Abstract
In liver, most metabolic pathways are under circadian control, and hundreds of protein-encoding genes are thus transcribed in a cyclic fashion. Here we show that rhythmic transcription extends to the locus specifying miR-122, a highly abundant, hepatocyte-specific microRNA. Genetic loss-of-function and gain-of-function experiments have identified the orphan nuclear receptor REV-ERBα as the major circadian regulator of mir-122 transcription. Although due to its long half-life mature miR-122 accumulates at nearly constant rates throughout the day, this miRNA is tightly associated with control mechanisms governing circadian gene expression. Thus, the knockdown of miR-122 expression via an antisense oligonucleotide (ASO) strategy resulted in the up- and down-regulation of hundreds of mRNAs, of which a disproportionately high fraction accumulates in a circadian fashion. miR-122 has previously been linked to the regulation of cholesterol and lipid metabolism. The transcripts associated with these pathways indeed show the strongest time point-specific changes upon miR-122 depletion. The identification of Pparβ/δ and the peroxisome proliferator-activated receptor α (PPARα) coactivator Smarcd1/Baf60a as novel miR-122 targets suggests an involvement of the circadian metabolic regulators of the PPAR family in miR-122-mediated metabolic control.
Keywords: Circadian, miRNA, miR-122, metabolism, clock, PPAR
Light-sensitive organisms possess a circadian timekeeping system that serves to synchronize gene expression and physiology with geophysical time (Reppert and Weaver 2002; Gachon et al. 2004). Current models of the mammalian molecular clocks are based on two interlocked transcriptional feedback loops (Sato et al. 2006): a positive limb, in which the heterodimeric BMAL1:CLOCK transcription factor mediates the transcriptional activation of cryptochrome (Cry1 and Cry2) and period genes (Per1 and Per2), and a negative limb, in which PER:CRY complexes repress the BMAL1:CLOCK-mediated transcription of their own genes. Coordination between the two limbs is accomplished by nuclear receptors of the REV-ERB and ROR families (Preitner et al. 2002; Reppert and Weaver 2002; Sato et al. 2004). Cyclic Rev-erbα transcription is regulated by the mechanisms described above for Cry and Per genes, and the circadian accumulation of the repressor REV-ERBα results in the rhythmic repression of target genes, such as Bmal1, carrying retinoid-related orphan receptor elements (ROREs) (Ueda et al. 2002). In addition to these transcriptional feedback loops, numerous post-translational modifications of core clock proteins are known to contribute to the rhythm-generating clockwork circuitry (Gallego and Virshup 2007).
The cyclic expression of clock output genes can be governed directly by core clock components via E-box or RORE sequences (Ueda et al. 2002), or transcription factors such as PAR bZip proteins whose genes are regulated by these mechanisms (Gachon et al. 2004). However, despite the similar molecular makeup of the core oscillator in different organs, its outputs vary substantially between tissues (e.g., Storch et al. 2002). Gene expression profiling in liver has suggested that, depending on the algorithms used for the identification of cyclically expressed genes, 2%–10% of the transcriptome may be under circadian control (Panda et al. 2002; Storch et al. 2002; Kornmann et al. 2007a; Miller et al. 2007). Many of these genes are involved in hepatocyte-specific metabolic pathways.
In part, the synergistic activation of genes by circadian and tissue-specific transcription factors may account for the rhythmic expression of cell type-specific transcripts. However, tissue-specific post-transcriptional regulation of gene expression may also participate in this endeavor. It is estimated that in mammals ∼30% of all mRNAs are subject to regulation by microRNAs (miRNAs) (Lewis et al. 2005), and miRNAs have been implicated in the post-transcriptional control of cellular proliferation, development, and differentiation (Bushati and Cohen 2007). miRNAs are short (∼22 nucleotides [nt]), endogenous RNAs that promote translational repression and/or destabilization of target mRNAs (Bushati and Cohen 2007; Liu 2008). Target recognition occurs via base-pairing interactions with the 3′ untranslated region (UTR). Usually the 5′ portion of the miRNA forms a perfect hybrid with a 6- to 8-nt seed site, whereas the remainder of the miRNA undergoes interactions of only partial complementarity with the 3′UTR of its target mRNA (Lewis et al. 2005). The mismatches and gaps between miRNA and mRNA duplexes render the de novo prediction of miRNA targets challenging. Generally, a given miRNA can be expected to fine-tune the production of large sets of proteins within the cell (Baek et al. 2008; Liu 2008; Selbach et al. 2008).
Given the large fraction of mRNAs targeted by miRNAs, it is likely that miRNAs also modulate clock and clock output functions (Cheng et al. 2007; Xu et al. 2007; Yang et al. 2008). We wished to examine this conjecture and initiated our studies with miR-122, a miRNA that has been proposed to constitute up to 70% of all miRNA molecules in hepatocytes (Lagos-Quintana et al. 2002). The knockdown of miR-122 expression in mice and monkeys has previously been recognized to result in a down-regulation of cholesterol and lipid metabolizing enzymes and a reduction in plasma cholesterol levels (Krutzfeldt et al. 2005; Esau et al. 2006; Elmen et al. 2008a,b). Both lipid and cholesterol metabolism are well known for their daytime-dependent regulation, similar to many other hepatic functions that require coordination of food intake with nutrient-processing and energy homeostasis (Panda et al. 2002).
Here, we show that transcription of the miR-122 locus is under circadian control, involving the transcriptional repressor REV-ERBα. Thus, pri- and pre-miRNA precursors oscillated about fourfold to 10-fold in abundance during the day but accumulated at nearly constant levels in the livers of Rev-erbα knockout mice. However, due to its high stability mature miR-122 levels were virtually constant throughout the day. Despite the apparent invariable temporal accumulation of miR-122, the identification of its target mRNAs suggested that miR-122 nevertheless participates in the circadian control of many transcripts involved in hepatic metabolism. Among the miR-122 targets we found the mRNAs encoding peroxisome proliferator-activated receptor β/δ (PPARβ/δ) and SMARCD1/BAF60a, which are themselves circadian regulators of metabolism (Yang et al. 2006; Seedorf and Aberle 2007; Li et al. 2008).
Results
The miR-122 locus is transcribed in a circadian fashion
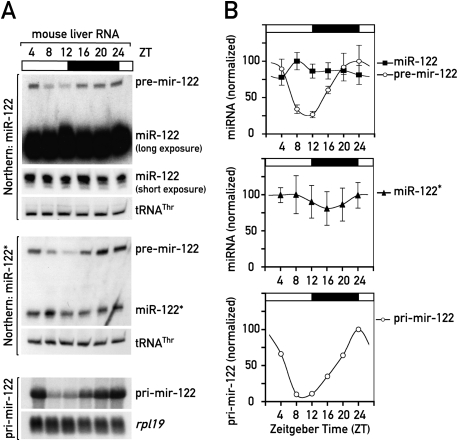
In a search for miRNAs that could shape the circadian expression of target mRNAs, we analyzed the expression of various miRNAs in mouse liver at different time points (Zeitgeber time, ZT) around the day. Several miRNAs (miR-19, miR-20, miR-22, miR-24, miR-30, miR-92, miR-126-3p), some of which had been predicted to target clock components (Lewis et al. 2005), only showed modest, if any, circadian changes in expression, as judged by Northern blot analysis (Supplemental Fig. 1). However, analysis of miR-122, the most abundant miRNA in liver, revealed that pre-mir-122 oscillated with an approximately fivefold daily amplitude in abundance, whereas mature miR-122 levels remained nearly constant over the day (Fig. 1A,B). Pre-mir-122 is a 66-nt hairpin-shaped precursor molecule from which the endonuclease Dicer cleaves the mature 22-nt miR-122. The mature miRNA is then incorporated into the RNA-induced silencing complex (RISC). The same expression pattern for pre-mir-122 was detected with a probe recognizing the strand complementary to the miRNA (known as the miRNA* sequence) (Fig. 1). The observed circadian changes in pre-mir-122 levels could be the result of either circadian synthesis or circadian processing into mature miRNA. To distinguish between these possibilities we analyzed the circadian levels of the miR-122 primary transcript, pri-mir-122, a ∼5-kb precursor (Chang et al. 2004), from which the pre-miRNA is cleaved by the Drosha-containing microprocessor complex.
Figure 1.
miR-122 precursors are circadian in mouse liver. (A) Northern blot analysis of miR-122 and its precursor RNAs using whole-cell RNA from male C57BL/6 mice sacrificed at the indicated ZT values around the clock. An RNA pool from three mice was used per time point, tRNAThr and rpl19 mRNA served as loading controls in denaturing polyacrylamide (top and middle panels) and agarose gel electrophoresis (bottom panels), respectively. (Top panels) miR-122 and pre-mir-122. (Middle panels) pre-mir-122 and miR-122*. miR-122* is the antisense “passenger strand” that is incorporated into RISC at low levels. (Bottom panels) pri-mir-122. (B, top and middle panels) miR-122, miR-122* and pre-mir-122 levels, normalized to tRNAThr, from Northern blots in which single animals were analyzed (data not shown). Mean values ± SEM. (Bottom panel) Quantification of pri-mir-122 levels, normalized to the circadianly invariant rpl19, from the Northern blot shown in A.
As shown in Figure 1A (bottom panels), pri-mir-122 accumulation was highly circadian (∼10-fold amplitude), showing a similar phase as pre-mir-122 (i.e., minimal levels at ZT8-12 and maximal levels at ZT24). We wanted to test if high-amplitude circadian precursors were specific for miR-122 or were a common feature of miRNAs. Two other loci tested, pri-mir-17-92 and pri-mir-22, did not show the circadian pattern observed for pri-mir-122 (Supplemental Fig. 1C,D). This suggested that specifically the miR-122 locus was transcribed in a circadian fashion. The two intermediates in miR-122 biogenesis can be expected to be short-lived and reflect the rate at which the gene is transcribed. In contrast, the absence of cyclic expression at the level of mature miR-122 was probably due to its high metabolic stability. Indeed, based on Northern blot experiments, we estimated that the ratio of miR-122/pre-mir-122 steady-state levels (which is largely determined by the ratio of the half-lives of the two species) is in the range of 400:1. If one assumes that the pre-mir-122 half-life is a few minutes, this means that the miR-122 half-life is probably well beyond 24 h.
The orphan nuclear receptor REV-ERBα drives circadian mir-122 transcription
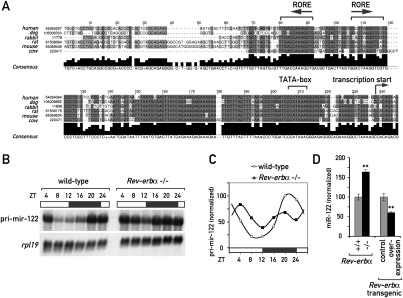
We wished to study the molecular mechanism accounting for circadian mir-122 transcription. The phase of pri-/pre-mir-122 expression suggested that the circadian transcriptional repressor REV-ERBα might be involved: REV-ERBα protein expression peaks at around ZT8, leading to minimal transcript levels for REV-ERBα target genes at around ZT12 (Preitner et al. 2002; Ueda et al. 2002). Consistent with the hypothesis of miR-122 being a REV-ERBα target gene, the mir-122 promoter contains two conserved ROREs ∼120–160 base pairs (bp) upstream of the transcriptional start site (Fig. 2A; see also Supplemental Fig. 2 for an alignment of the promoter region in 32 mammalian species). More importantly, the amplitude of cyclic pri-mir-122 accumulation was severely blunted in the livers of Rev-erbα knockout animals (Fig. 2B,C), and mature miR-122 accumulated to 1.6-fold higher levels (Fig. 2D). The residual amplitude in mir-122 transcription was possibly caused by REV-ERBβ, a highly related paralog of REV-ERBα (Preitner et al. 2002). A second mouse model, in which REV-ERBα was overexpressed specifically in hepatocytes (Kornmann et al. 2007a), showed the converse effect (i.e., 1.7-fold reduced miR-122 levels). In summary, these findings supported a model according to which the miR-122 locus is regulated by the circadian clock component REV-ERBα.
Figure 2.
REV-ERBα is involved in circadian control of the miR-122 locus. (A) Alignment of the genomic sequence upstream of the predicted transcriptional start site of pri-mir-122 in six mammalian species (extracted from the University of California at Santa Cruz alignment; see Supplemental Fig. 3). The predicted ROREs, TATA-box, and transcriptional start site are indicated. (B) Northern blot analysis of pri-mir-122 in total RNA samples from Rev-erbα knockout and littermate control mice sacrificed at the indicated ZT values around the clock. For each time point, an RNA pool of three female mice was used. (C) Quantification of the Northern blot shown in B; values are pri-mir-122 normalized to rpl19. (D) miR-122 levels in total liver RNA from individual animals (mixed ZTs) of the indicated genotypes were quantified by Northern blot (data not shown). Control animals were set to 100%. Data are mean ± SEM (n = 36 for Rev-erbα−/− vs. Rev-erbα+/+ and n = 18 for REV-ERBα overexpression vs. control); (**) P < 5*10−5 (two-tailed Student's t-test).
Does the miR-122 locus specify multiple functional RNAs?
Since the accumulation of pri-mir-122, but not that of mature miR-122, was rhythmic, we considered that this locus produced additional biologically active RNAs with shorter half-lives than miR-122. In fact, several pri-miRNAs are polycistronic and produce multiple miRNAs (Sewer et al. 2005). Although mature miR-122 shows a sequence conservation of 100% from fish to humans (Gerlach et al. 2009), its pri-miRNA gene structure is conserved only in mammals. In these organisms, the transcription start site is flanked by elements of a classical RNA polymerase II (PolII)-dependent promoter, which drives transcription of the ∼5-kb capped and polyadenylated pri-mir-122 containing the pre-mir-122 hairpin at its 3′-end (Fig. 2A; Supplemental Figs. 2, 3A; Chang et al. 2004). Overall, the pri-mir-122 sequence is poorly conserved, and we did not detect additional potential miRNAs (or conserved open reading frames) within the primary transcript. A thorough bioinformatics search for conserved RNA secondary structures within the pri-mir-122 genomic locus in the genomes of six mammalian species also failed to identify additional RNA structures that could carry a function (Supplemental Fig. 3). Thus, it appeared likely that a potential biological function associated with the circadian control of pri-mir-122 transcription was mediated by miR-122 itself.
Genome-wide identification of miR-122 targets
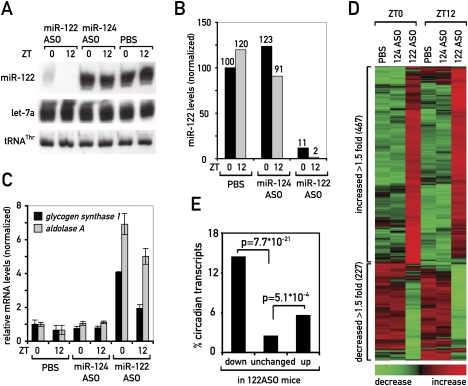
As miR-122 was produced in a circadian fashion, we wondered whether it might assume rhythmic functions despite its long half-life. We decided to approach this question in an unbiased way by identifying putative miR-122 targets. In particular, we wished to determine whether there are targets whose daily rhythms are influenced by miR-122. To deplete miR-122, we injected antisense oligonucleotides (ASOs) intraperitoneally into mice (termed 122ASO in the following sections) and used genome-wide Affymetrix oligonucleotide arrays to determine the impact this had on hepatic mRNA levels. As controls, we used animals treated with ASOs targeting a miRNA not expressed in liver (miR-124; samples 124ASO in the following sections) or with PBS alone. Mice were sacrificed at time-points ZT0 and ZT12, when pri-mir-122 transcription was highest and lowest, respectively.
The efficiency of miR-122 depletion was between 89% and 99% as judged by Northern blot hybridization (Fig. 3A,B). Residual miR-122 levels were consistently lower for mice sacrificed at ZT12, when miR-122 production was low, suggesting that miRNA stability was decreased by 122ASOs. Importantly, the abundance of the unrelated miRNA let-7a remained unchanged (Fig. 3A), demonstrating the specificity of the ASO. To functionally assess if miR-122 was sufficiently depleted to derepress its targets, we determined the mRNA levels of the formerly suggested targets glycogen synthase 1 (Gys1) and aldolase A (AldoA) by quantitative RT–PCR (qPCR). Similar to what had been observed previously (Krutzfeldt et al. 2005; Esau et al. 2006), these mRNAs were up-regulated twofold to sevenfold (Fig. 3C).
Figure 3.
Analysis of miR-122 targets at two time points ZT0 and ZT12. (A) Northern blot analysis of miR-122, let-7a, and tRNAThr of mice treated with miR-122 ASO, miR-124 ASO, or PBS. Pools of RNA of three mice were loaded per lane. (B) Quantification of Northern blot shown in A. (C) qPCR analysis of RNAs from individual mice treated with the ASOs or PBS, as indicated. Probes used were for the known miR-122 targets glycogen synthase 1 and aldolase A, normalized to 45S pre-rRNA. Values are mean ± SEM (n = 3). (D) Heat map of the probe sets up- and down-regulated in 122ASO-treated animals relative to both control groups, 124ASO- and PBS-treated animals (cutoff 1.5). The heat scale at the bottom of the panel represents changes on a linear scale, where green and red represent minimal and maximal expression, respectively. (E) Enrichment for circadian transcripts in the up- and down-regulated fractions in 122ASO mice. P-values were determined by a χ2 test.
miRNAs initially have been proposed to mediate translational repression of their target mRNAs. This is often accompanied by a decrease in mRNA abundance (Baek et al. 2008; Selbach et al. 2008). Transcriptomal profiling using microarrays is therefore a convenient means to identify potential miRNA targets. Obviously, this technology is unable to detect miRNA targets whose translational attenuation is not accompanied by increased degradation.
Using Affymetrix microarray hybridization, we detected signals for a total of 22,384 probe sets, representing 11,638 transcripts. Among these, we found 343 transcripts (represented by 467 probe sets) that were up-regulated, and 188 transcripts (227 probe sets) that were down-regulated at at least one of the two time points in 122ASO-treated animals, when we applied a 1.5-fold expression change cutoff (Fig. 3D). We next analyzed whether transcripts up-regulated in 122ASO livers were enriched for potential miR-122 targets. For the prediction of potential miR-122-binding sites we applied a model that takes into account both the presence of miRNA seed sites and the energy of miRNA:mRNA duplexes, ensuring that energetically stable miRNA–target interactions are considered. Using this thermodynamic model (with an energy cutoff of −15 kcal/mol), we observed that 52% of transcripts in the up-regulated fraction contained a predicted miR-122-binding site (Supplemental Fig. 4). With only 22% of transcripts in the unchanged and 14% in the down-regulated fraction, this enrichment in the up-regulated fraction was statistically highly significant (up vs. unchanged: P-value ∼10−39; up vs. down: P-value ∼10−17). The differences between the unchanged and down-regulated fractions, however, were barely significant (P-value ∼0.02). With a less elaborate model that only considers seed site presence, the enrichment for potential miR-122 targets in the up-regulated fraction was significant as well (Supplemental Fig. 4).
We next wished to determine, whether transcripts showing a time point-specific regulation by miR-122 could be clustered into particular metabolic pathways. To this end, we selected the transcripts that showed regulation upon 122ASO treatment exclusively at one of the two time points. Genome ontology (GO) analyses in the down-regulated fraction revealed that the genes involved in lipid and cholesterol metabolism (which had been reported previously to be most responsive to miR-122 depletion) also showed the strongest temporal regulation (P ∼ 10−10). Thus, the down-regulation of these mRNAs was significantly stronger at ZT12 than at ZT0 (Supplemental Fig. 5A). For up-regulated genes, transcripts belonging to GO:9607 “response to biotic stimuli” were most overrepresented (P ∼ 10−7). Their up-regulation occurred mainly at ZT0 and less so at ZT12 (Supplemental Fig. 5B). These observations suggested a considerable amount of cross-talk between circadian gene expression and miR-122, and encouraged us to analyze the effect of miR-122 depletion on circadian gene expression in greater detail.
Circadian transcripts are highly enriched among miR-122 targets
We wished to focus on transcripts that were direct potential targets of miR-122 and that showed circadian expression. For the genome-wide analysis of cyclic transcripts, we used previously reported transcriptome profiling experiments (Kornmann et al. 2007a). This work analyzes the hepatic transcriptome in a transgenic mouse model in which REV-ERBα can be conditionally overexpressed in liver in a doxycycline-dependent manner (tet-off system). In the presence of doxycycline, the hepatic circadian clock is functional in these animals, as the Rev-erbα transgene is constitutively repressed. The gene expression profiles from these animals, sampled over a 48-h period (with a resolution of 4 h), have been used to identify the circadian hepatic transcriptome using stringent algorithms (Kornmann et al. 2007a,b). In the absence of doxycycline, REV-ERBα overexpression arrests the endogenous liver clock. Thus, most circadian genes lose rhythmicity, with the notable exception of a small fraction of transcripts whose rhythms are driven by systemic cues rather than local oscillators (Kornmann et al. 2007a,b). In these mice, REV-ERBα overexpression also led to reduced miR-122 levels (Fig. 2D). It may thus be assumed that the derepression of miR-122 targets contributed to the gene expression changes observed upon REV-ERBα overexpression. We therefore compared the gene expression changes common to REV-ERBα overexpression and 122ASO administration. Of the transcripts whose abundance changed under both conditions, the majority (79.2%) indeed showed regulation in the same direction and only few (20.8%) showed reverse regulation (Supplemental Fig. 6). These observations lend further support to a role of REV-ERBα in miR-122 regulation.
We next analyzed the probe sets representing transcripts with circadian accumulation. Using stringent algorithms, these corresponded to ∼2.8% of the liver transcriptome (Kornmann et al. 2007a,b). We found that the up- and down-regulated fractions in the 122ASO mice were significantly enriched for circadian transcripts: 14.4% of the down-regulated, 5.5% of the up-regulated, but only 2.4% of the unchanged mRNAs were among those classified as circadian (Fig. 3E). We thus concluded that the effects of depleting miR-122 were biased toward a misregulation of circadian transcripts. Since the enrichment was particularly high in the down-regulated fraction that contained indirect miR-122 targets, there were possibly common circadian regulatory mechanisms in control of this group. Indeed, almost a quarter of the transcripts in this fraction belonged to lipid/cholesterol metabolizing enzymes.
Identification of circadian mRNAs with functional miR-122-binding sites
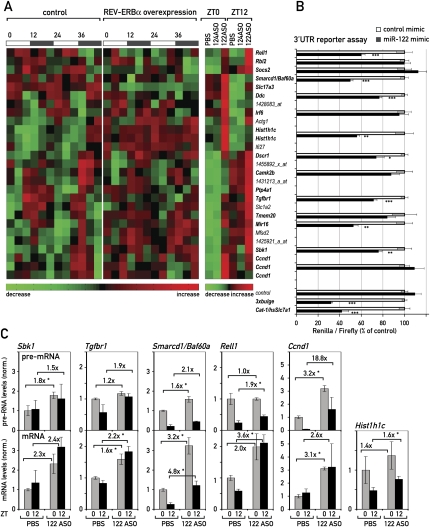
We next investigated in more detail the group of transcripts with circadian accumulation that were up-regulated upon miR-122 depletion, as this subset was likely to contain the direct miR-122 targets (Fig. 4A). Within this group, 16 transcripts (specified by 19 probe sets) contained potential miR-122-binding sites in their 3′UTRs and were therefore candidates for circadianly expressed miR-122 targets (Fig. 4A, bold type). Many of them were also up-regulated in REV-ERBα-overexpressing animals (Fig. 4A, middle panel). We wished to verify that the changes in mRNA abundance detected by microarray analysis were potentially the direct result of miR-122 derepression, as opposed to more complicated indirect effects. Therefore, we tested the impact of miR-122 on the 3′UTRs of several candidate transcripts in cotransfection experiments. To this end, we cloned the candidate 3′UTRs into a vector carrying a renilla luciferase reporter gene, and transfected these constructs together with synthetic miRNA mimics into Hela cells, which do not express endogenous miR-122. We then measured the ability of a miR-122 mimic to inhibit the expression of luciferase when its open reading frame was followed by a particular 3′UTR. Two 3′UTRs known to be regulated by miR-122 served as positive controls: an artificial 3′UTR containing three optimized miR-122-binding sites (3xbulge) (Pillai et al. 2005), and the 3′UTR of Cat-1/human Slc7a1, a well-known miR-122 target (Chang et al. 2004; Bhattacharyya et al. 2006). These two 3′UTRs mediated a miR-122-dependent repression by about 68% and 58%, respectively. In contrast, luciferase expression from reporters harboring the vector-based 3′UTR devoid of miR-122 seed sites was not affected (Fig. 4B; Supplemental Fig. 7). Of the circadian transcripts up-regulated in 122ASO mice, we found that the 3′UTRs of Rell1 (receptor expressed in lymphoid tissues-like 1), Smarcd1/Baf60a (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 1/BRG1-associated factor 60a), Ddc (dopa decarboxylase), Hist1h1c (histone cluster 1, H1c), Dscr1 (down syndrome critical region protein 1), Tgfbr1 (TGF-β receptor type 1), Mir16 (membrane-interacting protein of RGS16), and Sbk1 (SH3-binding kinase 1) conferred sensitivity toward miR-122 (Fig. 4B). A complete compilation of the >30 3′UTRs we tested, including those of several newly identified miR-122 targets, is given in Supplemental Figure 7.
Figure 4.
Circadian genes are miR-122 targets. (A) Heat map of the circadian probe sets (left and middle panel; taken from Kornmann et al. 2007b) that are up-regulated in 122ASO mice (right panel). Smarcd1/Baf60a was just below the stringent criteria used for circadian expression in the microarray data of Kornmann et al. (2007b), but was also included in the figure as it was confirmed as robustly circadian by qPCR (see Fig. 5). Heat scales at the bottom of the panels represent changes on a linear scale with green and red representing minimal and maximal expression, respectively. Transcripts in bold type contain potential miR-122 seed sites in their 3′UTRs. (B) The effect of miR-122 mimics in a 3′UTR luciferase assay. Control has only the vector 3′UTR, containing no seed sites. 3xbulge and Cat-1/hsSlc7a1 are positive controls for 3′UTRs known to be regulated by miR-122. Values are mean ± SEM (n ≥ 6 per transfection). (*) P < 10−2; (**) P < 10−3; (***) P < 10−4 (two-tailed Student's t-test). (C) qPCR analysis in 122ASO mice and PBS controls of pre-mRNA (top panels) and mRNA (bottom panels) levels of selected transcripts from A. Hist1h1c is an intron-less gene; hence, pre-mRNA levels were not measured. Note that Ccnd1 is also changed on the pre-mRNA level and is hence probably up-regulated by an indirect, transcriptional effect. Data are mean values of three mice per condition ±SEM. (*) P < 10−2 (two-tailed Student's t-test).
miR-122 contributes to circadian mRNA expression
For some selected targets, we wanted to verify that their up-regulation in the 122ASO mice was indeed caused by post-transcriptional, rather than indirect transcriptional mechanisms. Since miRNAs are thought to act on processed mRNAs, a derepression mediated by the 122ASO should manifest itself on the level of the mature mRNA, but not on that of its pre-mRNA. Indirect effects, however, can be expected to occur through changes in transcription rates, caused by the up-regulation of activators or repressors whose production depends on miR-122. These changes should also be visible on the pre-mRNA level. Hence, we designed qPCR probes enabling us to measure mRNA and intron-containing pre-mRNA levels of several of the identified targets. Our analyses showed that the up-regulation of mature mRNA levels for the transcripts Sbk1, Tgfbr1, Smarcd1/Baf60a, Rell1, and Hist1h1c was similar, or even greater, than assessed by the microarray analysis. The effects of the 122ASO on pre-mRNA levels, however, were less pronounced (Fig. 4C). In contrast, a transcript such as Ccnd1 fulfills the criteria for being indirectly affected. Thus, while Ccnd1 was also circadian and up-regulated in 122ASO mice (Fig. 4A), it did not confer sensitivity to miR-122 in the 3′UTR assay (Fig. 4B). In keeping with this observation, the changes in Ccnd1 expression were already observed on the level of pre-Ccnd1 mRNA accumulation (Fig. 4C).
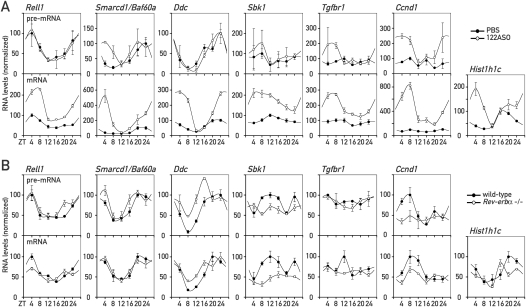
To evaluate more precisely which influence miR-122 had on shaping the rhythmic accumulation of these transcripts, we extended our analyses to 122ASO mice that had been sacrificed at six time points around the clock. Using RNA pools from three to four animals per time point and for both control and 122ASO mice (see Supplemental Fig. 8), we observed similar increases in target mRNA accumulation as in the previous two time point experiments (Fig. 5A; Supplemental Fig. 8D). In addition, it was apparent that miR-122 depletion had striking effects on the circadian amplitude (Smarcd1/Baf60a, Ddc, Hist1h1c), magnitude (Rell1) and phase (Smarcd1/Baf60a, Hist1h1c, and Ddc) of accumulation (Fig. 5A, bottom panels). For several transcripts (Smarcd1/Baf60a, Ddc, and Hist1h1c) we also observed that derepression caused an especially strong up-regulation at around ZT4 (Fig. 5A, bottom panels). This time point corresponds to a few hours after maximal mir-122 transcription (see Fig. 1B). Moreover, despite a particularly efficient miR-122 depletion at ZT12 (Fig. 3A,B; Supplemental Fig. 8), derepression clearly had a milder effect at this time point (Fig. 5A, bottom panels). For some of the miR-122 targets, these time-dependent effects were already observed in the microarray data (Fig. 4A). Due to their low abundance, the detection of the corresponding pre-mRNAs was less robust than that of the mature transcripts (Fig. 5A, top panels). Nevertheless, it was evident that (with the exception of Ccnd1) differences between 122ASO and control mice could not be accounted for by different transcription rates. These findings indicated that miR-122 probably assumes rhythmic functions despite its constant levels (see the Discussion). Importantly, the circadian clock per se did not appear to be affected by 122ASO treatment, as the mRNA levels of core clock and clock output genes were essentially unchanged in 122ASO mice (Supplemental Fig. 9).
Figure 5.
miR-122 targets in 122ASO and Rev-erbα knockout mice around the clock. (A) pre-mRNA (top panels) and mRNA (bottom panels) levels for the indicated transcripts in 122ASO and PBS-injected control mice around the clock. For each data point, transcript levels were measured in triplicate by qPCR using a pool of total liver RNA isolated from three to four mice. Due to low abundance, the detection of pre-mRNA levels was less robust, as indicated by generally larger error bars (standard deviations) in the qPCR analysis. (B) As in A, pre-mRNA (top panels) and mRNA (bottom panels) levels measured around the clock in Rev-erbα knockout and wild-type littermate animals, using a pool of whole-cell liver RNA isolated from five female mice.
As shown in Figure 2, the circadian amplitude of mir-122 transcription was blunted in Rev-erbα knockout mice (Fig. 2B,C), leading to ∼1.6-fold higher miR-122 levels (Fig. 2D). We wanted to examine whether these alterations in miR-122 production were sufficient to perturb the rhythmic expression of any of the targets analyzed above. When measuring mRNA and pre-mRNA abundances around the clock in Rev-erbα knockout and control animals, we observed the strongest post-transcriptional perturbations of rhythms for Rell1, a ubiquitously expressed member of the tumor necrosis factor (TNF) receptor family (Cusick et al. 2006). The amplitude of Rell1 mRNA, but not that of its pre-mRNA, was blunted in the Rev-erbα knockout (1.7-fold amplitude) as compared with the control mice (2.6-fold amplitude) (Fig. 5B). This further supported our conclusion that Rell1 was an example for an mRNA whose circadian rhythm was partially shaped by post-transcriptional mechanisms. In view of its up-regulation in 122ASO mice on the mRNA but not the pre-mRNA level (Figs. 4C, 5A) and the fact that the Rell1 3′UTR conferred sensitivity to miR-122 in the reporter assay (Fig. 4B), it is likely that miR-122 was directly implicated in this process. The cyclic accumulation of other miR-122 targets, such as Smarcd1/Baf60a, was unchanged in Rev-erbα knockout mice, whereas some changes already occurred on the pre-mRNA level (Fig. 5B).
Cross-talk between miR-122 and PPARs
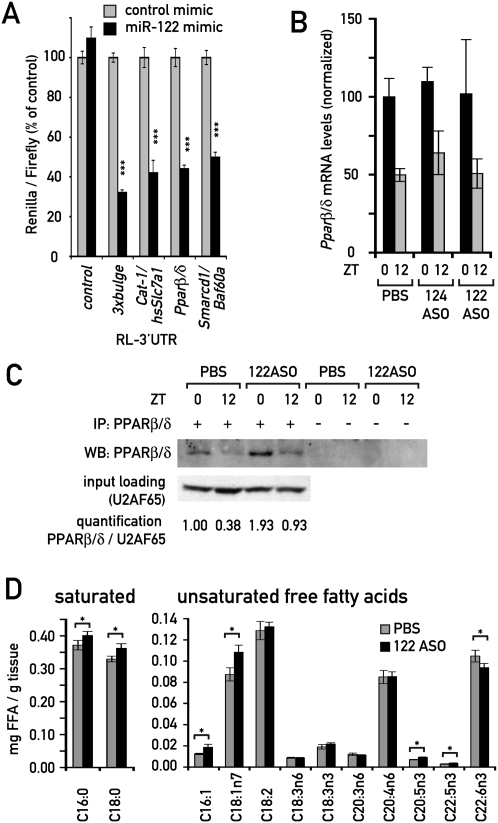
The down-regulation of enzymes associated with lipid and cholesterol metabolism (see Krutzfeldt et al. 2005; Esau et al. 2006; Elmen et al. 2008a; this study) in miR-122-depleted mice implies that the corresponding mRNAs are regulated by indirect mechanisms. However, the direct miR-122 targets responsible for these control mechanisms remained to be identified. We suspected that these direct targets were also expressed in a circadian manner, since the down-regulation of mRNAs encoding lipid and cholesterol enzymes was daytime-dependent (Supplemental Fig. 5A). Interestingly, recent work has suggested that SMARCD1/BAF60a, a component of the SWI/SNF chromatin-remodeling complex, specifically regulates hepatic lipid metabolizing genes (Li et al. 2008). In our experiments, Smarcd1/Baf60a mRNA appeared as a circadian and direct miR-122 target, as it was robustly up-regulated in 122ASO mice (Figs. 4A,C, 5A) and as its 3′UTR was responsive to miR-122 in our cotransfection experiments (Figs. 4B, 6A). Li et al. (2008) further demonstrated that SMARCD1/BAF60a interacts and cooperates with the metabolic regulator PPARα, and that SMARCD1/BAF60a and PPARα share a large number of target genes.
Figure 6.
Cross-talk between miR-122 and PPAR receptors. (A) The effect of the miR-122 mimic in a 3′UTR luciferase assay as in Fig. 4B, using the Pparβ/δ and Smarcd1/Baf60a 3′UTRs. Values are mean ± SEM (n ≥ 9 per transfection). (***) P < 10−5 (two-tailed Student's t-test). (B) Expression levels of Pparβ/δ mRNA quantified from Northern blots. Data are mean ± SEM (n = 3 animals per condition). (C) Immunoprecipitation-Western blot of PPARβ/δ protein from 122ASO and PBS-treated mice, as described in the Materials and Methods. Each immunoprecipitation was performed from a pool of extracts from three mice. U2AF65 protein levels in the input of the same pool served as a loading control. (D) FFA levels in liver pieces from 122ASO- and PBS-injected animals, as determined by GC/MS. Values are mean ± SEM (n = 6). (*) P < 0.05 (two-tailed Student's t-test).
PPARs belong to the nuclear hormone receptor superfamily and are well-known metabolic regulators. They are activated upon binding to their mainly amphipathic ligands, which are mostly derived from dietary fat or endogenous fatty acid metabolism. Of the three PPAR isotypes, PPARα, and the less-studied PPARβ/δ, serve predominantly catabolic functions, whereas PPARγ mainly promotes lipid storage in adipose tissue. In liver, all PPARs show circadian expression (Yang et al. 2006). Although we did not find PPAR transcripts misregulated using microarrays with RNA from 122ASO mice, we noticed that the Pparβ/δ 3′UTR contained four miR-122 seed sites that could be predicted to confer strong targeting by miR-122. We therefore tested if the Pparβ/δ 3′UTR showed sensitivity to miR-122 in our cotrans-fection experiments. Indeed, this 3′UTR caused a miR-122-dependent reduction of luciferase activity by 56%, which was among the highest down-regulation effects we observed in these assays. Only the two positive controls, the artificial 3xbulge and the Cat-1/human Slc7a1 3′UTR showed a slightly stronger repression (Fig. 6A; Supplemental Fig. 7). Consistent with these findings, we observed that whereas Pparβ/δ mRNA levels remained unchanged upon miR-122 depletion (Fig. 6B), the protein was up-regulated around twofold to threefold, as judged by Western blot experiments with 122ASO liver extracts (Fig. 6C). These findings strongly suggested that Pparβ/δ was a bona fide miR-122 target that thus far had been overlooked, supposedly because it is not regulated on the level of mRNA stability.
Unsaturated fatty acids are probably the most important endogenous PPAR ligands, and their levels are known to be tightly regulated in vivo. The perturbation of lipid metabolism associated with miR-122 depletion may thus also lead to changes in PPAR ligand availability. We therefore determined the concentrations of free fatty acids (FFAs) in livers from 122ASO and control mice by GC/MS. Several unsaturated FFA species were indeed significantly changed, including palmitoleic acid (C16:1; up by 51% in 122ASO mice) and vaccenic acid (C18:1n7; up by 24%) (Fig. 6D). The latter constitutes a significant proportion of the total unsaturated FFA pool and has previously been proposed as a PPARβ/δ ligand (Fyffe et al. 2006). We therefore deemed it likely that PPAR activity in 122ASO mice was additionally modulated by changes in ligand concentration.
We conclude that miR-122 has several ties to the PPAR family of nuclear receptors, via Pparβ/δ, Smarcd1/Baf60a, and possibly ligand availability. Given the important functions PPARs possess in regulating metabolism in liver, these connections are very likely to contribute to the overall metabolic phenotype observed in 122ASO mice.
Discussion
Circadian mir-122 transcription and function
In the present study, we show that miRNA miR-122 expression and function are embedded in the output system of the circadian clock. Thus, we found that the miR-122 locus was transcribed in a circadian manner, manifesting itself in rhythmic pri-mir-122 and pre-mir-122 expression. Based on genetic loss-of-function and gain-of-function experiments we concluded that the orphan receptor REV-ERBα is the dominant regulator of circadian mir-122 transcription. On a genome-wide scale, we observed that the portion of the transcriptome sensitive to miR-122 depletion was highly enriched for circadian mRNAs, and it appeared that these were biased toward specific circadian phases (Supplemental Fig. 10). This temporal gating was particularly evident for mRNAs encoding cholesterol and lipid metabolizing enzymes, which were identified previously as indirectly regulated miR-122 targets (Krutzfeldt et al. 2005; Esau et al. 2006; Elmen et al. 2008a,b). Further analyses of individual up-regulated transcripts around the clock enabled us to identify several circadian transcripts that were likely candidates for direct miR-122 targets. The rhythmic accumulation of these mRNAs showed changes in amplitude (Smarcd1/Baf60a, Ddc, and Hist1h1c), magnitude (Rell1), and phase (Smarcd1/Baf60a and Hist1h1c) upon miR-122 depletion. In Rev-erbα knockout animals, miR-122 synthesis was nearly constant over the day and steady-state miR-122 levels were 1.7-fold elevated. REV-ERBα regulates many clock-controlled genes directly by repressing their transcription in a cyclic manner (see also Supplemental Fig. 6; G Le Martelot, T Claudel, O Schaad, B Kornmann, G Lo Sasso, A Moschetta, and U Schibler, in prep.). Irrefutable evidence that changes in miR-122 levels and/or production account for the circadian misregulation of target transcripts in Rev-erbα knockout mice is therefore difficult to obtain. However, as indicated by our analysis of pre-mRNA and mRNA expression, miR-122 misregulation is likely to be responsible for the altered circadian amplitude of Rell1 mRNA accumulation in Rev-erbα knockout mice. Interestingly, Rev-erbα knockout mice show a cholesterol- and lipid-related phenotype opposite to 122ASO mice (G Le Martelot, T Claudel, O Schaad, B Kornmann, G Lo Sasso, A Moschetta, and U Schibler, in prep.). Again, REV-ERBα probably regulates these pathways mainly by more direct, transcriptional mechanisms, but miR-122 up-regulation is likely to contribute to these phenotypes as well.
The regulation of lipid metabolism by miR-122 may involve PPAR receptors
The direct miR-122 targets involved in hepatic lipid metabolism have not yet been identified. The decrease in hepatic fatty acid and cholesterol synthesis and the increase in hepatic fatty acid oxidation are paralleled by an increased activation of AMP-activated protein kinase (AMPK) in 122ASO mice (Esau et al. 2006). Thus, miR-122 may act through the modulation of this central sensor of metabolism. Our experiments also uncovered several connections of miR-122 to the nuclear receptors of the PPAR family, which are well-known regulators of metabolism. Specifically, we found that upon miR-122 inactivation, PPARβ/δ protein was up-regulated by around twofold to threefold. The Pparβ/δ 3′UTR contains seed sites for miR-122, and among the >30 3′UTRs we tested, it conferred one of the strongest levels of miR-122-mediated repression. In liver, PPARα and PPARβ/δ, the two PPARs executing catabolic functions, are both expressed in a circadian manner with a phase difference of ∼8 h (Yang et al. 2006). Since PPARα is the predominant isoform in this organ, hepatic functions of PPARβ/δ have not yet been studied in detail. Although PPAR functions can vary in different tissues, it is interesting to note that recently an interaction between the PPARβ/δ and AMPK pathways was shown in muscle. Thus, a constantly active VP16-PPARβ/δ transgene led to constitutive AMPK stimulation (Narkar et al. 2008). Therefore, it is tempting to speculate that at least in part the AMPK activation (Esau et al. 2006) could be the result of higher PPARβ/δ protein levels in the livers of miR-122-depleted mice.
The newly identified miR-122 target Smarcd1/Baf60a provides a second link to PPARs. Smarcd1/Baf60a is a core subunit of the SWI/SNF chromatin remodeling complexes and was very recently identified in a screen for transcription factors whose activity is augmented by PPARγ coactivator-1α (PGC-1α) (Li et al. 2008). In this study, SMARCD1/BAF60a overexpression in hepatocytes was shown to have surprisingly specific effects on the transcriptional activation of genes involved in fatty acid oxidation, and many of these were also activated by a synthetic PPARα agonist. In addition, SMARCD1/BAF60a was found to physically interact with PPARα and to be required for its function. Both proteins are corecruited with PGC-1α to PPAR response element (PPRE)-containing promoters.
A third connection of miR-122 to PPARs was provided by the observation that the livers of miR-122-depleted animals contained higher levels of FFAs, known to serve as PPAR ligands. All in all, our results suggest that PPARs might act as mediators to link miR-122 function to the control of circadian gene expression and hepatic lipid metabolism, although the detailed genetic and biochemical dissection of this network will require many additional experiments.
Speculations about the circadian action of miR-122
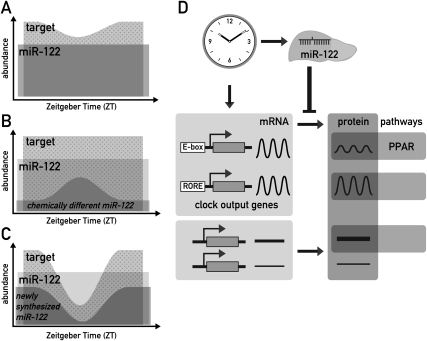
Generally, RISC-bound miRNAs are thought to be long-lived (Lee et al. 2003; Lund et al. 2004), and we estimated the miR-122 half-life to exceed 24 h. In the light of the large, stable steady-state pool of miR-122 the question thus arises of how circadian miR-122 production could nevertheless have an impact on its targets. Below we present three possible mechanisms through which miR-122 could modulate circadian gene expression on the post-transcriptional level. The first involves the attenuation of basal mRNA accumulation and/or translation by invariant miR-122 activity, the second implies chemically different subpopulations of miR-122 with distinct purposes, and the third a different availability of newly assembled and old RISC complexes for being loaded on target mRNAs.
Strictly speaking, there is no requirement for a miRNA to be circadian itself in order to contribute to the circadian accumulation of a target transcript. For example, the constant repression of basal levels of translation from such mRNA could strongly increase the circadian amplitude of the produced protein, as depicted in Figure 7A. Such a mechanism could have the additional benefit of conferring robustness to low protein expression in the trough: Rather than relying on very low transcription rates, which inevitably contain a stochastic component, low expression levels might thus be achieved more precisely by simultaneously producing a mRNA and its inhibiting miRNA, which will partially annul each other. A related role for miRNAs in denoising and conferring robustness to gene expression has previously been suggested in developmental timing (Stark et al. 2005; Cohen et al. 2006; Li et al. 2006, 2009). With regard to liver-specific miR-122, this mechanism could also represent a way of modulating the circadian rhythm of outputs in a tissue-specific manner.
Figure 7.
Models for how miR-122 could impart on circadian gene expression of its targets. (A) Even constant miR-122 levels (dark gray) could shape the circadian rhythm of a target (light gray) by constantly repressing basal levels of translation (represented by the overlap of the two areas). Only the amount of target mRNA represented by the dotted area would be available for translation into protein. As shown in the cartoon, this mechanism could increase the amplitude of cycling and convert a low-amplitude mRNA rhythm into a higher amplitude protein rhythm. In addition, this regulation could confer robustness to low protein expression levels in the trough, as described in the Discussion. The mechanism depicted in this cartoon would require a high affinity of the miRNA–target interaction and an excess of targets over the miRNA. Considering that miR-122 probably has hundreds of targets, of which many contain several seed sites, this assumption is quite plausible even for this highly abundant miRNA (B) Conceivably, chemically distinct, short-lived miR-122 subpopulations (dark gray) could exist within the pool of bulk miR-122. If these distinct miR-122 species also had specific functional properties, this speculative model would imply that target mRNAs would be subject to circadian repression. Consequently, the transcript available to produce protein (dotted area) would show circadian oscillations. (C) Conceivably, newly assembled RISC complexes could immediately get committed to their target mRNAs and remain stably associated with them, as described in the Discussion. The availability of such newly assembled miR-122 RISC would be expected to closely follow circadian miR-122 production (dark gray). If targets are transcribed circadianly as well, the phase relationship of the two rhythmic processes will determine to what extent a target will encounter miR-122 RISCs in the cell, and what influence this has on the circadian amplitude, magnitude, and phase of the produced protein (dotted area). As in A, this model would demand that the miRISC–mRNA affinity be high and that the targets are in excess. (D) Model for the integration of miR-122 in circadian hepatic gene expression. MiR-122 is depicted as a tissue-specific modulator of circadian output genes, with PPAR-dependent regulation of gene expression as one of the regulated output pathways.
As mentioned above the high metabolic stability of miR-122 prevents its cyclic accumulation. However, our experiments do not exclude that functionally distinct, less stable subpopulations exist within the large pool of miR-122 molecules. This speculative scenario is schematically depicted in Figure 7B (see the figure legend for explanation). Although we currently have no direct evidence for such distinct miR-122-containing RISC subpopulations, they could be produced by miRNA editing, RISC protein composition or subcellular localization. It is interesting to note in this context that miR-122 was recently shown to undergo cytoplasmic 3′ adenylation, affecting miR-122 stability (Katoh et al. 2009). Hence, different miR-122 subpopulations with varying metabolic stabilities may indeed coexist. Nocturnin, a rhythmically expressed deadenylase, is also involved in the regulation of lipid metabolism (Green et al. 2007). Although bona fide target mRNAs have not yet been identified for this enzyme, the regulation of poly(A) length is known to contribute to translational repression also in the case of miRNA-mediated mechanisms (Liu 2008; Eulalio et al. 2009). Circadian deadenylation may thus also contribute to the post-transcriptional control of protein synthesis. It should be emphasized in this context that almost half of the cycling liver proteins identified by mass spectrometry are translated from stably expressed mRNAs (Reddy et al. 2006).
Recent work has suggested that the ternary RISC–miRNA–target complex is remarkably stable, allowing for the immunopurification of RISC-bound targets (e.g., Beitzinger et al. 2007; Karginov et al. 2007). One might therefore speculate that mainly uncommitted, “fresh” miRNA-loaded RISCs are available for the silencing of newly synthesized targets, whereas “old” RISCs, which are already engaged in silencing, are less so (Fig. 7C). Since “fresh” miR-122 RISC is produced in a circadian fashion, the extent of target capture and silencing may well be daytime-specific. If targets are transcribed circadianly as well, it becomes evident that the phase relationship of the two rhythmic processes will determine to what extent a target will encounter miR-122 RISCs in the cell. For several of the circadian miR-122 target profiles we determined in 122ASO livers (e.g., Smarcd1/Baf60a and Ddc; see Fig. 5A), the factor of up-regulation upon miR-122 depletion was indeed especially high around ZT4, just after the peak of miR-122 production. These transcripts were transcribed in phase with miR-122, and miR-122 could function to buffer against and counteract too extreme target oscillations. Future experiments will need to address whether and to what extent the three miR-122-related mechanisms contribute to the post-transcriptional regulation of circadian output rhythms.
Materials and methods
Animal care and treatment
Animal studies were conducted in accordance with the regulations of the veterinary office of the State of Geneva. Mice were maintained under standard animal housing conditions (12-h light/12-h dark cycles; free access to food/water). Rev-erbα knockout/transgenic mice have been described (Preitner et al. 2002; Kornmann et al. 2007a). ASO treatment was performed in 11-wk male C57BL/6 mice (Elevage Janvier) by intraperitoneal injection. ASOs were chimeric 2′-fluoro/2′-O-methoxyethyl-modified oligonucleotides with a completely modified phosphorothioate backbone. The exact chemistry is available on request. Mice received four doses of 20 mg of ASO per kilogram of body weight in 150 μL, or 150 μL of saline alone (PBS control), over the course of 2 wk. Two days to 3 d after the last injection, animals were sacrificed at the respective ZTs, and livers were snap-frozen in liquid nitrogen.
RNA analysis
RNA was prepared as in Kornmann et al. (2007a), except that the LiCl wash was omitted to prevent loss of small RNAs. mRNA Northern blots were performed as in Kornmann et al. (2007a). Single-stranded 32P-labeled DNA probes were generated by linear PCR using standard methods. Templates were obtained by PCR amplification from liver cDNA or genomic DNA using gene-specific oligonucleotides (Supplemental Table 1). For miRNA Northern blots 10–30 μg of total RNA per sample were separated by 15% denaturing PAGE/1× TBE, electroblotted (36 min; 3.3 mA/cm2; 0.5× TBE; 4°C) to Genescreen Plus (NEN) membrane, and immobilized by UV and baking. Hybridizations with radioactively labeled oligonucleotide probes were performed overnight in 5× SSC, 20 mM Na phosphate at pH 7.2, 7% SDS, 2× Denhardt's solution at 50°C, followed by four 15-min washes (3× SSC, 25 mM Na phosphate at pH 7.5, 5% SDS, 10× Denhardt's) and a 5-min wash with 1× SSC and 1% SDS. The sequences of oligonucleotide probes are listed in Supplemental Table 1. Quantification of Northern blots was performed by phosphorimaging using Quantity One Software (Bio-Rad).
Global transcriptome profiling using Affymetrix oligonucleotide microarrays
Whole-cell liver RNAs from ASO-injected mice (ZT0 and ZT12) were analyzed individually on a total of 18 microarrays. Five micrograms of RNA were employed for the synthesis of biotinylated cRNA, of which 8.75 μg were hybridized to Affymetrix Mouse Genome 430 2.0 arrays according to the supplier's instructions. To identify differentially expressed transcripts, pairwise comparisons were carried out using Affymetrix GCOS 1.2 software. Transcripts were considered as expressed if they were detectable in at least two of three replicates in at least one of the experimental conditions. To compare two experimental conditions, each of the triplicates of one condition was compared with the triplicates of the other condition, resulting in nine pairwise comparisons. This approach is based on the Mann-Whitney pairwise comparison test, and allows the ranking of results by concordance and the calculation of significance (P-value) for each identified change in gene expression (Hubbell et al. 2002; Liu et al. 2002). Genes whose concordance in the pairwise comparisons exceeded the imposed threshold of 77% (seven of nine comparisons) were considered to be statistically significant. Transcripts were considered as up- or down-regulated in 122ASO samples when their accumulation had an average change of at least 1.5-fold with regard to both control samples, 124ASO, and PBS. The extraction of circadian genes from Affymetrix data sets (Fig. 4A) has been described previously (Kornmann et al. 2007b). The ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress) accession number for the microarray data is E-TABM-692.
qPCR analysis
cDNA was synthesized from 2 μg of DNase-digested whole-cell RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen) following the supplier's instructions. cDNAs were PCR-amplified (7900HT Sequence Detection Systems, Applied Biosystems) using TaqMan Universal Master Mix, No AmpErase UNG (Applied Biosystems), and raw threshold cycle (Ct) values were calculated with SDS 2.0 software (Applied Biosystems). Mean levels were calculated from triplicate PCR assays for each sample and normalized to those obtained for the control transcripts Eef1a1, Gapdh, GusB, and 45S pre-rRNA. RT− samples were included to exclude contaminations with genomic DNA. For primers and probes, see Supplemental Table 1.
miR-122 target predictions and enrichment statistics
We relied on the Ensembl version 50 mapping of the Affymetrix probes to transcripts. Up-regulated, down-regulated, and unchanged transcripts were selected as described above. The seed sequence of the miRNA was defined as 6–8 bases from the second position of the miRNA 5′-end, not allowing mismatches except a single G:U in 7-mers and 2 G:U in 8-mers. Duplex energies were computed with the cofolding function from the RNA Vienna Package (Hofacker 2003). The statistical significance of the putative miR-122 target site enrichment in the up, equal, and down fractions was evaluated using a χ2 test.
Plasmids, clonings, and analysis of 3′UTRs
3′UTR sequences were amplified by PCR from mouse liver cDNA or genomic DNA with specific oligonucleotides (Supplemental Table 1) and cloned 3′ to the renilla luciferase (RL) sequence in vector pRL-control. The identity of the UTRs was verified by sequencing. Plasmids pRL-control and pRL-Cat-1 are as in Bhattacharyya et al. (2006) and pRL-3xbulge is similar to the homonymous plasmid in Pillai et al. (2005), except that bulges match miR-122 instead of let-7. For normalization, a CMV-driven firefly luciferase-expressing plasmid on the basis of pEGFP-C1 was used. Details on all plasmids are available on request. For 3′UTR assays, 2 ng of pRL, 40 ng of FL plasmid, and 10 pmol of miRNA mimic (miR-122 and control mimic cel-miR-67 from Dharmacon) were transfected into 104 HeLa cells per well of a 96-well plate by reverse transfection using Lipofectamine 2000 (Invitrogen) according to the supplier's instructions. Transfection mixes were replaced by normal growth medium after 6 h. Luciferase activities were measured 28 h after transfection with the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase signals were normalized to firefly luciferase and for each 3′UTR construct set to 100% for the cotransfection with the control mimic. Each transfection was repeated at least six times. Growth medium was DMEM, 10% FCS, 1% PSG (Gibco).
Immunoprecipitation-Western blotting
Liver pieces of 122ASO and control mice, ZT0 and ZT12 (triplicates) were homogenized in three volumes of RIPA (150 mM NaCl, 1% NP40, 0.5% Na-deoxycholate, 0.1% SDS, 50 mM Tris-HCl at pH 8.0, protease inhibitors) using a motorized hand tool (Xenox). Insoluble material was removed by centrifugation (15 min, 20,000g, 0°C). Supernatants were kept at −80°C. For the immunoprecipitation, extracts were further diluted to 5 vol of RIPA per volume of liver and adjusted to 0.2% SDS. After another spin (as above), equal amounts of protein extract from the triplicates of the same experimental condition were pooled (∼600 μg protein/liver). An aliquot was kept for the input sample, and immunoprecipitation was performed from the remaining pool using standard protocols with a rabbit polyclonal antibody to PPARβ/δ (ab8937, Abcam) and protein A-agarose (Roche). Immunoprecipitated complexes and inputs were analyzed by SDS-PAGE/Western blotting using antibodies to PPARβ/δ and U2AF65 (Sigma, U4758). Semiquantitative analysis of Western blots was performed using Quantity One Software (Bio-Rad).
FFA analysis
Liver homogenates in MeOH (0.1% BHT) were spiked with heptadecatrienoate, TAG (17:0/17:0/17:0) and heptadecanoic acid (FFA C17:0) and extracted by chloroform after addition of 0.9% sodium chloride. The lower organic phase was separated, evaporated under nitrogen flow, and dissolved into petroleum ether (bp 40°C–60°C). The samples were transesterified with sodium methoxide (NaOMe, 0.5 M in MeOH), acidified (15% NaHSO4 in H2O) and extracted with petroleum ether. The organic phase containing fatty acid methyl esters (FAME) from bound fatty acids and FFAs was separated, evaporated under nitrogen flow, and redissolved into hexane. Two-microliter aliquots were used for GC injection (splitless 1 min) at 280°C and the analyses were performed on an FFAP fused silica capillary column (25 m, i.d. 0.32 mm) by using helium as the carrier gas (pressure program). The oven temperature was increased from 70°C to 240°C at 7°C per minute, and the fatty acids were detected by flame ionization detector (FID, 300°C). Identification was based on retention times and GC/MS spectra of reference substances.
Acknowledgments
We thank Suvendra Bhattacharyya and Witek Filipowicz for plasmids, staff at the NCCR Genomics Platform for help with microarray/qPCR experiments, Tuulikki Seppänen-Laakso for help with FFA measurements, Nicolas Leuenberger and Walter Wahli for discussions and communication of unpublished data, members of the Schibler laboratory for comments on this manuscript, and Nicolas Roggli for help with artwork. This research was supported by the Swiss National Science Foundation (through an individual research grant to U.S., and the National Center of Competence in Research Program Frontiers in Genetics, and grant SNSF PDFM33-118375 to E.Z.), the State of Geneva, the Louis Jeantet Foundation of Medicine, the Bonizzi-Theler-Stiftung, and the 6th European Framework Project EUCLOCK. D. Gatfield received and gratefully acknowledges long-term fellowships from The Federation of European Biochemical Societies (FEBS) and The International Human Frontier Science Program Organization (HFSP).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1781009.
Supplemental material is available at http://www.genesdev.org.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding—A new role for microRNAs. Genes & Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- Cusick JK, Xu LG, Bin LH, Han KJ, Shu HB. Identification of RELT homologues that associate with RELT and are phosphorylated by OSR1. Biochem Biophys Res Commun. 2006;340:535–543. doi: 10.1016/j.bbrc.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008a;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008b;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyffe SA, Alphey MS, Buetow L, Smith TK, Ferguson MA, Sorensen MD, Bjorkling F, Hunter WN. Recombinant human PPAR-β/δ ligand-binding domain is locked in an activated conformation by endogenous fatty acids. J Mol Biol. 2006;356:1005–1013. doi: 10.1016/j.jmb.2005.12.047. [DOI] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM. miROrtho: Computational survey of microRNA genes. Nucleic Acids Res. 2009;37:D111–D117. doi: 10.1093/nar/gkn707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007a;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Reinke H, Saini C, Schibler U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb Symp Quant Biol. 2007b;72:319–330. doi: 10.1101/sqb.2007.72.041. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARδ agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf U, Aberle J. Emerging roles of PPARδ in metabolism. Biochim Biophys Acta. 2007;1771:1125–1131. doi: 10.1016/j.bbalip.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]