA in a sequence-specific manner in gel shift assays, and therefore it is thought to require recruitment by a DNA-binding protein in order to establish sequence-specific localization on chromatin (Pai et al. 2004). Analysis of the ChIP–chip data indicates that about half of the CP190 sites do not contain either Su(Hw) or dCTCF. This suggests that there must be at least one, and perhaps several, additional DNA-binding proteins that can recruit CP190 to the DNA. We therefore analyzed all of the CP190 sites not bound by Su(Hw) or dCTCF for the presence of any additional sequence motifs. From this detailed sequence analysis, we identified CGATA as a consensus motif present at these sites. This sequence is the binding motif for the Drosophila insulator protein BEAF (Zhao et al. 1995), suggesting that BEAF may be a third protein that can recruit CP190. In order to confirm the colocalization between CP190 and BEAF, we performed ChIP–chip analysis with α-BEAF antibody using the same method previously used for the other insulator proteins. This analysis revealed 2995 BEAF localization sites genome-wide using the more stringent cutoff of <1% FDR. Of these sites, 71% were found to colocalize with CP190, a more striking colocalization than that observed for either Su(Hw) or dCTCF (Fig. 1C; Supplemental Table S6). The scs' insulator is included in these sites of BEAF and CP190 colocalization. Interestingly, although Su(Hw) and BEAF show colocalization at only 103 sites, dCTCF and BEAF colocalize at 541 sites (i.e., 24% of dCTCF sites and 18% of BEAF sites). This suggests some cross-talk between the dCTCF and BEAF insulator subclasses. Taking Su(Hw), dCTCF, and BEAF together at a 1% FDR cutoff, 85% of CP190 sites colocalize with at least one of the three DNA-binding proteins. Using the less stringent cutoff for each data set of a 5% FDR, 91% of CP190 sites colocalize with either Su(Hw), dCTCF, or BEAF (Supplemental Table S7). This finding suggests that we have identified the DNA-binding proteins that recruit the majority of CP190 to chromatin, but does not eliminate the possibility that there exist additional CP190 recruiting proteins.
Sequence analysis of BEAF-binding sites reveals that increased CGATA clustering enhances the likelihood of BEAF binding
Previous studies have experimentally identified a few genomic BEAF-binding sites and determined that they contain clustered CGATA motifs, but no other sequence specificity or pattern was reported (Zhao et al. 1995; Hart et al. 1997; Cuvier et al. 1998). However, a recent report using a computational approach identified roughly 1700 putative BEAF-binding sites genome-wide called dual-core elements based on the occurrence of five or six CGATA motifs organized in two clusters separated by a 200-base-pair (bp) AT-rich sequence, and showed evidence that some of these sites bind BEAF in vivo (Emberly et al. 2008). The genome-wide ChIP–chip data presented here show BEAF binding to only 35% of these dual-core elements. One explanation for this discrepancy is that the dual-core elements that were found to be unoccupied in Kc cells may represent sites that are bound by BEAF in other cell types or in response to specific stimuli. Additionally, >2000 in vivo BEAF-binding sites were identified in the Kc cell ChIP–chip data that do not contain dual-core elements, suggesting that this dual-core arrangement of CGATA motifs may represent only a subset of the genome-wide BEAF-binding sites.
We performed consensus sequence analysis with the ChIP–chip BEAF data and found no enriched motifs in addition to the CGATA motif at genomic BEAF sites. Additional consensus sequence analysis was done with these sequences after removing all CGATA motifs, and no additional overrepresented sequences were identified. We therefore searched the collection of BEAF-binding sites for CGATA motifs. Although this analysis did not reveal any pattern in the spacing or orientation of CGATA motifs within BEAF localization sites, we found that 68% of the identified sites contained at least three CGATA motifs. In order to understand the DNA sequence requirements to achieve BEAF binding, we identified all the CGATA clusters in a 1000-bp window genome-wide and determined which ones are bound by BEAF according to the ChIP–chip data (Table 1). This analysis revealed that the majority of bound sites contain four to seven CGATA motifs. Additionally, as the number of CGATA motifs in a 1000-bp cluster increases, so does the percentage of sites that are bound, and this value plateaus around 50% for nine or more CGATA motifs. These data suggest an additive effect, in which the more CGATA motifs clustered together, the more likely that the site is bound by BEAF. This also suggests that CGATA motifs may not be sufficient to recruit BEAF, as a large number of CGATA clusters do not bind BEAF. Therefore, secondary chromatin structures, nucleosome occupancy, and/or regulatory proteins may also be important for the ability of BEAF to bind chromatin.
Table 1.
Number of bound BEAF consensus site clusters genome-wide
Genomic distribution of insulator proteins suggests differential roles for insulator subclasses in genome regulation
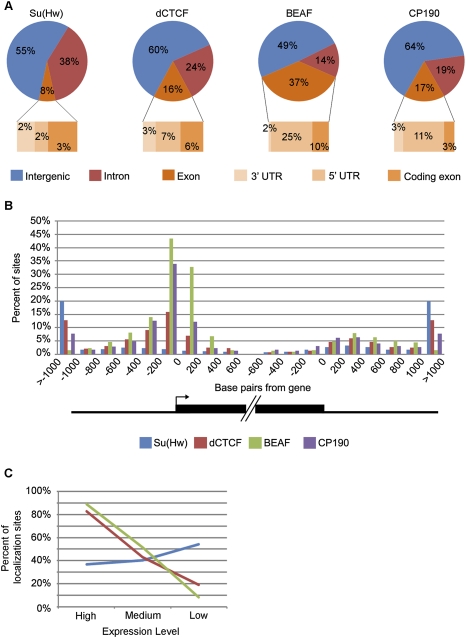
Su(Hw)-, dCTCF-, and BEAF-binding sites are found throughout the Drosophila genome. In order to understand the possible functional significance of their distributions and decipher the basis for the existence of these various subclasses of CP190 sites, we examined the location of insulator protein-binding sites in relation to genes. We first analyzed the distribution based on whether sites are located within introns, exons, or intergenic regions of the genome (Figs. 1A, 2A). Although it is not understood how the transcription machinery deals with the presence of insulator sites within genes, it is known that a Su(Hw)-binding site can be placed within an intron without disruption of transcription (Geyer and Corces 1992). Therefore, it was not surprising that we found a substantial number of intronic sites for each protein. As expected for regulatory elements, Su(Hw), dCTCF, and CP190 were all found to be preferentially excluded from exonic regions (P < 1 × 10−12), with 8%, 16%, and 17% of sites found within exons, respectively, compared with the 21% of the genome that represents exons. However, it was unexpected to find any insulator sites within exons, and, surprisingly, BEAF sites were found to be enriched in exons over the fraction of the genome represented by exons (P = 3 × 10−86). Therefore, we dissected the exonic region into the 3′ untranslated regions (UTR), 5′UTRs, and protein coding regions. This analysis revealed that 5′UTRs are the most occupied subgroup of exons for dCTCF (7%), BEAF (25%), and CP190 (11%) and that these proteins are highly enriched in this region over the 2% of the genome that represents 5′UTRs (P < 5 × 10−53). Conversely, all four insulator proteins are excluded from protein coding regions of exons (P < 6 × 10−19). The localization of dCTCF and BEAF, but not Su(Hw), in the 5′UTRs of genes suggests that these three insulator subclasses may have different roles in the regulation of chromatin organization and gene expression.
Figure 2.
Genomic distribution of insulator protein-binding sites. (A) Pie charts representing the distribution of Su(Hw), dCTCF, BEAF, and CP190 in relation to intergenic regions, introns, and exons. Exonic distribution is then further broken down into protein-coding exons, 5′UTRs, and 3′UTRs. (B) The distribution of the insulator proteins in relation to the 5′ and 3′ ends of annotated genes. The graph depicts 200-bp fragments encompassing1 kb upstream of and downstream from the start and end of genes as well as 600 bp into the start and end of genes. (C) Percent of insulator protein-binding sites within 200 bp of the 5′ end of genes that localize to genes with high, medium, and low/no expression. Line colors correspond to the same proteins as denoted in B.
In order to further dissect the distribution of insulator protein-binding sites, we extended our analysis to sequences in and around genes. For each annotated gene, we analyzed the number of insulator protein-binding sites in 200-bp segments within the 1-kb flanking regions and the 600-bp regions inside the 5′ and 3′ ends (Fig. 2B; Supplemental Fig. S1). In the case of Su(Hw), the distribution is similar at the 5′ and the 3′ ends of genes and fairly uniform with increasing distance from genes. Few Su(Hw)-binding sites are found in the 1-kb regions flanking genes. However, dCTCF, BEAF, and CP190 show a distribution that is highly skewed toward the 5′ end of genes and is notably enriched in the first 200 bp just upstream of the transcription start site (TSS). This distribution is most dramatic for BEAF. These findings suggest that dCTCF, and to a greater extent BEAF, may have a role in regulating the expression of single, specific genes whereas Su(Hw) may have a more general role in chromatin organization.
This differential distribution of insulator subclasses around genes led us to explore the relationship between insulator protein-binding sites and levels of gene expression. To do this we determined gene expression levels using NimbleGen gene expression microarrays that contain 15,634 genes (Supplemental Fig. S2). We then divided the genes into three groups based on their expression signal (i.e., high, medium, and low/no expression) (Supplemental Table S8). We could then ask what percent of the insulator protein-binding sites that localize within 200 bp of the 5′ ends of genes are associated with each of these three expression groups (Fig. 2C; Supplemental Fig. S1). For this analysis, we only used sites with both a DNA-binding protein and CP190 since CP190 is necessary for insulator function (Pai et al. 2004; Gerasimova et al. 2007; Mohan et al. 2007). We found that 83% of dCTCF sites and 89% of BEAF sites at the 5′ end of genes localize to genes with a high expression signal. However, Su(Hw)-binding sites showed less of a preference for any group of genes, but were most often found near genes with a low expression signal (i.e., 54% of sites). We also performed this analysis for insulator sites that localize to the 3′ end of genes, the UTRs, and introns and found similar distributions for each of the three proteins, though the trends are stronger at the 5′ end of genes than at the 3′ ends (Supplemental Fig. S3). Considering localization at these various gene regions, it is clear that BEAF shows a stronger association with genes with a high expression signal than dCTCF. This provides further evidence that although all three insulator subclasses contain CP190, they may play different roles in regulating gene expression.
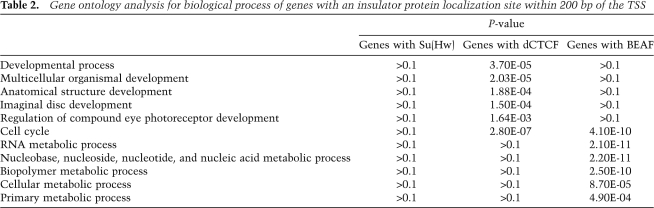
To further investigate the significance of these distribution patterns, we performed gene ontology analysis based on biological process. Using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Dennis et al. 2003), we investigated whether a subset of genes could be identified that requires insulator protein localization near the TSS. We found that genes containing dCTCF in the 200-bp upstream of their TSS are mostly involved in developmental processes, while genes containing BEAF in this region are mostly involved in metabolic processes (Table 2; Supplemental Tables S9–S11). Both dCTCF- and BEAF-containing genes also show an enrichment for cell cycle genes, whereas Su(Hw)-containing genes show little significant clustering based on biological process. Together, the genome distribution analysis shows that Su(Hw), dCTCF, and BEAF have different patterns of localization around genes and they appear to be located adjacent to functionally different types of genes with different levels of expression, suggesting divergent roles for these three subclasses of insulators in chromatin organization and gene regulation.
Table 2.
Gene ontology analysis for biological process of genes with an insulator protein localization site within 200 bp of the TSS
Cell type-specific distributions of insulator proteins suggest a role for insulators in regulating differential gene expression
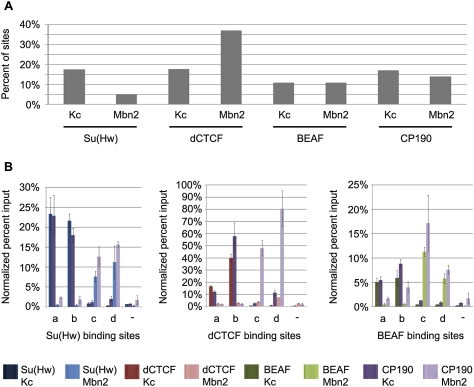
The different distributions of the three subclasses of insulator proteins around developmentally regulated genes suggest that there may be cell type-specific differences in insulator protein localization that have a role in regulation of gene expression. If this is the case, we would expect to find differences in the distributions of insulator proteins when comparing cell lines originating from two different tissues. To test this possibility we performed ChIP–chip analysis using Mbn2 cells, a tumorous hematopoietic cell line derived from D. melanogaster larvae (Gateff 1978), for each of the four insulator proteins used in this study; data were then compared with those obtained with embryonic Kc cells derived from neuronal tissue (Cherbas et al. 1977). Comparison between the two cell lines revealed that while many sites are constant, a fraction of the localization sites for each of the four insulator proteins are different between the two lines (Fig. 3A; Supplemental Fig. S4). At a 1% FDR, 18% of Su(Hw) sites in Kc cells and 5% of Su(Hw) sites in Mbn2 cells were found to be cell type-specific. For dCTCF, we found 18% of sites in Kc cells and 37% in Mbn2 cells to be unique, whereas the number of cell type-specific BEAF sites is 11% in Kc cells and 11% in Mbn2 cells. In the case of CP190, which is found at all three insulator subclasses, 17% of sites present in Kc cells and 14% in Mbn2 cells were found to be cell type-unique. These percentages vary between 9% and 46% for the four proteins when we used the data sets generated with the less stringent cutoff of 5% FDR (Supplemental Table S12). Therefore, regardless of the cutoff used to define the data sets, a subset of the localization sites was found to be cell type-specific for all four insulator proteins. Additionally, dCTCF showed more cell type-specific DNA binding than Su(Hw) or BEAF, again suggesting that these three subclasses of insulators may play different roles in the regulation of gene expression.
Figure 3.
Insulator protein DNA binding is a cell type-specific process. (A) Summary of the whole-genome comparison between Su(Hw), dCTCF, BEAF, and CP190 in Kc and Mbn2 cells. Bars represent the percentage of binding sites that are cell type-unique using the data sets defined with the more stringent cutoff of a 1% FDR. (B) Validation of four Su(Hw) (left), four dCTCF (middle), and four BEAF (right) cell type-specific binding sites by real-time PCR. For each protein, two Kc cell type-specific binding sites (columns a,b) and two Mbn2 cell type-specific binding sites (columns c,d) were tested. Each binding site was analyzed by conventional ChIP for the DNA-binding protein as well as CP190 in each cell type. Percent input values were normalized to a well-known binding site for each insulator protein.
To confirm these findings, we used ChIP with real-time PCR analysis to test two cell type-specific binding sites in Kc and Mbn2 cells for Su(Hw), dCTCF, and BEAF (Fig. 3B). The results indicate that all the sites tested show more than a fivefold enrichment for both Su(Hw) and CP190, dCTCF and CP190, or BEAF and CP190 in one cell type over the other. Additionally, we compared the ChIP–chip data for Kc and Mbn2 cells reported here with ChIP–chip data for CP190 and dCTCF data from S2 cells (Bartkuhn et al. 2009, modENCODE). The S2 cell data from these two sources differs considerably; however, both S2 cell data sets showed that the majority of sites for both dCTCF and CP190 are shared with Kc and Mbn2 cells. Nevertheless, there are unique dCTCF- and CP190-binding sites for each of the three cell types (Supplemental Fig. S5). This confirms that a subset of insulator protein-binding sites varies between different cell types, suggesting that regulation of the interaction between insulator proteins and DNA may be used to modulate insulator-dependent chromatin organization.
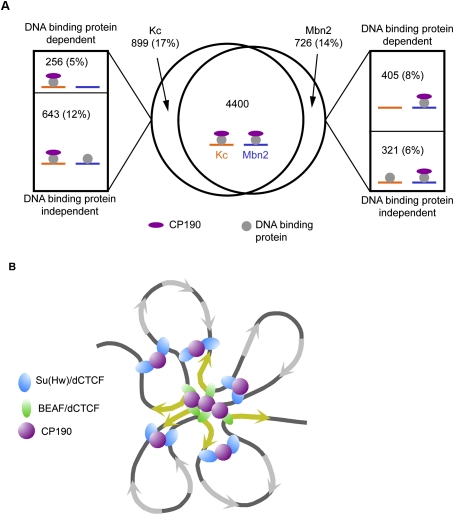
In addition to regulation at the level of DNA binding, the presence of Su(Hw), dCTCF, and BEAF sites without CP190 implies that recruitment of CP190 may also be a regulated process. We examined whether this type of regulation takes place by comparing the distribution of CP190 between the two cell types. We found that 12% of CP190 sites in Kc cells and 6% of CP190 sites in Mbn2 cells are cell type-unique and colocalize with Su(Hw), dCTCF, or BEAF sites that are present in both cell lines (Fig. 4A). This observation suggests an additional level of regulation in which cells control the interaction between CP190- and DNA-binding proteins to modulate insulator activity. Therefore, insulator activity may be regulated at multiple levels between different cell types, and together these forms of regulation may generate changes in chromatin organization necessary to establish different gene expression profiles during cell specification.
Figure 4.
The role of CP190 insulators in chromatin organization. (A) Venn diagram depicting the overlap of CP190-binding sites between Kc cells and Mbn2 cells. Roughly 17% of CP190 sites in Kc cells and 14% of sites in Mbn2 cells were found to be cell type-specific. Of these cell type-specific binding sites, a subset was found to be dependent on differential DNA-binding protein localization (top boxes) and a subset was found to be independent of DNA-binding protein localization (bottom boxes). Cartoons indicate the state of DNA-binding protein and CP190 localization in Kc cells (orange) and Mbn2 cells (blue) at sites represented by each region. (B) Model depicting the functional specialization of CP190 insulator subclasses. Su(Hw) and dCTCF may orchestrate the primary level of chromatin organization. BEAF and dCTCF could then fine-tune this organization around highly expressed genes (yellow arrows). The interactions between insulator protein-binding sites may be facilitated by CP190.
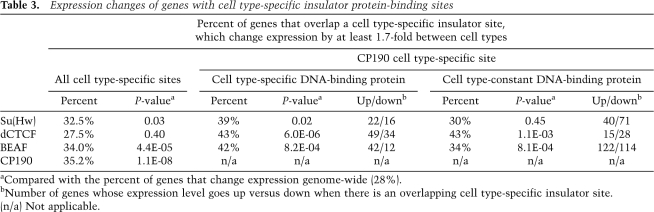
Since insulator localization sites change between these two cell lines, we asked how these differences correlate with changes in gene expression. To do this, we performed gene expression analysis for Mbn2 cells and found 4422 genes that change by at least 1.7-fold between Kc cells and Mbn2 cells (Supplemental Fig. S2). We then identified all of the genes that have a cell type-specific insulator site within the gene or 1 kb upstream of or downstream from the gene, and determined the percent of these genes that change expression (Table 3). To determine statistical significance, we compared the percent of genes with a cell type-specific insulator site that change expression with the percent of genes that change expression genome-wide (28%). We found that although there is little enrichment for genes that change expression at sites with cell type-specific DNA-binding proteins, genes found near cell type-specific CP190 showed a significant enrichment for those that change in gene expression. Therefore, we refined our analysis to only those genes that have CP190 cell type-specific binding, and divided these genes into those with a cell type-specific DNA-binding protein and those with a cell type-constant DNA-binding protein (Table 3). This analysis revealed that genes with a site that is CP190 and Su(Hw), dCTCF, or BEAF cell type-specific showed significant enrichment for genes that change expression. Those genes with a CP190 cell type-specific site and a constant DNA-binding protein showed significant enrichment for genes that change expression at dCTCF and BEAF sites. Therefore, CP190 cell type-specific sites, especially those with cell type-specific DNA-binding proteins, may regulate the gene expression of nearby genes. Interestingly, even though dCTCF and BEAF associate with genes with a high expression signal and Su(Hw) with genes with low or no expression signal, this analysis revealed that genes with a cell type-specific site nearby show both increased and decreased expression between cell types (Table 3). Therefore, although these insulator sites associate with genes of a certain expression level, they may both positively and negatively affect transcription of these genes. This suggests that insulator sites may play a role in regulating gene expression between cell types and that CP190 is a key component of this regulation.
Table 3.
Expression changes of genes with cell type-specific insulator protein-binding sites
aCompared with the percent of genes that change expression genome-wide (28%).
bNumber of genes whose expression level goes up versus down when there is an overlapping cell type-specific insulator site.
(n/a) Not applicable.
Discussion
The Drosophila insulator proteins Su(Hw) and dCTCF have been shown previously to form the DNA-binding component of two different subclasses of an insulator defined by the presence of CP190 (Gerasimova et al. 2007; Mohan et al. 2007). In this study, we identify BEAF as a third DNA-binding protein that colocalizes with CP190 and, therefore, defines a third subclass of the CP190 insulator family. Su(Hw), dCTCF, and BEAF together account for 91% of CP190 sites in Kc cells, indicating that these are the three major DNA-binding components of this insulator.
Su(Hw), dCTCF, and BEAF have all been implicated in chromatin loop formation (Blanton et al. 2003; Byrd and Corces 2003; Gerasimova et al. 2007; Mohan et al. 2007), and the interaction of these different DNA-binding proteins with CP190 could have functional implications for the arrangement of the chromatin fiber within the nucleus. The work presented here provides critical insight into the genome-wide distribution of these four insulator proteins and is a first, crucial step toward understanding the role that they play in chromatin organization and the regulation of gene expression.
Although insulator elements containing Su(Hw), dCTCF, and BEAF could, in principle, play similar roles, we found that they have very different distribution patterns with respect to gene location. Only 20% of Su(Hw) sites are located within 1 kb of the 5′ or 3′ ends of genes. On the other hand, 47% of dCTCF sites and 84% of BEAF sites are found within 1 kb of gene ends, and their distributions are highly skewed toward the 5′ end of highly expressed genes. dCTCF and BEAF appear to display further functional compartmentalization in their roles, since BEAF tends to be present at the 5′ end of genes involved in metabolic processes and dCTCF is enriched near genes involved in developmental processes. This could indicate that BEAF plays a specific role in the regulation of gene units consisting of metabolic genes, whereas Su(Hw) may play a more general role by setting the foundation for chromatin organization. dCTCF, which shows an intermediate distribution compared with Su(Hw) and BEAF, may sometimes function in large-scale organization and sometimes work at the level of individual developmental gene units. Together, the three CP190 insulator subclasses could create a chromatin web that is part of the framework organizing DNA in the nucleus (Fig. 4B).
Insulators have been typically characterized as sequences capable of regulating interactions between transcriptional regulatory sequences and/or chromatin states (Gaszner and Felsenfeld 2006). This function can easily be envisioned in the case of Su(Hw) and dCTCF sites located far from genes, where these sites could flank a group of transcription units that would then represent a domain of coregulated genes. If this is the case, what is the function of the remaining dCTCF and BEAF sites located close to the 5′ and 3′ ends of genes? This distribution is surprising in the context of what we normally think of as insulator function; however, when CTCF-binding sites were mapped in the human genome, a similar distribution pattern was observed (Kim et al. 2007; Cuddapah et al. 2009). This is suggestive of a wider role for insulator proteins than just the establishment of chromatin domains, and, in fact, alternative insulator protein functions have been suggested. For example, CTCF in humans is present in the Igh locus in many of the VH as well as DH and JH exons, suggesting a role in V(D)J recombination (Degner et al. 2009). Additionally, this study provides evidence that insulator proteins near genes play a role in the regulation of expression of specific genes and suggests that the mechanism behind this regulation differs from classic transcription factors, since the same insulator complexes were seen to both activate and repress transcription. These functions could be a consequence of the ability of these proteins to both interact with each other and mediate intra- and interchromosomal loops. Bringing together various insulator protein-binding sites could facilitate localization to either transcriptionally active or transcriptionally repressed regions of the nucleus depending on the genomic context of the sites.
Comparison of the genome-wide distribution of the three insulator subclasses in two different cell lines allows us to gain insights into possible mechanisms employed during cell differentiation to establish different patterns of gene expression. Overall, the analysis suggests that cells may control insulator function at multiple levels and that these forms of regulation occur throughout the genome. Regulation of insulator function seems to begin at the level of DNA binding, as we observed differential binding at 5%–37% of sites for Su(Hw), dCTCF, and BEAF between two different cell lines even with the most conservative statistical analysis. Similar percentages of cell type-specific binding sites were observed for vertebrate CTCF between different cell lines (Kim et al. 2007; Cuddapah et al. 2009). Previous analysis of Su(Hw) binding in various tissues has not revealed any significant tissue-specific binding sites (Parnell et al. 2006; Adryan et al. 2007), perhaps because only a small number of sites was analyzed in these studies. Alternatively, the discrepancy could be due to the use of whole tissues in previous studies that contain multiple cell types, making it difficult to detect cell type-specific sites.
After Su(Hw), dCTCF, and BEAF bind DNA, they are thought to recruit other proteins such as CP190. We also observed regulation at this level throughout the genome, where a subset of the Su(Hw), dCTCF, and BEAF sites recruit CP190 in a cell type-specific manner. The additional Su(Hw), dCTCF, and BEAF sites that do not recruit CP190 in either Kc or Mbn2 cells may do so in other cell types or other growth conditions not tested in this study. This idea is supported by the two dCTCF sites in the bithorax region that were found to contain CP190 in third instar larvae brains (Mohan et al. 2007) but not in the data sets collected in Kc cells or Mbn2 cells. Although further study is needed to determine which sites of insulator protein localization participate in chromatin organization, we expect that sites lacking CP190 do not, since mutations in CP190 are known to disrupt insulator body formation and only those sites that recruit CP190 seem to affect gene expression. Therefore, these sites may represent insulators that are poised for incorporation into chromatin loops upon recruitment of CP190. On the other hand, these sites could function through the recruitment of an alternative cofactor and in this way represent a functionally distinct subset of Su(Hw)-, dCTCF-, and BEAF-binding sites.
An additional layer of regulation may then occur at the level of protein–protein interactions mediated by CP190. This type of regulation cannot be gleaned from ChIP–chip data, but other experiments have shown that sumoylation of insulator proteins is able to inhibit protein–protein interactions affecting Su(Hw) insulator body formation but not association of insulator proteins with DNA (Capelson and Corces 2006). Similarly, vertebrate CTCF insulator function has been linked to poly(ADP-ribosyl)ation (PARlation), and it has been suggested that PARlation facilitates CTCF self-interaction (Yu et al. 2004; Klenova and Ohlsson 2005). Furthermore, the presence of RNA and RNA-binding proteins may also contribute to the formation or maintenance of insulator bodies required to create chromatin loops (Lei and Corces 2006). Finally, insulator bypass that results in the inactivation of insulator activity through pairing of nearby insulator elements, and specialized sequences such as the promoter targeting sequences (PTS), can allow an enhancer to bypass an insulator (Schweinsberg et al. 2004; Gruzdeva et al. 2005; Kyrchanova et al. 2007, 2008; Rodin et al. 2007). These forms of regulation may alter the ability of insulator proteins to interact with one another to regulate insulator loop formation.
We expect that these various forms of regulation including DNA binding, CP190 recruitment, and loop formation result in changes in gene expression between different cell lines. However, transcription analysis with insulator proteins is difficult since insulator elements are thought to control regulatory elements such as enhancers and silencers that can be found far away from their target promoters. Therefore, determining which genes are controlled by an insulator site is not a trivial process. In our transcription analysis, we only considered genes with a cell type-specific insulator site within the gene or the 1 kb surrounding region. Despite this limitation, we still saw a significant enrichment for genes that change expression between cell types when they have a cell type-specific insulator site nearby, supporting the idea that insulator proteins are involved in the regulation of gene expression. Genes that did not change expression despite being located near a cell type-specific insulator protein-binding site may not be the actual target genes of the insulator sites; therefore, this analysis probably greatly underestimates the effect of insulator proteins on gene expression. Additionally, we found that insulator protein-binding sites that localize to genes are enriched at genes with certain expression signals, high expression for dCTCF and BEAF, and low expression for Su(Hw). However, comparison between the two cell lines revealed that expression can be either positively or negatively regulated by sites with each DNA-binding protein. Therefore, although an insulator protein associates with a highly expressed gene, it may lead to either an increase or decrease in transcription of this gene. The observed level of expression may be an additive effect of many different regulatory elements, including multiple insulator sites. Different mechanisms may be used to regulate a highly transcribed gene versus a gene with low levels of transcription, and therefore the different insulator subclasses may target these different mechanisms.
The transcription analysis in this study suggests that insulator proteins play a role in the regulation of gene expression, but has just begun to explore the depth of their effect. Numerous steps at which insulator activation can be subject to regulation allow for a vast amount of variation between different cell types and could play a major role in establishing the diverse patterns of chromatin organization necessary for cell type-specific gene expression. The different CP190 insulator subclasses might have distinct roles in this cell type-specific nuclear organization. In vertebrates, CTCF is the only insulator known thus far, and an important question to address in the future is the apparent disparity between genome complexity and insulator diversification between flies and vertebrates. It is possible that vertebrates have insulator subclasses represented by DNA-binding proteins other than CTCF that have not yet been identified. Alternatively, it is possible that vertebrate CTCF has acquired all the functions of the various Drosophila insulator subclasses. The distribution pattern of dCTCF suggests that it can play a role in both global organization and in the regulation of individual genes, making it the most likely candidate of the three Drosophila subclasses to play this overarching organizational role in vertebrates. Therefore, vertebrates may use methods other than variant DNA-binding proteins to distinguish insulator subclasses, such as recruitment of different CTCF interaction partners at different insulator sites (Wallace and Felsenfeld 2007). This would make it difficult to distinguish between the various layers of insulator control in the vertebrate genome. If this is the case, Drosophila could provide a powerful model system to dissect the various functions and levels of regulation of chromatin insulators.
Materials and methods
Antibodies
Rabbit α-Su(Hw) (Gerasimova et al. 1995), α-dCTCF (Gerasimova et al. 2007), and α-CP190 (Pai et al. 2004) antibodies have been validated previously. For BEAF antibody production, an N-terminal His-tagged protein consisting of amino acids 1–83 of the BEAF-32B gene was purified from Escherichia coli on a nickel-agarose column and was used to immunize rabbits using standard procedures. The specificity of the resulting BEAF antibody was validated by staining polytene chromosomes and showing the absence of staining in mutant flies (G Baraka and V Corces, unpubl.).
ChIP–chip analysis
ChIP was performed with 3 × 107 to 5 × 107 Kc or Mbn2 cells at ∼80% confluency. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Nuclear lysates were sonicated to generate 200- to 1000-bp DNA fragments. ChIP was then performed with 6 μL of rabbit α-Su(Hw), α-dCTCF, α-CP190, or α-BEAF-32B antibody. For microarray analysis, samples were amplified two times using the GenomePlex Complete Whole Genome Amplification kit (Sigma, WGA2) to obtain a sufficient amount of DNA. Sample labeling, hybridization, and peak analysis was then performed by NimbleGen using whole-genome tiling arrays (dm2 genome annotation). Three biological replicates were performed for Su(Hw), dCTCF, and CP190, and we used NimbleGen peak analysis software to define peaks for each replicate. Cutoffs were then used to create less stringent and more stringent data sets. The less stringent data sets included all peaks with less than a 5% FDR, and the more stringent data sets included all peaks with less than a 1% FDR. These data sets were then further refined by requiring the presence of each peak in two out of three of the biological replicates to be included in the final data set. The final log2 value for each peak was defined as the average height of the peaks in the biological replicates. For BEAF we performed two biological replicates and the remaining analysis was performed as described above for the other insulator proteins, except that a peak had to be found in both biological replicates to be included in the final data set. BED files containing peak data that can be uploaded to the UCSC genome browser are included in the Supplemental Material. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO accession number GSE15661.
Real-time PCR analysis
Real-time PCR analysis for random peak validation was performed in duplicate with independent ChIP samples than were used for microarray analysis. Thermo Scientific Absolute Blue QPCR SYBR Green ROX Mix (AB-4163) was used and percent input was calculated with a three-point standard curve from the input sample. For cell type-specific peak validation, real-time PCR analysis was performed in triplicate with independent ChIP samples, different from those used for the microarray analysis, using a five-point standard curve from the input sample. Primers used for this analysis are listed in Supplemental Table S13. In order to account for variation between experiments, all Su(Hw) and CP190 percent input values were normalized by setting the percent input at the 1A-2 locus to 1, dCTCF data were normalized by setting the Fab-8 locus to 1, and BEAF data were normalized by setting the scs' locus to 1.
Motif sequence identification
All motif enrichment analyses were conducted using the motif discovery program Weeder (Pavesi et al. 2004). For all the analyses, the following Weeder parameters were used: The length of the motif sought and the degree of approximation allowed were set to “large,” both strands of the input sequences were considered, the motif does not have to appear in all sequences, and the motif can appear more than once per sequence. Using these parameters, enrichment motifs for CP190 and BEAF ChIP–chip data were conducted. The top 100 peaks from each of the ChIP-chip data sets analyzed were also used with the motif discovery programs MEME (multiple em for motif elicitation) (Bailey and Elkan 1994), BioProspector (Liu et al. 2001), and SCOPE (Suite for Computational identification Of Promoter Elements) (Carlson et al. 2007) to verify Weeder results. These additional analyses revealed similar enrichment motifs. In addition, the top 100 peaks from the ChIP–chip data sets underwent secondary motif enrichment analysis where each of the sequences were randomly shuffled, keeping GC content unchanged. There was no enrichment for the BEAF consensus in the shuffled sequences, giving credence to the sequences used and not to random GC content for the specificity of the BEAF motif.
Genome distribution and overlapping peak analysis
Genome distribution analysis was performed with Galaxy (http://main.g2.bx.psu.edu). To determine the position of a peak relative to genomic elements, the middle of a peak was used as its location in the genome, and gene annotations were obtained from the April 2004 UCSC database. To relate peaks to gene expression data between the two cell types, a gene was considered the gene plus 1 kb upstream and downstream. Overlapping peak analysis to determine insulator protein overlap and cell type-specific insulator sites was performed using a 500-bp window on either side of a defined peak. Enriched regions for CP190 and dCTCF from modENCODE data for S2 cells were obtained from the modENCODE project website (http://www.modencode.org). Pair files from Bartkuhn et al. (2009) (GEO accession no. GSE12749) were subject to ChIP Compute Ratio Files analysis and ChIP-Find Peaks analysis with NimbleScan software (http://www.nimblegen.com/products/software/index.html) using default settings and a 1% FDR cutoff.
Gene expression analysis
Microarray analysis was carried out using D. melanogaster 60-mer expression arrays (NimbleGen catalog no. A4351001-00-01) that contain 15,634 genes. RNA was purified from Kc cell and Mbn2 cells with an RNeasy kit (Qiagen) and cDNA synthesis was performed with a High-Capacity cDNA Reverse Transctipion kit (Applied Biosystems). Sample labeling, hybridization, and data normalization were performed by NimbleGen. Experiments were conducted in duplicate for each cell type (Supplemental Fig. S2) and data analysis was performed with ArrayStar software. All 15,634 genes were ranked by expression signal and then divided into three equal groups of ∼5211 genes each. Genes in the top one-third, with the highest expression signal from the microarray analysis, were considered to have a high expression signal, the next one-third were considered to have a medium expression signal, and the bottom one-third were considered to have low expression signal. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO accession number GSE15660.
Gene ontology analysis
Gene ontology analysis for genes with Su(Hw), dCTCF, or BEAF sites within 200 bp upstream of the TSS was performed with DAVID (http://david.abcc.ncifcrf.gov). Flybase IDs were used to determine statistically enriched biological process categories on the basis of a background list of all annotated genes in the Drosophila genome.
Acknowledgments
We thank the members of the Corces laboratory for invaluable discussions and suggestions. We also thank Drs. Caline Karam, Elissa Lei, and Jennifer Phillips for advice and critical reading of the manuscript and the BimCore Facility at Emory University for help in data analysis. This work was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1798209.
Supplemental material is available at http://www.genesdev.org.
References
- Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 2007;8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. EMBO J. 2006;25:1906–1914. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, DeZiel CE, Gross RH. SCOPE: A Web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas P, Cherbas L, Williams CM. Induction of acetylcholinesterase activity by β-ecdysone in a Drosophila cell line. Science. 1977;197:275–277. doi: 10.1126/science.877552. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Laemmli UK. Identification of a class of chromatin boundary elements. Mol Cell Biol. 1998;18:7478–7486. doi: 10.1128/mcb.18.12.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Kas E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–2910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Golovnin A, Biryukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003;130:3249–3258. doi: 10.1242/dev.00543. [DOI] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hart CM, Zhao K, Laemmli UK. The scs' boundary element: Characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle. 2005;4:96–101. doi: 10.4161/cc.4.1.1398. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Liu X, Brutlag DL, Liu JS. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001;2001:127–138. [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: Boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci. 2003;100:13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–2349. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. New properties of Drosophila fab-7 insulator. Genetics. 2007;177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes & Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: Insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–888. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]