Abstract
Activated oncogenic signaling is central to the development of nearly all forms of cancer, including the most common class of primary brain tumor, glioma. Research over the last two decades has revealed the particular importance of the Akt pathway, and its molecular antagonist PTEN (phosphatase and tensin homolog), in the process of gliomagenesis. Recent studies have also demonstrated that microRNAs (miRNAs) may be responsible for the modulation of cancer-implicated genes in tumors. Here we report the identification miR-26a as a direct regulator of PTEN expression. We also show that miR-26a is frequently amplified at the DNA level in human glioma, most often in association with monoallelic PTEN loss. Finally, we demonstrate that miR-26a-mediated PTEN repression in a murine glioma model both enhances de novo tumor formation and precludes loss of heterozygosity and the PTEN locus. Our results document a new epigenetic mechanism for PTEN regulation in glioma and further highlight dysregulation of Akt signaling as crucial to the development of these tumors.
Keywords: microRNA, miR-26a, PTEN, glioma, Akt
Glioma represents the most prevalent diagnostic category of primary brain tumor in the adult population. Many of its variants, including its most malignant form, glioblastoma multiforme (GBM), demonstrate striking refractiveness to even aggressive treatment regimens. Recent advances have implicated a defined set of oncogenic pathways in the underlying biology of this tumor group (Baehring 2005; Funari et al. 2007). Among these crucial signaling networks, the Akt pathway and its negative regulator PTEN (phosphatase and tensin homolog) have emerged as being particularly important in glioma pathogenesis. Mutation or deletion of PTEN, often by way of complete loss of its locus on chromosome 10q, is found in a large percentage of GBM (James 2005; The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008) and is correlated with poor prognosis in multiple glioma subtypes (Sasaki et al. 2001; Smith et al. 2001). Furthermore, PTEN loss dramatically enhances gliomagenesis in a number of murine model systems (Hu et al. 2005; Wei et al. 2006; Kwon et al. 2008). Given the central role of PTEN in glioma biology, the identification of alternative mechanisms for PTEN regulation in this tumor entity would be of considerable interest.
MicroRNAs (miRNAs) are a recently discovered class of small (∼22-nucleotide) noncoding RNAs that, in mammals, regulate genes at the translational level by binding loosely complimentary sequences in the 3′-untranslated regions (UTRs) of target mRNAs (Zamore and Haley 2005). The relatively low stringency requirements for functional miRNA/mRNA interactions give each miRNA the capacity to regulate numerous mRNAs. Additionally, the 3′-UTRs of regulated mRNAs frequently contain numerous miRNA recognition sites, further increasing the potential complexities of this regulatory mechanism. Several miRNAs and polycistronic miRNA clusters have been implicated in human cancer (Dalmay 2008; Fabbri et al. 2008), and a smaller group has been specifically linked to glioma biology. miR-221, for instance, has been found to be up-regulated in GBM and directly targets the tumor suppressor p27 (Ciafre et al. 2005; le Sage et al. 2007). Conversely, miR-7, whose confirmed targets include the epidermal growth factor receptor (EGFR), frequently demonstrates decreased levels in GBM, and its forced expression reduces proliferation, survival, and invasiveness in cultured glioma cells (Kefas et al. 2008). miR-124 and miR-137, both down-regulated in high-grade glial neoplasms, also inhibit glioma cell proliferation in vitro and induce neuronal differentiation (Silber et al. 2008). miR-21 is almost invariably overexpressed in GBM along with a number of other tumor types (Calin and Croce 2006), and targets a wide variety of genes involved in cell motility, migration, and apoptosis (Chan et al. 2005; Corsten et al. 2007; Zhu et al. 2007, 2008; Gabriely et al. 2008). Antisense knockdown of miR-21 in glioma cell lines leads to reduced viability in murine xenografts (Corsten et al. 2007). Interestingly, miR-21 appears to directly regulate PTEN expression in hepatocellular carcinoma (HCC) (Meng et al. 2007), although the importance of this interaction in gliomagenesis has not been confirmed. On a similar note, miR-19a and miR-214 have also been shown to target PTEN (Pezzolesi et al. 2008; Yang et al. 2008), but neither appears to be up-regulated in glioma. No miRNA has been definitively shown to either enhance or suppress the process of glioma formation in vivo.
We were initially interested in miRNAs with potential effects on glioma development, specifically those that might regulate relevant tumor suppressor genes like PTEN. In this report, we show that the miRNA miR-26a is overexpressed in a subset of high-grade gliomas and directly targets the PTEN transcript. We also demonstrate that overexpression of miR-26a in glioma primarily results from amplification at the miR-26a-2 locus, a genomic event strongly associated with monoallelic PTEN loss. Finally, we show that mir-26a reduces PTEN levels and facilitates glioma formation in a well-characterized murine model system, and that overexpression of the miRNA can functionally substitute for loss of heterozygosity at the PTEN locus.
Results
miR-26a is overexpressed in high-grade glioma and directly targets PTEN
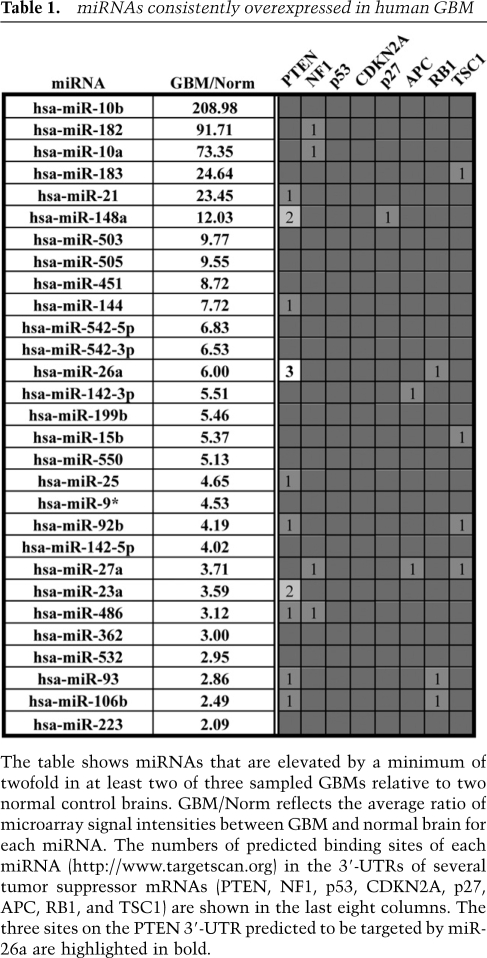
To identify miRNAs potentially involved in glioma pathogenesis, we extracted total RNA from three human GBM samples and performed microarray analysis. Two RNA extracts derived from unaffected human cerebral cortex served as negative controls. After further limiting our study to miRNAs whose expression was elevated by a minimum of twofold in at least two of three GBM samples relative to both normal controls, we were left with 29 miRNAs exhibiting some degree of overlap with those identified in previous reports (Table 1; Chan et al. 2005; Ciafre et al. 2005). Using published results and the Targetscan bioinformatics algorithm (Lewis et al. 2003, 2005), we were then able to assess which glioma-implicated tumor suppressor genes (PTEN, NF1, RB1, p27, APC, TSC1, CDKN2A, and P53) harbored predicted binding sites for the individual miRNAs on our list. Notably, the PTEN 3′-UTR was found to contain potential binding sites for no less than 10 of the miRNAs elevated in GBM, more than twice as many as any other analyzed tumor suppressor (Table 1, PTEN). We then chose to narrow our focus to miRNA/mRNA pairings characterized by multiple 3′-UTR-binding sites. This criterion, which has emerged as an independent factor in the accurate prediction of functional miRNA/mRNA interactions (Bartel 2009), directed our attention to three PTEN-targeting miRNAs (Table 1). Interestingly, one member of this group, miR-26a, was found to have three potential binding sites in the PTEN 3′-UTR (Fig. 1A, B1–B3), making it a particularly attractive target for further investigation.
Table 1.
miRNAs consistently overexpressed in human GBM
The table shows miRNAs that are elevated by a minimum of twofold in at least two of three sampled GBMs relative to two normal control brains. GBM/Norm reflects the average ratio of microarray signal intensities between GBM and normal brain for each miRNA. The numbers of predicted binding sites of each miRNA (http://www.targetscan.org) in the 3′-UTRs of several tumor suppressor mRNAs (PTEN, NF1, p53, CDKN2A, p27, APC, RB1, and TSC1) are shown in the last eight columns. The three sites on the PTEN 3′-UTR predicted to be targeted by miR-26a are highlighted in bold.
Figure 1.
miR-26a directly targets PTEN. (A) Schematic of the PTEN 3′-UTR showing binding sites for miR-26a. Nucleotide changes for binding site mutants are indicated in red. (B) NIH-3T3 or human glioblastoma (LN-18) cells were retrovirally transduced with empty vector (−), miR-26a, or control miRNA (miR-Con). Western blots for PTEN, pAkt, pS6RP, and actin are shown. (C) miR-26a targets PTEN directly as measured by luciferase reporter assay. A luciferase reporter plasmid containing the PTEN 3′-UTR was transfected into HEK 293 cells along with miRNA-expressing constructs (transfected miR; n = 3 for each). Cells were lysed 24–48 h later, and luciferase activity was quantified. (D) Mutagenesis of the three potential miR-26a-binding sites in the PTEN 3′UTR either singly or in combination (M1, M2, M3, M123, M23) in the context of this assay demonstrated that sites B2 and B3 are functional. (*) P < 0.01.
miR-26a was found to significantly repress PTEN in cell culture, an effect that, notably, was not seen with any other miRNA predicted to interact directly with the tumor suppressor, whether through one or two potential binding sites (data not shown). Western blotting demonstrated that miR-26a overexpression achieved a ∼50% knockdown of PTEN protein in both NIH-3T3 cells and LN-18 human glioblastoma cells (Fig. 1B). The observed decrease in PTEN expression was accompanied by enhanced Akt pathway signaling, as was evident by concomitant increases in pAkt and pS6RP protein levels in both cell types (Fig. 1B). As a side note, we were also able to confirm in glioblastoma cells the miR-26a-mediated knockdown of EZH2 and SMAD1, both of which are known targets of the miRNA (Supplemental Fig. 1a; see the Discussion).
We then assessed whether miR-26a directly targets the PTEN mRNA transcript using a reporter construct harboring the entire murine PTEN 3′-UTR downstream from a luciferase gene. Transfection of this reporter into HEK-293 cells led to robust expression of luciferase whose activity could be readily quantified in cell lysates. Subsequent cotransfection with miR-26a resulted in a 39% reduction in luciferase levels, exceeding the knockdown achieved with the known PTEN regulator miR-19a (Pezzolesi et al. 2008). No drop in luciferase activity was seen with coexpression of miR-124a, which does not target PTEN (Fig. 1C). To further confirm that miR-26a binds the PTEN 3′-UTR, we generated luciferase reporters with mutated miR-26a-binding sites both singly and in combination (Fig. 1A). While disrupting the B1 site alone failed to restore any luciferase activity, mutating either the B2 or B3 site yielded partial rescue (Fig. 1D, mut1, mut2, and mut3). Additionally, mutating all three binding sites or only the B2 and B3 sites in combination led to an almost complete restoration of luciferase activity (Fig. 1D, mut123 and mut23). These findings indicated that the glioma-implicated miRNA miR-26a directly binds to the B2 and B3 sites in the 3′-UTR of PTEN, mediating translational repression and reduced steady-state levels of the protein.
miR-26a is amplified in a subset of high-grade gliomas, and its overexpression is strongly associated with monoallelic PTEN loss
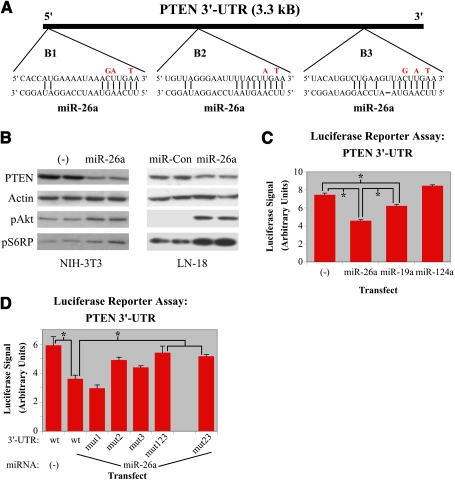
To ascertain the consistency with which miR-26a is up-regulated in high-grade gliomas, we performed quantitative RT–PCR (QRT–PCR) for the miRNA on a larger cohort of six tumor samples. This analysis revealed miR-26a to be significantly overexpressed in only a subset of high-grade gliomas (Fig. 2A). Given that GBM-associated oncogenes, such as EGFR, are frequently amplified (Wong et al. 1992; The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008), we next determined whether either of the two miR-26a loci present in the human genome (mir-26a-1 and miR-26a-2) exhibited elevated copy number in our tumor set. Q-PCR performed on extracted genomic DNA demonstrated amplification of miR-26a-2 on chromosome 12q in the same tumors where miR-26a itself was up-regulated (Fig. 2A), suggesting that, in high-grade gliomas, miR-26a overexpression is primarily mediated by copy number increases at the genomic level.
Figure 2.
miR-26a is amplified in glioma. (A) QRT–PCR and Q-PCR performed on total RNA and DNA extracted from human high-grade gliomas (G340–G349) and normal cerebral cortex (Norm). RNA (blue) and DNA (red) levels are expressed as fold increase relative to the normal control. (B) Schematic of TCGA data showing GBMs with amplification of the miR-26a-2 locus on chromosome 12q (n = 25 of 233). Each tumor corresponds to a horizontal bar across the chart. Red and green indicate copy number increases and decreases, respectively. The locations of miR-26a-2 and CDK4 are indicated by the blue line and the dashed black line, respectively. Tumors with chromosome 12 amplicons containing <30 genes (defined as focal in this study) are presented above the solid black line. (C) Schematic of TCGA data demonstrating that miR-26a overexpression (Y-axis) is almost invariably associated with miR-26a-2 amplification (X-axis). Array signal intensity for miR-26a is normalized to the mean of all samples (0 on Y-axis). (D) Schematic of TCGA data demonstrating the association of miR-26a overexpression (Y-axis) with PTEN status (X-axis). (C,D) Amplified tumors are indicated in red with focal amplifications indicated as solid red circles.
We then sought to expand our analysis of miR-26a amplification in GBM using the cancer genome atlas (TCGA), a publicly available repository that has accumulated comparative genomic hybridization (CGH), gene expression, and DNA methylation analyses for ∼200 GBM samples to date (for complete results, see Supplemental Table 1; The Cancer Genome Atlas Research Network 2008). Using this database, we were able to confirm amplification of miR-26a-2 in a subset of GBMs (11%) that also tended to exhibit elevated miR-26a (Fig. 2B,C). No amplification events were identified at the miR-26a-1 locus. Additionally, we found that miR-26a-2 is coamplified in all cases with a defined set of genes, including that of the oncoprotein CDK4. Indeed, only ∼80 kB separate the CDK4 and miR-26a-2 genes (Fig. 2B). CDK4 amplification has been repeatedly associated with a significant minority of GBMs and is thought to contribute to oncogenesis via its inhibitory affects on the RB tumor suppressor (Henson et al. 1994; Funari et al. 2007).
The presence of miR-26a-2 within a previously characterized oncogenic amplicon raised the possibility that miR-26a overexpression in GBM simply represents a functionally insignificant epiphenomenon associated with CDK4 amplification. To address this question, we correlated TCGA data for both miR-26a and PTEN and found, importantly, that miR-26a overexpression was absent from all tumors characterized by homozygous PTEN deletion. Additionally, amplification of the miRNA segregated, for the most part, with tumors harboring monoallelic deletion at the PTEN locus (Fig. 2D). When nonfocal amplicons—those containing >30 genes—were excluded from our analysis, this correlation became even stronger, with only one exception falling into the PTEN+/+ subgroup.
The association of miR-26a overexpression with monoallelic PTEN loss suggests that amplification of the miRNA serves to silence residual PTEN transcript in PTEN+/− tumors, analogous to a loss of heterozygosity event. Our data indicate that 12% of PTEN+/− GBMs (21 out of 170) are characterized by miR-26a amplification. Interestingly, we also found that the vast majority (25 out of 28) of GBMs pairing monoallelic deletion with inactivating mutations at their PTEN loci—a group that accounts for 26% of PTEN+/− gliomas with available sequencing data (28 out of 109)—do not exhibit miR-26a amplification (Supplemental Table 1). This suggests that miR-26a amplification and PTEN mutation represent alternative PTEN silencing mechanisms that operate, for the most part, in nonoverlapping sets of monoallelic tumors. Furthermore, while PTEN mutation appears to be more frequent (26% vs. 12%) than miR-26a amplification in PTEN+/− tumors, both occur at considerably higher rates than deletion of the remaining PTEN allele, given that only 4% of TCGA GBM samples (10 out of 233) exhibit homozygous PTEN deletion (Supplemental Table 1). Taken together, these bioinformatics correlations are consistent with a functional role for the miR-26a/PTEN interaction in glioma biology regardless of the miRNA's linkage with CDK4.
miR-26a enhances tumorigenesis in a murine glioma model
To further demonstrate the physiological relevance of miR-26a in glioma pathogenesis, we incorporated miRNA expression into the RCAS/tv-a system. RCAS/tv-a technology uses a specific avian leukosis virus (RCAS) to mediate somatic gene transfer into cells transgenic for its receptor (tv-a) (Holland and Varmus 1998; Holland et al. 1998). We showed previously that localized delivery of RCAS retrovirus encoding the human PDGF-B gene (RCAS-PDGF) into the brains of newborn mice expressing a nestin-driven tv-a retroviral receptor (Ntv-a) yields gliomas by 3–4 mo with high penetrance (Dai et al. 2001; Shih et al. 2004). Elevated PDGF signaling and/or PDGF receptor amplification has been found in a variety of low- and high-grade glioma subtypes (Funari et al. 2007). Of the 24 GBMs in the TCGA study group harboring PDGFR amplification, 25% (six out of 24) and 79% (19 out of 24) are characterized by miR-26a amplification and PTEN loss, respectively. Additionally, 15 of 26 miR-26a-amplified tumors fall within the ∼39% of TCGA GBMs (91 out of 233) whose transcriptional profiles cluster with PDGFR-amplified tumors (CW Brennan, H Momota, D Hambardzumyan, T Ozawa, A Tandon, A Pedraza, and EC Holland, in prep.), an enrichment (P = 0.053) that further underscores the relevance of PDGF-driven gliomagenesis in the present study.
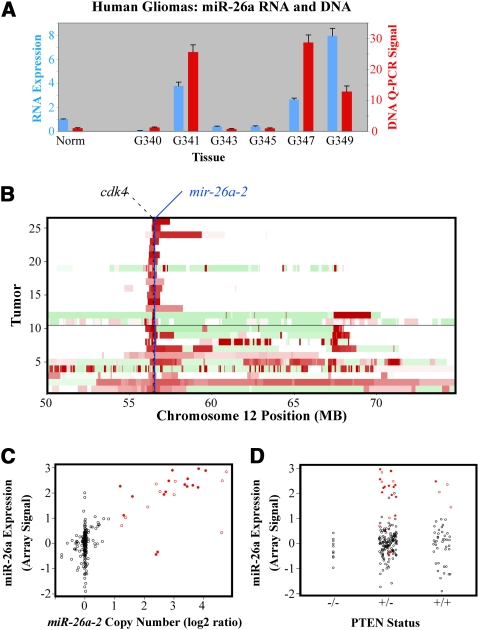
We developed a new series of RCAS vectors expressing single miRNA constructs downstream from a GFP reporter (Fig. 3A). As a negative control, one such construct was designed bearing a short hairpin sequence targeting the Lac-Z transcript (miR-Z). Additionally we used a murine Ntv-a strain containing homozygous floxed PTEN alleles (Ntv-a/PTENfl/fl) (Hu et al. 2005), such that the RCAS-mediated delivery of cre-recombinase (RCAS-cre) in these mice results in complete loss of PTEN expression in nestin-positive cells.
Figure 3.
miR-26a-expressing murine gliomas (P/26a). (A) Schematic of RCAS-miR constructs; CMV denotes CMV promoter, GFP denotes GFP cassette, and miR denotes miRNA domain. (B) Overexpression of miR-26a was verified in lysates of crudely dissected tumors by QRT–PCR. (C,D) A grade III murine glioma with forced miR-26a expression. H&E staining (C) and LNA-in situ hybridization (LISH; D) for miR-26a are shown. (D) For the latter, a grade III tumor with no forced miR-26a expression (P/Z) is also shown (bars, 100 μm).
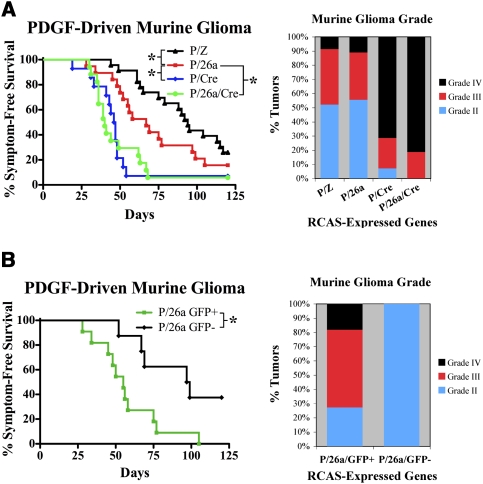
Coinjection of RCAS-PDGF and RCAS-miR-26a (P/26a) into Ntv-a/PTENfl/fl mice led to a mixture of low- and high-grade gliomas over the course of 4 mo that were morphologically indistinguishable from those generated previously using RCAS-PDGF as the primary oncogenic driver (Fig. 4A; Dai et al. 2001; Shih et al. 2004). Retrovirally mediated miR-26a overexpression was documented in these tumors by either QRT–PCR, immunohistochemistry for GFP, or miR-26a-specific LNA in situ hybridization (LISH) (Fig. 3B–D; Supplemental Fig. 1b). P/26a mice were then compared with analogous cohorts injected with either RCAS-PDGF and RCAS-miR-Z (P/Z) or RCAS-PDGF and RCAS-cre (P/Cre). While the deletion PTEN in P/Cre mice led to dramatically reduced symptom-free survival, consistent with previous reports (Hu et al. 2005; Wei et al. 2006; Kwon et al. 2008), P/26a mice exhibited an intermediate degree of disease-free latency relative to both the P/Z and P/Cre cohorts (Fig. 4A). The employment of two independent vectors in the generation of P/26a mice resulted in a mixed population of tumors, with only some actually infected by RCAS-miR-26a virus. Using detectable GFP immunostaining as a marker, we were able to extract this group and compare them with GFP-nonexpressing counterparts. Subsequent Kaplan-Meier analysis revealed that symptom-free survival was more strongly reduced in mice with confirmed miR-26a expression (Fig. 4B).
Figure 4.
miR-26a facilitates gliomagenesis in vivo. (A) Kaplan-Meier curves demonstrating symptom-free survival for P/Z, P/26a, P/Cre, and P/26a/Cre mice (n = 23, 19, 14, and 17 respectively; [*] P < 0.05). The percentages of tumors exhibiting WHO grade II, grade III, and grade IV histological features are also shown for each genotype. (B) Kaplan-Meier curves demonstrating symptom-free survival for P/26a mice with and without detectable immunostaining for GFP reporter (n = 11 and 8 respectively; [*] P < 0.05). Histological grades for tumors in the P/26a GFP+ and P/26a GFP− groups are also shown.
Histological grades were also assigned to the generated tumors across all three murine genotypes. While our initial compilation demonstrated similar grading profiles between the P/Z and P/26a cohorts, with both exhibiting fewer floridly high-grade features than P/Cre tumors, examination of the P/26a group after GFP stratification revealed a plurality of high-grade lesions in GFP+ mice and only grade II tumors in GFP− mice (Fig. 4A,B). Taken together, these findings demonstrate that miR-26a contributes to gliomagenesis in vivo. Furthermore, the less malignant phenotype exhibited by P/26a mice relative to their P/Cre counterparts is consistent with the notion that PTEN activity, while not entirely lacking in these mice, is sufficiently reduced so as to facilitate oncogenesis.
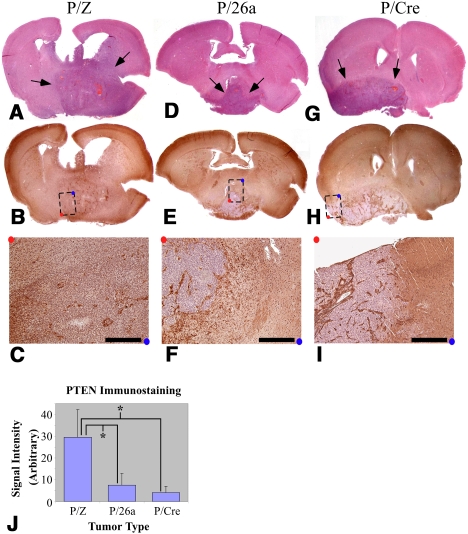
In order to assess whether down-regulation of PTEN represented the primary tumorigenic mechanism of miR-26a in our murine model system, we generated gliomas using RCAS-PDGF, RCAS-miR-26a, and RCAS-cre (P/26a/Cre). We reasoned that miR-26a-mediated repression of additional non-PTEN mRNA targets could augment gliomagenesis even in the absence of PTEN provided those interactions were consequential to the oncogenic process. However, we observed a phenotypic enhancement that was minimal at best when measured by either disease-free latency or tumor grade (Fig. 4A). These data, while not formally excluding the existence of other pathologically relevant miR-26a–target interactions in glioma biology, are consistent with the miR-26a/PTEN repression event being the predominant effect of the miRNA in this tumor type. Furthermore, we confirmed that forced miR-26a expression actually reduced PTEN levels in our murine gliomas by immunostaining multiple grade IV tumors derived from P/Z, P/26a, and P/Cre mice for PTEN protein. We found minimal PTEN labeling in P/26a gliomas, whose staining pattern was strikingly similar to that seen in P/Cre tumors (Fig. 5D–J). This effect notably exceeded the more modest (∼50%) reduction in PTEN expression achieved in vitro (Fig. 1B). Furthermore, PTEN expression in P/Z tumors was relatively retained (Fig. 5A–C,J). In all cases, nonneoplastic tumor vasculature stained strongly for PTEN serving as an excellent internal control for signal quantification. These findings demonstrate that, in addition to enhancing tumorigenesis, miR-26a effectively represses endogenous PTEN protein in a relevant glioma model system.
Figure 5.
miR-26a represses PTEN expression in murine gliomas. Grade IV tumors from P/Z (A–C), P/26a (D–F), and P/Cre (G–I) mice. H&E staining is shown in A, D, and G; immunostaining for PTEN is shown in B, C E, F, H, and I. C, F, and I are enlargements at 5× magnification of the regions designated in by dashed boxes in B, E, and H, respectively (bars, 400 μm). Arrows indicate tumors on H&E staining. Red and blue dots are orientation guides. (J) Quantification of PTEN immunostaining for two tumors in each genotype (see the Materials and Methods); units are arbitrary. (*) P < 0.05.
As an aside, it bears mentioning that expression of several other glioma and cancer-implicated miRNAs in this experimental paradigm, including miR-21, miR-10b, and miR-19a, yielded no significant effects on gliomagenesis (Supplemental Fig. 1c). Additionally, the invariable coamplification of miR-26a with CDK4 in human GBM prompted us to assess the functional impact of CDK4 in our murine model system. Surprisingly, however, RCAS-mediated CDK4 expression in PDGF-driven gliomas failed to increase either tumor grade or the rate of tumor formation (Supplemental Fig. 2).
miR-26a overexpression functionally substitutes for loss of heterozygosity at the PTEN locus
As stated above, our analysis of TCGA data suggests a role for miR-26a in the epigenetic silencing of residual PTEN protein following monoallelic loss at the genomic level. To address this possibility experimentally, we bred Ntv-a/PTENfl/fl mice with Ntv-a/PTEN+/+ counterparts to generate Ntv-a/PTEN+/fl heterozygotes and coinjected them either with RCAS-PDGF, RCAS-cre, and RCAS-miR-Z (P/Z/Cre), or with RCAS-PDGF, RCAS-cre, and RCAS-miR-26a (P/26a/Cre). We reasoned that if miR-26a served to functionally eliminate residual PTEN activity in heterozygous mice, its overexpression in this context might result in enhanced gliomagenesis. Furthermore, given that PDGF-driven PTEN+/− gliomas are prone to genomic loss of their remaining PTEN allele (E Fomchenko and EC Holland, in prep.), we anticipated that miR-26a-expressing tumors, as a result of robust miRNA-induced PTEN silencing, would exhibit a decreased tendency to develop loss of heterozygosity at the PTEN locus, and that such an effect would ultimately be reflected in the relative retention of PTEN at the genomic level in P/26a/Cre tumors.
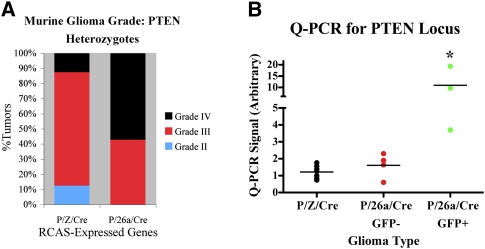
We generated equal numbers of P/Z/Cre and P/26a/Cre gliomas in Ntv-a/PTEN+/fl mice and found a somewhat higher percentage of grade IV tumors in the P/26a/Cre group (Fig. 6A), although no effect on symptom-free survival was observed (data not shown). More importantly, however, Q-PCR analysis of genomic DNA from each glioma revealed that P/26a/Cre PTEN+/− tumors exhibited a relative maintenance of their remaining PTEN allele compared with P/Z/Cre PTEN+/− counterparts, and that this effect closely mirrored miR-26a expression as assessed by GFP staining (Fig. 6B, GFP+ and GFP−). These findings indicate that miR-26a overexpression can functionally substitute for and, consequently, preclude PTEN loss of heterozygosity in the context of gliomagenesis in vivo, and thus correlate well with our bioinformatics associations between miR-26a amplification and monoallelic PTEN loss in GBM.
Figure 6.
miR-26a overexpression precludes PTEN loss of heterozygosity in PDGF-driven PTEN+/− murine gliomas. P/Z/Cre and P/Z/26a gliomas were generated in Ntv-a/PTEN+/fl mice (n = 7 and 7, respectively). (A) P/Z/26a Ntv-a/PTEN+/fl mice exhibited more grade IV gliomas. (B) Additionally, genomic DNA was extracted from paraffin sections of all PTEN+/− tumors and analyzed for PTEN by Q-PCR. GFP+ P/26a/Cre tumors exhibited a significantly higher level of retained PTEN than GFP− P/26a/Cre and P/Z/Cre tumors. (*) P < 0.05. Means are designated by horizontal lines.
Discussion
Since the discovery of miRNAs as a new class of gene regulators, numerous studies have emerged correlating altered expression levels of particular miRNAs with the neoplastic phenotype (Iorio et al. 2005, 2007; Lu et al. 2005; Volinia et al. 2006; Dalmay 2008; Fabbri et al. 2008). Precise classification of specific miRNAs as oncogenes or tumor suppressors, however, necessitates a deeper analysis of their functional roles in vivo. In the present study, we identified miR-26a as a negative regulator of PTEN, documented its overexpression in human glioma, and demonstrated its oncogenic potential in a relevant murine model system. We initially found miR-26a to be overexpressed in high-grade gliomas using miRNA microarrays. A similar array-based analysis did not reveal miR-26a to be consistently up-regulated in either GBM samples or glioblastoma cell lines (Ciafre et al. 2005). This discrepancy most likely reflects the stochastic nature of miR-26a overexpression in high-grade glioma, which seems to depend heavily on amplification at the miR-26a-2 locus.
miR-26a has been sparsely studied to date, and its relevance to the oncogenic process is only beginning to be elucidated. We found miR-26a to repress PTEN in cell culture both by Western blot and luciferase reporter assay. As mentioned briefly above, PTEN is a known target of multiple miRNA species including, most notably, the glioma-implicated miR-21 (Meng et al. 2007; Pezzolesi et al. 2008; Yang et al. 2008). Despite repeated efforts, however, we were unable to demonstrate that overexpression of miR-21 reduces PTEN levels in vitro (data not shown). Our findings may simply reflect differences in cell type; we note that the prior study linking miR-21 with PTEN used primarily hepatocytes and HCC-derived cell lines (Meng et al. 2007). This discrepancy also raises the possibility that the reported oncogenic effects of miR-21 in glioma cell lines may result from the targeting of other proteins (Chan et al. 2005; Corsten et al. 2007; Zhu et al. 2007, 2008; Gabriely et al. 2008; Papagiannakopoulos et al. 2008). As indicated in the main text, incorporation of miR-21 into our RCAS/tv-a model system did not significantly enhance gliomagenesis. However, miR-21 levels were found to be significantly up-regulated in our PDGF-driven murine gliomas even without RCAS-mediated miRNA overexpression (data not shown), potentially implying a saturation of relevant mRNA interactions prior to the forced addition of the miR-21. Alternatively, miR-21 might only enhance gliomagenesis in the context of different oncogenic drivers (i.e., not PDGF).
The physiologic relevance of miR-26a in gliomagenesis was demonstrated using a well-established murine model system. Coexpression of miR-26a with the oncogenic driver PDGF impacted both disease-free latency and tumor grade, although these effects were not as dramatic as those seen with cre-mediated PTEN deletion. These findings correlated well with our in vitro data demonstrating partial, but not complete, knockdown of PTEN protein with miR-26a overexpression. Additionally, we were able to document PTEN repression within P/26a murine gliomas. It remains quite possible that miR-26a mediates its tumorigenic effects by regulating a number of targets not limited to PTEN; a potential interaction with the RB1 transcript is particularly intriguing, though as of yet unverified in our hands (Table 1; data not shown). Indeed, two large recent studies have further solidified the notion that individual miRNAs impact cellular processes by modestly repressing the levels of numerous proteins (Baek et al. 2008; Selbach et al. 2008). However, the failure of miR-26a to significantly enhance murine gliomagenesis in the absence of PTEN strongly underscores the particular importance of the miR-26a/PTEN interaction in this process, a conclusion that is further supported by both our bioinformatics analysis and the ability of miR-26a overexpression to preclude PTEN loss of heterozygosity in vivo (see below).
On a related note, miR-26a has been shown to directly target both the SMAD1 transcription factor and the histone methyltransferase EZH2 (Luzi et al. 2007; Wong and Tellam 2008), with the latter of these interactions actually appearing to have tumor-suppressive effects in both lymphoma and rhabdomyosarcoma (Sander et al. 2008; Ciarapica et al. 2009). Coupled with our own findings, these reports suggest that the pro-oncogenic properties of miR-26a in glioma may largely reflect the cellular context of the miRNA's overexpression and the array of potential mRNA targets, including PTEN, at its disposal. Our studies indicate that miR-26a overexpression in LN-18 cells does decrease EZH2 and SMAD1 levels, implying that the transcripts of both proteins could be effectively targeted by the miRNA during gliomagenesis. Whether such repression is functionally relevant in this tumor type, however, remains unclear. The fact that miR-26a overexpression enhances gliomagenesis in vivo argues that the miRNA's effects on EZH2 and SMAD1 are either irrelevant in this tumor type or minor enough to be overcome by the physiologic impact of substantial PTEN repression. Interestingly, one recent study has directly linked EZH2 to glioma, specifically regarding a potential role for the protein in the maintenance of the tumor initiating phenotype in cells derived from human GBM (Lee et al. 2008). These findings, however, were documented in a single glioblastoma cell line (out of four tested) that itself was not characterized by miR-26a amplification, implying a fundamental difference in the biology of this particular glioma subset and further underscoring, perhaps, the diversity of pro-oncogenic mechanisms at work in the disease entity as a whole.
We investigated the scope of miR-26a overexpression and amplification in high-grade glioma using the extensive resources of the TCGA. Analysis of their data set revealed that miR-26a amplification is a relatively frequent occurrence in human GBM and is correlated with monoallelic PTEN deletion. In contrast, no association was seen between miR-26a up-regulation and either PTEN+/+ or PTEN−/− tumors. These correlations not only support a functional, tumor-promoting role for the miR-26a/PTEN interaction but also suggest a temporal sequence to the molecular evolution of miR-26a-amplified gliomas, with PTEN loss most likely preceding miR-26a-2 copy number gain. Subsequent repression of the remaining PTEN allele by miR-26a would then, presumably, eliminate the driving force for formal loss of heterozygosity. The additional finding that miR-26a amplification tends not to occur in the setting of coupled PTEN deletion and mutation provides further support for such an oncogenic mechanism. More significantly, our in vivo glioma modeling has effectively recapitulated this sequence of events, with miR-26a expression in genetically engineered PTEN+/− mice precluding loss of their remaining PTEN allele.
Our bioinformatics studies also revealed the invariable coamplification of the mir-26-2 locus with the known oncogene CDK4. Such clustering of cancer-related genes is hardly unprecedented, with the CDKN2A locus, containing the INK4A, INK4B, and ARF tumor suppressors, being a prime example (Gil and Peters 2006). The reciprocal relationship between miR-26a and PTEN discussed above, coupled with the very tangible impact of expressing the miRNA both in vitro and in vivo, demonstrates that increased copy number at the miR-26a-2 locus, in all likelihood, carries with it a biological significance independent of its proximity to CDK4, a conclusion that is all the more compelling in light of the failure of CDK4 overexpression to significantly alter PDGF-driven gliomagenesis in mice. Nevertheless, the close physical association of miR-26a-2 and CDK4 does imply a cooperative relationship between the two genes, and additional in vivo studies should provide further insight into the functional significance of this association. The distinct lack of amplification at either the miR-26a-1 locus or highly homologous miR-26b locus in glioma might be seen as additional evidence of a synergistic link between miR-26a and CDK4. However, this discrepancy could simply reflect basic differences in genomic fragility at the respective chromosomal positions, the determinants of which are still very much unclear (Glover et al. 2005).
Akt pathway activation has been shown to occur in a plurality of GBMs, most frequently resulting from modulation of its primary negative regulator PTEN (The Cancer Genome Atlas Research Network 2008). In this report, we document a novel epigenetic mechanism by which PTEN activity is regulated in glioma. While deletion and mutation represent the two most common avenues to PTEN loss, alternative means do exist. Promoter methylation has been documented as a cause for reduced PTEN expression in a variety of glioma subtypes (Baeza et al. 2003; Wiencke et al. 2007). Much like miR-26a expression, PTEN promoter methylation is, for the most part, mutually exclusive with PTEN mutation/deletion and, interestingly, tends to occur in lower-grade gliomas that subsequently evolve in to GBMs (so-called secondary GBMs) (Wiencke et al. 2007). This latter finding is reminiscent of our mouse modeling studies showing that miR-26a expression yields lower grade tumors than those associated with complete PTEN loss, perhaps reflecting a more modest degree of PTEN knockdown.
Epigenetic mechanisms for PTEN repression, such as promoter methylation and miR-26a amplification, enrich the spectrum of molecular abnormalities with the potential to drive glioma progression. Our data suggest that approximately one-eighth of GBMs already lacking one PTEN allele achieve silencing of their remaining copy through overexpression of miR-26a. While PTEN mutation or deletion appears to serve the same functional purpose in an additional 26% and ∼4% of PTEN+/− tumors, respectively, it remains to be seen what portions of this glioma subgroup employ PTEN promoter methylation or other still undiscovered epigenetic means to mediate similar effects. High-quality promoter methylation data for GBMs has yet to be released by TCGA on a large scale. Regardless, it appears almost certain that human gliomas use a variety of strategies to functionally eliminate PTEN and, in this way, activate oncogenic Akt signaling.
In conclusion, we identified the first miRNA species that facilitates the process of glioma formation in vivo, along with a mechanism for its overexpression in human tumors. Furthermore, the repression of PTEN by miR-26a provides an intriguing link to an oncogenic pathway whose role in glioma biology is already well established, and, in this way, strengthens this case for therapeutic inhibition of Akt signaling. Additional characterization of this and other glioma-implicated miRNAs, along with their mRNA targets, should greatly enhance the understanding of this novel epigenetic regulatory mechanism as it relates to cancer biology in general and gliomagenesis in particular.
Materials and methods
Cell culture, transfections, and Western blots
Cells were cultured, lysed, and processed for Western blotting by standard methods. Transfections were performed using the Fugene 6 reagent. The following primary antibodies were used for Western blots: PTEN (Cascade Bioscience); Actin (Santa Cruz Biotechnologies); pAkt, pS6RP, and EZH2 (Cell Signaling Technology); and SMAD1 (Abcam). Western blots were developed using ECL Detection Reagents (GE Healthcare), and bands were quantified by densitometry with ImageJ 1.40 (NIH).
Retroviral transduction
miR-Vec constructs were generated as described previously (Voorhoeve et al. 2006). Retrovirus was produced in the Plat-E packaging cell line (Cell Biolabs) and harvested 48 h after miR-Vec transfection. NIH-3T3 cells were infected using virus-containing supernatant supplemented with 5 μg/mL polybrene and selected with blasticidin. RCAS-PDGF and RCAS-Cre constructs were generated as described previously (Shih et al. 2004; Hu et al. 2005). RCAS-miR constructs were initially assembled in the pcDNA6.2-GW/EmGFP-miR vector (Invitrogen) by inserting PCR-amplified human or murine miR sequences (250–500 base pairs [bp]) downstream from the EmGFP expression cassette. A larger sequence including the CMV promoter, EmGFP cassette, and miRNA domain was then PCR-amplified and cloned into the RCASY plasmid as between the NotI and ClaI restriction sites. RCAS virus was produced in DF-1 packaging cells (American Type Culture Collection) as described previously (Holland and Varmus 1998; Holland et al. 1998).
Luciferase reporter assay
The PTEN 3′-UTR luciferase reporter construct (PCPTEN) was generated by PCR-amplifying the murine PTEN 3′-UTR from genomic DNA. The fragment was then subcloned into pYAP6 to obtain appropriate restriction sites before insertion into the psiCHECK-2 vector downstream from the renilla luciferase cassette. Predicted miR-26a-binding sites were mutated using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). HEK-293 cells were transfected with PCPTEN and RCAS-miR constructs, lysed 24–48 h later, and processed for luciferase assay using the Dual-Luciferase Reporter Assay System (Promega). Assays were read in a Veritas Microplate Luminometer (Turner Biosystems) normalizing renilla luciferase signal to firefly luciferase signal.
Q-PCR and QRT–PCR
DNA was extracted from human brain tissue using the DNeasy Blood and Tissue Kit and from paraffin sections of murine tumors using the QIAamp DNA FFPE Tissue Kit (both from Qiagen). Total RNA was extracted from cells and/or tissue using the Trizol reagent and protocol (Invitrogen). Q-PCR and QRT–PCR was performed using miRNA-specific primer sets, reagents, and protocols from Applied Biosystems in a 7900 HT Fast Real-Time PCR System (Applied Biosystems).
Microarray analysis
Total RNA extracted from brain tumors and normal controls was enriched for miRNA and labeled by the Agilent protocol followed by hybridization to the human mRNA version 1.0 array (Agilent Technologies). Raw data were normalized to total signal intensity prior to comparative analysis.
Analysis of TCGA data
miRNA expression, gene expression, array CGH, and mutational analysis data from GBM samples were collected as previously described (The Cancer Genome Atlas Research Network 2008). Processed data sets were obtained directly form the public access data portal (http://cancergenome.nih.gov/dataportal/data/about; Supplemental Material). Data sets were cross-referenced using tumor-specific identification numbers: 233 samples had miRNA expression, gene expression, and array-CGH. Post-segmentation array-CGH data were used for local copy number estimation. Amplification of mir-26a was defined by regional log2 ratio >1.0. PTEN locus was classified by minimal log2 ratio (log2R) across the gene as follows:
“euploid” = −0.2 < log2R < 0.4; “single copy loss” = −1.0 < log2R < −0.2; “homozygous deletion” = log2R < −1.5; and “indeterminate” = intermediate log2R. PTEN mutations were derived from the TCGA data as described in the Supplemental Material. Tumor IDs, source samples, and derived data values are given in Supplemental Table 1.
Generation of murine gliomas
Ntv-a/PTENfl/fl mice, Ntv-a/PTEN+/+ mice, and procedures for RCAS-mediated gliomagenesis have been described previously (Holland and Varmus 1998; Holland et al. 1998; Hu et al. 2005). After injection of relevant RCAS viruses during the newborn period, mice were aged until they developed symptoms of disease (lethargy, poor grooming, weight loss, macrocephaly).
Statistics
Error bars in all cases indicate standard deviation. Kaplan-Meier analysis of symptom-free survival in murine gliomas was performed using the Gehan-Breslow-Wilcoxon test in Prism 5 software (GraphPad). Luciferase reporter assays and Q-PCR data for PTEN were analyzed by two-tailed t-test in either Excel 2004 (Microsoft) or Prism 5, respectively. Correlations between tumor groups identified in TCGA data were analyzed by Fisher's exact test (R package:stats, http://www.cran.org).
Histology and immunohistochemistry
Mouse brains were prepared for either paraffin or frozen sectioning and stained with hematoxylin and eosin as described previously (Hu et al. 2005; Becher et al. 2008). Tumor grade was established using World Health Organization (WHO) criteria (Louis et al. 2007). Briefly, grade II tumors exhibited increased cellularity with predominantly monomorphic cellular features and minimal mitotic activity; grade III tumors exhibited more marked cellular atypia, increased mitoses, and mild microvascular proliferation; and grade IV tumors exhibited florid microvascular proliferation and foci of pseudopalisading necrosis. Immunohistochemical staining was performed as previously described (Hambardzumyan et al. 2008). The following antibodies were used: GFP (Invitrogen) at 1:200; PTEN (Cell Signaling Technology) at 1:100. Immunohistochemical staining for PTEN was quantified using Metamorph software (Molecular Devices). Two tumors from each genotype (P/Z, P/26a, and P/Cre) were used for the analysis. Eight 20× images from each tumor were loaded into Metamorph (Molecular Devices). Threshold signal intensity was established in each case using staining from nonneoplastic tumor vasculature. The amount of signal reaching threshold was then quantified in identically sized rectangles placed over solid areas of tumor cells in each micrograph.
LISH
In situ hybridization was carried out using a DIG-labeled locked nucleic acid probe directed against miR-26a ([A][G]CC[T]A[T]CC[T]GG[A]TT[A]C[T]T[G][A]A, locked residues marked with brackets) as described previously (Supplemental Material; Pena et al. 2009).
Acknowledgments
We thank Yevgeniy Romin and Katia Manova for their assistance in the quantification of immunohistochemical staining, Agnes Viale for running the miRNA microarrays, and Robert “Jim” Finney for technical support. We also thank Massimo Squatrito, Nduka Amankulor, Elena Fomchenko, Sohail Tavazoie, and Anne-Marie Bleau for their helpful discussions, and Andrea Ventura for his critical reading of the manuscript. This work was supported by a STARR Cancer Consortium Award (to T.T. and E.C.H) and the David Tetenbaum Hope American Brain Tumor Association Research Fellowship (to J.T.H).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1777409.
Supplemental material is available at http://www.genesdev.org.
References
- Baehring JM. An update on oligodendroglial neoplasms. Curr Opin Neurol. 2005;18:639–644. doi: 10.1097/01.wco.0000189877.31637.74. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol. 2003;106:479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher OJ, Hambardzumyan D, Fomchenko EI, Momota H, Mainwaring L, Bleau AM, Katz AM, Edgar M, Kenney AM, Cordon-Cardo C, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 2008;68:2241–2249. doi: 10.1158/0008-5472.CAN-07-6350. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, Giordano A. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T. MicroRNAs and cancer. J Intern Med. 2008;263:366–375. doi: 10.1111/j.1365-2796.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- Funari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes & Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet. 2005;14:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & Dev. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JW, Schnitker BL, Correa KM, von Deimling A, Fassbender F, Xu HJ, Benedict WF, Yandell DW, Louis DN. The retinoblastoma gene is involved in the malignant progression of astrocytomas. Ann Neurol. 1994;36:714–721. doi: 10.1002/ana.410360505. [DOI] [PubMed] [Google Scholar]
- Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7:356–368. doi: 10.1593/neo.04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- James CD. Molecular genetics of tumors of the central nervous system. In: Ali-Osman F, editor. Brain tumors. Humana Press; Totowa, NJ: 2005. pp. 19–32. [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, et al. Epigenetic-mediated dysfunction of the bone morphogenic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Luzi E, Marini F, Carbonell Sala S, Tognarini I, Galli G, Luisa Brandi M. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting the SMAD1 transcription factor. J Bone Miner Res. 2007;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-M, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berniger P, Zavolan M, et al. miRNA in situ hybridization in formalidehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolesi MG, Platzer P, Waite KA, Eng C. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet. 2008;82:1141–1149. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Zlatescu MC, Betensky RA, Ino Y, Cairncross JG, Louis DN. PTEN is a target of chromosome 10q loss in anaplastic oligodendrogliomas and PTEN alterations are associated with poor prognosis. Am J Pathol. 2001;159:359–367. doi: 10.1016/S0002-9440(10)61702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64:4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O'Fallon JR, Schaefer PL, Scheithauer BW, James CD, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, et al. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–7437. doi: 10.1158/0008-5472.CAN-06-0712. [DOI] [PubMed] [Google Scholar]
- Wiencke JK, Zheng S, Jelluma N, Tihan T, Vandenberg S, Tamguney T, Baumber R, Parsons R, Lamborn KR, Berger MS, et al. Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neuro-oncol. 2007;9:271–279. doi: 10.1215/15228517-2007-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-genome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]