Abstract
Protein phosphorylation-mediated signaling networks regulate much of the cellular response to external stimuli, and dysregulation in these networks has been linked to multiple disease states. Significant advancements have been made over the past decade to enable the analysis and quantification of cellular protein phosphorylation events, but comprehensive analysis of the phosphoproteome is still lacking, as is the ability to monitor signaling at the network level while comprehending the biological implications of each phosphorylation site. In this review we highlight many of the technological advances over the past decade and describe some of the latest applications of these tools to uncover signaling networks in a variety of biological settings. We finish with a concise discussion of the future of the field, including additional advances that are required to link protein phosphorylation analysis with biological insight.
Keywords: Electrospray ionization-tandem mass spectrometry, Protein phosphorylation, Signal transduction profiling, Spectrometry
1 Introduction
In the postgenomic era rapid advancement in the characterization of new genes and their protein products has driven an increased demand to functionally classify these proteins. Classical genetic, biochemical, and protein chemical approaches have been historically used to tackle this challenge for selected biomolecules, but these methods tend to be time-consuming, laborious, and usually require large amounts of material. Application of these approaches to characterize thousands of proteins is therefore unrealistic. However, recently developed proteomic methods, quickly improving with technical advancements in equipment, permit large-scale protein analysis while maintaining molecular resolution. While these large-scale methods do not directly provide functional characterization, they can be used to generate hypotheses regarding the function of selected proteins. Follow-on biochemical studies can then be performed on these proteins to validate hypotheses.
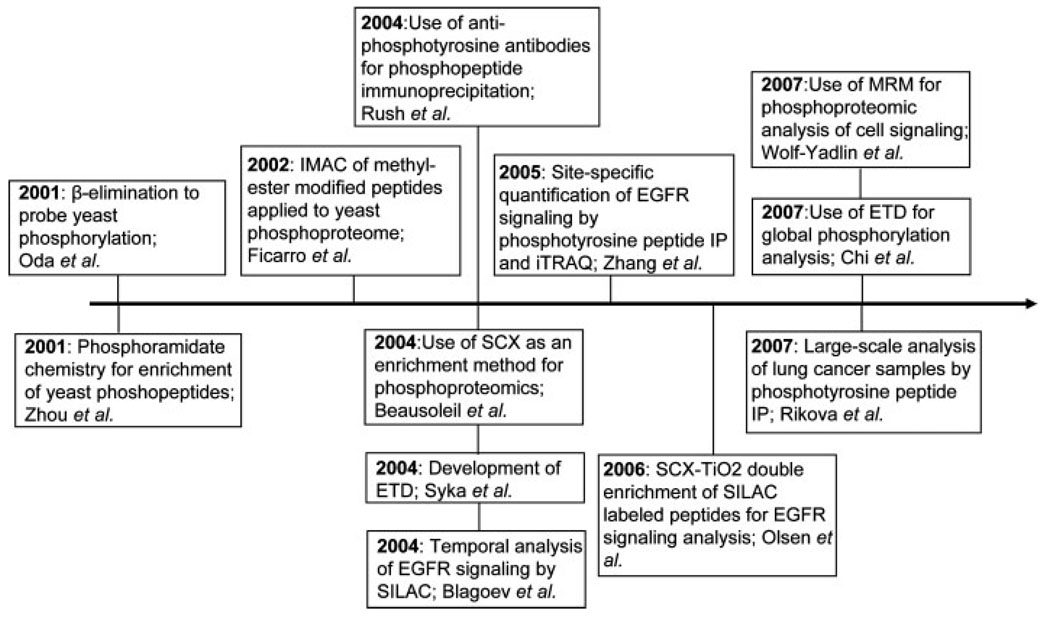
Functional classification is further complicated by protein PTMs, which can modify enzymatic activity, binding affinities, and protein conformation. Among PTMs, phosphorylation is perhaps the best studied due to the association between dysregulated phosphorylation and human pathologies [1]. Protein phosphorylation on Ser (~90%), Thr (~10%), and Tyr (~<0.05% of protein phosphorylation) residues is reversible and its dynamic addition can produce fast and precise changes in protein properties, which in turn affect many critical processes, such as protein–protein interactions, cell signaling, cytoskeleton remodeling, cell cycle events, and cell–cell interactions [2]. Protein phosphorylation analysis is still very challenging, although breakthrough developments over the past decade have now enabled the identification and quantification of thousands of sites from given biological samples. To put these advancements in the field of phosphoproteomics into perspective, Fig. 1 high-lights several of the most significant publications over the past 7 years.
Figure 1.
Timeline of selected milestones in quantitative phosphoproteomics during the last decade. Each publication has been selected based on implementation of a new method or application of recently developed methodology to uncover novel aspects of signaling networks.
Our focus in this review is on quantitative phosphoproteomics by MS. Here we discuss the latest developments in the field, including instrumentation, reagents, and enrichment techniques. Selected applications are highlighted to demonstrate the capabilities of these methods, with an eye toward quantification of signaling networks and use of this information for drug target discovery (for recent reviews, see ref. [3, 4]).
2 Challenges of phosphoproteomics
Phosphoproteomic analysis is plagued by the same challenges facing all proteomic experiments: complexity, dynamic range, and temporal dynamics. The true complexity of the phosphoproteome has yet to be determined, but the Phosphosite database (http://www.phosphosite.org) now lists >30 000 phosphorylation sites on >17 000 proteins, and this number is steadily increasing as each large-scale phosphorylation analysis continues to identify a large number of novel sites. With so many of the proteins in the cell being phosphorylated, the dynamic range of the phosphoproteome is similar to that of the proteome (i.e., ~109), but is further increased by substoichiometric modification. In addition, the temporal dynamics of protein phosphorylation regulate the rapid activation and deactivation of cellular signaling networks, further complicating analysis of the phosphoproteome. So the challenge is not simply to identify and catalog all of the phosphorylation sites, but rather to identify the site, quantify the stoichiometry, and monitor the temporal change in phosphorylation in response to a variety of cellular perturbations. Performing this task on a large number of phosphorylation sites across a broad swath of the signaling network is especially challenging, but is required to understand the mechanisms by which protein phosphorylation controls cell biology.
3 Enrichment methods
Phosphorylated proteins span the gamut of protein expression level, from hundreds of millions to a few copies per cell. However, many of the phosphorylation events associated with canonical cellular signaling pathways occur on proteins expressed at relatively low levels. Since phosphorylation of these proteins is often substoichiometric and transient, phosphopeptides obtained from these proteins after proteolytic digest are nearly impossible to detect in the whole cell lysate or tissue sample, which can generate potentially millions of peptides. Selective enrichment of phosphorylated peptides and proteins is required and has been accomplished in a number of ways, including antiphosphotyrosine antibodies [5], immobilized metal affinity chromatography (IMAC) [6], chemical modification, and strong cation exchange chromatography (SCX) [7].
Immunoprecipitation (IP) of tyrosine phosphorylated proteins and peptides with high affinity antiphosphotyrosine antibodies [8] provides good yield and specificity and has been demonstrated on a broad variety of applications [9–12]. Several reliable antiphosphotyrosine antibodies are sold commercially. These antibodies primarily recognize phosphotyrosine, but each has some bias toward the surrounding amino acids, and therefore performing the IP with multiple antibodies may increase coverage of the tyrosine phosphoproteome. Since the fraction of tyrosine phosphorylated protein to total protein may vary significantly from sample to sample, experimental optimization of conditions, including relative amount of antibody to total sample protein, is often necessary to reduce nonspecific binding while maximizing yield for the particular sample. It is worth noting that while IP has been succesfully implemented for tyrosine phosphorylation, anecdotal evidence indicates that the analogous pan-specific antibodies against phosphoserine and phosphothreonine tend to be of lower affinity, and therefore yield unsatisfactory enrichment for these subsets of phosphorylated peptides. However, recent work by Matsuoka et al. [13] has demonstrated the potential of using multiple phosphospecific antibodies recognizing ATM/ATR substrate phosphorylation sites to identify and quantify hundreds of serine and threonine phosphorylation sites matching the ATM/ATR kinase motif. Since many phospho-specific antibodies have off-target affinity, it may be that this strategy could be applied to a variety of serine/threonine kinases, effectively supplementing the need for high affinity pan-specific phospho-serine/threonine antibodies, and enabling network analysis of serine/threonine phosphorylation, one motif at a time.
For many applications, the goal is to generate a global view of serine, threonine, and tyrosine phosphorylation within the sample rather than focusing specifically on a selected subset of phosphorylated peptides. Perhaps the most common technique to enrich for global phosphorylation is IMAC, which is based on the high affinity of phosphate groups for metal ions such as Fe3+ Zn2+ and Ga3+ One of the main limitations associated with IMAC-based phosphopeptide enrichment has been the nonspecific retention of nonphosphorylated acidic peptides, due to the weak affinity between negatively charged carboxylates and positively charged metal ions. However, conversion of carboxylate groups to esters effectively eliminates nonspecific retention of nonphosphorylated peptides on the IMAC column [14]. This method has also been used in an automated platform involving online IMAC, nano-LC, and ESI-MS, enabling reproducible detection and identification of phosphopeptides in a low-femtomole range [15], and may be coupled with a stable-isotope labeling step for relative quantification [14]. Since different metal ions appear to enrich for slightly different subsets of phosphorylated peptides, maximal coverage of the phosphoproteome may be obtained by multiple analyses with different metals, or by mixing multiple metal ions in a single IMAC enrichment step.
Within the past couple of years, titanium dioxide (TiO2) has emerged as the most common of the metal oxide affinity chromatography (MOAC)-based phosphopeptide enrichment methods. This technique requires significantly shorter preparation time and offers increased capacity relative to IMAC resins with the same bed volume. Since this method exploits the same principle as IMAC, it is similarly prone to nonspecific retention of acidic nonphosphorylated peptides. However, loading peptides in 2,5-dihydroxybenzoic acid has been shown to reduce nonspecific binding to TiO2, thereby improving phosphopeptide enrichment without chemical modification of the sample [16]. Overall, TiO2 is often considered to be interchangeable with IMAC, in that similar sample levels (e.g., micrograms of protein) can be analyzed and hundreds of sites per sample can be identified when either technique is used as the sole enrichment method, although each method has demonstrated differential bias and selectivity.
As an alternative to metal-ion-based enrichment strategies, SCX has been successfully used to separate phosphorylated peptides from peptide mixtures for subsequent MS analysis [7, 16, 17]. In this technique, binding to the SCX column is dependent on columbic interaction between negatively charged resin and positively charged peptides. If sample loading is performed under strong acidic conditions (pH ~2.7), carboxylates are rendered neutral, while the phosphate group retains a negative charge. As a result, the total charge of phosphorylated tryptic peptides is reduced from + 2 to + 1, and the interaction strength with the SCX resin is correspondingly reduced. Elution with a gradient of increasing salt concentration thus allows phosphopeptides to elute earlier relative to nonphosphorylated peptides, providing semiselective enrichment [7]. To reduce the nonphosphopeptide background, a second, IMAC-based enrichment step has been performed on SCX fractions, enabling the identification of thousands of phosphorylated peptides from given samples [7, 17, 18]. As another variation and improvement of the SCX method, a mixed-bed resin comprised of a blend of anion and cation exchangers (ACE) has been recently proposed for phosphopeptide enrichment, increasing retention of acidic peptides, and reducing retention of basic and neutral peptides by the added anion-exchange resin, which in turn improved the identifications of phosphopeptides by 94% over SCX [19].
Phosphorylation enrichment by SCX-based fractionation, either solely or coupled with other enrichment steps, has successfully been applied to identify large numbers of phosphorylation sites (in the order of thousands). However, it is worth noting that the technique, as implemented to date, requires a large amount of starting material (tens of milligrams of protein) which makes it inapplicable to samples that are available in small or limited quantity. In addition, SCX fractionation decreases the complexity of the starting samples by dividing it into many fractions, each of which requires a separate MS analysis, leading to the possibility of up to 100 MS analyses for each biological replicate. The sample requirements, analysis time, and labor associated with each biological sample has unfortunately limited the application of this technique such that few studies have incorporated biological replicates.
Several laboratories have taken the approach of chemically modifying the phosphate to provide an affinity enrichment tag. For instance, the phosphate groups on serine and threonine can be removed by β-elimination and replaced by ethanedithiol coupled to a biotin tag, making it possible to purify modified peptides using an avidin affinity column [20]. The primary disadvantage of this approach is that tyrosine phosphorylation does not undergo β-elimination, and therefore these peptides are not enriched by this method. It is also possible to directly attach an affinity tag to the phosphate through phosphoramidate chemistry (PAC). Recent improvements in this approach have improved the yield by reducing the number of steps, making the approach much more user-friendly [21].
Different enrichment methods may yield different pools of phosphopeptides from the same peptide mixture, as recently shown in a comparative study conducted by the Aebersold group, where PAC, IMAC, and two types of TiO2 methods were employed to isolate phosphopeptides from a tryptic digest of Drosophila melanogaster Kc167 cells [22]. Performing multiple analyses with several complementary phosphopeptide enrichment methods may be the best way to maximize depth of coverage, albeit at the cost of increased sample consumption and reduced throughput.
It is often the case that any single enrichment step does not provide sufficient specificity when dealing with complex biological samples. Therefore, double enrichment, as in the above scenario with IMAC and SCX, is often required to improve phosphopeptide analysis. In another example, our laboratory has combined antiphosphotyrosine peptide IP with IMAC to analyze tyrosine phosphorylation in murine adipocytes [23], human Jurkat cells [24], and in the epidermal growth factor receptor (EGFR) signaling network in human mammary epithelial cells (HMECs) [25, 26].
4 Quantification approaches
To date much of the work in phosphoproteomics has focused on developing novel methods for the enrichment of phosphorylated peptides/proteins and subsequent application of these methods to identify large numbers of phosphorylation sites from given biological samples. Data generated in these “cataloging” studies may be informative for laboratories studying selected proteins whose phosphorylation sites appear in the catalog, but it is often difficult to link the information in these large-scale datasets to cellular signaling networks. In order to identify phosphorylation events that may be regulating biological response to cellular perturbation, quantification of phosphorylation pre- and post-cell stimulation is necessary. Several MS-based quantification methods have been implemented for phosphoproteomics, including stable-isotope labeling through chemical modification of peptides, stable-isotope labeling of amino acids in cell culture (SILAC), and label-free methods.
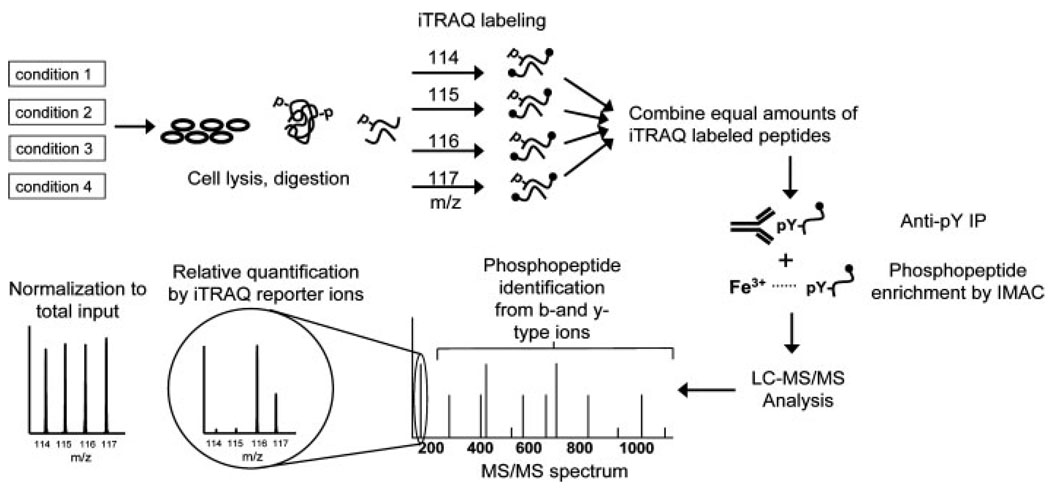
Multiple chemical modification protocols have been utilized to incorporate stable isotopes; among these methods, iTRAQ has become the most commonly used option due to its multiplex capability. The iTRAQ reagent consists of four isobaric isoforms which react with primary amines, thereby enabling quantitative comparison of four protein samples in parallel. Since the labels are isobaric, quantification is performed in MS/MS mode by comparing peak areas of the marker ions resulting from fragmentation of the iTRAQ label, so the same spectrum is used for quantification and sequence identification of the phosphopeptide. As demonstrated in Fig. 2, when coupled to phosphotyrosine peptide IP and IMAC, iTRAQ has been successfully applied for the quantification of phosphorylation states of differentially stimulated Jurkat cells [24], adipocytes [23], and for analysis of the temporal dynamics of the ErbB signaling network [25, 26]. Recently, the eight-plex version of iTRAQ has been developed and applied to proteome analysis [27], demonstrating the potential to further increase throughput in quantitative proteomic analysis, but this reagent has yet to be used for the quantification of protein phosphophorylation.
Figure 2.
Example workflow of the quantitative phosphotyrosine analysis experiment using iTRAQ labeling. Proteins are extracted from four biological samples (cell lines, stimulation time points, tissue samples), modified and digested. For quantification, the resulting peptides are labeled with iTRAQ reagent and combined. The mixture is then subjected to two steps of enrichment for tyrosine phosphorylated peptides: IP with antiphosphotyrosine antibodies and IMAC. Phosphorylated peptides eluted from the IMAC column are analyzed by LCMS/ MS, typically on a quadrupole TOF mass spectrometer. Each MS/MS spectrum contains both sequence-specific fragmentation events for identification of the peptide and phosphorylation site as well as the low-mass iTRAQ marker ions to quantify phosphorylation across the four samples. iTRAQ quantification is normalized to the supernatant to eliminate variation resulting from the preparation process.
For metabolic isotope labeling, cells are cultured in a medium where the natural form of an amino acid (typically arginine or lysine) is replaced with a stable isotope form, such that proteins expressed by the cell incorporate the heavier version of this amino acid and therefore alter their molecular mass (see ref. [28] for the detailed, updated review of the method). This technique is generally referred to as SILAC, and enables comparison of up to three samples in a single analysis. Initially, SILAC was developed for mammalian cells [29], but its use has been broadened to bacteria [30] and yeast [17]. There have also been reports of in vivo metabolic labeling in whole organisms (Plasmodium falciparum [31], plants [32], D. melanogaster, Caenorhabditis elegans [33], and rats [34]), but they require feeding labeled reagents to model organisms, which makes multiple experiments cost-ineffective.
There are advantages and disadvantages to both metabolic and chemical modification-based labeling methods. For SILAC, cells need to undergo multiple cell divisions in medium containing stable isotope-labeled amino acids to ensure sufficient isotope incorporation for reliable comparison between cell states. For this reason, it is not practical to apply SILAC to generate quantitative data from primary cells or to compare tumor tissue specimens directly. Moreover, culture conditions need to be carefully monitored to prevent interconversion between arginine and proline, which could negatively affect quantification accuracy. However, since cells can be mixed prior to cell lysis and sample processing, quantification error associated with differences in these steps can be avoided, potentially leading to higher accuracy. By comparison, postextraction methods permit quantitative analysis of a broader variety of samples, including animal tissues and human tumors, providing the opportunity to follow in vivo changes between healthy and diseased states, which in turn can lead to the discovery of new drug targets. Since labeling typically occurs following enzymatic digestion, sample handling needs to be carefully controlled to minimize variation introduced prior to mixing differentially labeled samples.
For many applications, quantification relative to an arbitrary state is not sufficient, and absolute quantification is desired. Typically, absolute quantification would have required the chemical synthesis of heavy-isotope coded peptides [35] to be added to the sample as internal standards. Recently, however, such peptides can be biologically expressed using the method (named QconCAT) introduced by Pratt et al. [36, 37], in which Escherichia coli is transfected with a modified gene containing the peptide of interest. The transfected E. coli is cultured in a medium containing heavy lysine and arginine and the protein is digested following purification, yielding the desired peptide, which can then be added to the sample. A recent review by Mirzaei et al. [38] summarizes the current techniques for production of isotope-labeled peptide standards.
Label-free quantification may be employed as a less expensive alternative. These analyses are typically performed either through direct comparison of two samples analyzed on the same platform, or by spiking the sample with standard peptides and quantifying in reference to these standards [39–41]. Unfortunately, label-free quantification does not provide the multiplex advantage associated with SILAC or chemical modification (e.g., iTRAQ), and therefore requires a separate MS analysis for each sample. Moreover, label-free analysis tends to have greater quantification error compared to analysis of stable isotope-labeled samples, due to inconsistent sample processing and chromatography across multiple analyses.
In the near future, MS-based proteomics should be increasingly focused on absolute quantification of protein expression level and stoichiometry of major PTMs. This information will make it feasible to directly compare data between experiments, conditions, and laboratories. Moreover, absolute quantification will enable the development of more complex, kinetic computational models describing the biological systems in much greater detail.
5 Instrument choice
As with all MS, optimal instrumentation for phosphoproteomic analysis is defined by the application. For instance, in the case of global phosphoproteomics, instrument choice may be influenced by the facile neutral loss of phosphoric acid (98 Da) from serine and threonine phosphorylation, often resulting in uninterpretable MS/MS spectra. To circumvent this problem, with a quadrupole ion trap (IT) it is possible to perform MS3 on the neutral loss peak from the MS/MS spectrum; this strategy has been successfully implemented for several large-scale phosphoproteomic studies [42]. The facile neutral loss problem may also be addressed by using a quadrupole TOF instrument, as the intensity of the neutral loss peak is diminished by multiple collisions in a high-pressure quadrupole, yielding an increase in sequence-specific fragmentation and improved phosphopeptide identification. More recently, electron capture dissociation (ECD) [43] and, especially, electron transfer dissociation (ETD) [44] have been demonstrated to be particularly useful for the analysis of labile PTMs including serine and threonine phosphorylation, providing good sequence coverage even for large peptides and proteins.
Compared to serine or threonine phosphorylation, the phosphate attached to tyrosine is relatively stable and usually does not produce a neutral loss in MS/MS mode, although loss of 80 Da may be seen from some tyrosine phosphorylated peptides. In fact, MS/MS spectra of tyrosine phosphorylated peptides tend to resemble nonphosphorylated peptides, although fragmentation N- and C-terminal to phosphotyrosine typically produces a characteristic immonium ion of m/z 216.0426. Since this fragment ion is specific to tyrosine phosphorylation, precursor ion scanning coupled with subsequent MS/MS analysis of the selected precursor ions has been used to identify these pTyr-containing peptides from complex mixtures.
Instrument choice is further affected by the chosen quantification method. SILAC experiments require high resolution and high mass accuracy because the number of species in the MS spectra are doubled (or tripled), leading to increased complexity and increased frequency of overlapping peaks. Although many of the initial SILAC experiments were analyzed on a quadrupole TOF instrument, most of these studies are now conducted on instruments with quadrupole IT fragmentation and Fourier-transform based detection (e.g., LTQ-FTMS or LTQ-Orbitrap) due to increased MS/MS acquisition speed in the IT and increased MS acquisition mass accuracy and resolution in the FT mass analyzer. Choice of instrumentation for iTRAQ-based quantification is more restrictive due to the low m/z ratio of the iTRAQ marker ions. Quadrupole IT instruments have traditionally not performed well for these experiments because fragmentation in a quadrupole IT is typically performed at a Q-value of 0.25–0.3, leading to loss of the low mass region of the MS/MS spectrum. The hybrid quadrupole TOF mass spectrometer has been the instrument of choice for iTRAQ-based quantitative phosphoproteomic analyses due to the high resolution and mass accuracy (low ppm range) in both MS and MS/MS mode, providing accurate detection of the charge state and unambiguous assignment of the monoisotopic mass. Importantly, high-resolution MS/MS spectra obtained on this instrument have improved quantification accuracy by separating iTRAQ marker ions from contaminant ions at the same nominal m/z ratio [45]. The recent development of C-Trap-based fragmentation on the LTQ-Orbitrap now enables triple quadrupole-like fragmentation and high resolution, high mass accuracy detection in the orbitrap mass analyzer, potentially providing a viable alternative to quadrupole TOF instruments for iTRAQ-based quantification of phosphorylated peptides [46].
Given the large array of available enrichment and quantification techniques and the possible combinations of these approaches with various types of MS, it is worth reviewing how these options have been implemented to interrogate the phosphoproteome.
6 Phosphoproteomics in the EGFR network
In canonical growth factor signaling, stimulation of cell surface receptors first triggers activation of the receptor and subsequently transmits the signal to a large number of intracellular effecter molecules. The EGFR network is one of the most extensively studied areas of signal transduction, and the one which best exemplifies oncogenic aberrations in cellular signaling [47]. EGFR is a member of ErbB family of RTKs which comprises four receptors (EGFR, HER2, HER3, and HER4) and 13 polypeptide ligands, each of which contains a conserved epidermal growth factor (EGF) domain. This complex signaling network has been one of the primary targets for phosphoproteomic analysis. In fact, one of the first studies to address quantitative dynamics of phosphorylation at the network level was performed in the EGFR system. In this study, a monoclonal antiphosphotyrosine antibody was used to immunoprecipitate SILAC-labeled tyrosine phosphorylated proteins and their binding partners. These proteins were enzymatically digested to peptides and analyzed by LC-MS/MS, resulting in the identification of ~80 signaling proteins, including many known EGFR substrates and several novel effectors [9]. The relative intensity of SILAC-labeled peptides was used to quantify temporal dynamics within the network following EGF stimulation of HeLa cells. However, since enrichment for tyrosine phosphorylation was performed at the protein level and enzymatic digestion produces a broad variety of tryptic peptides, most of which represent nonphosphorylated sections of the immunoprecipitated proteins, very few phosphorylation sites were identified in this study, and therefore much of the key signaling information is missing. In fact, since phosphorylation often happens at multiple tyrosine residues within a single protein, and different phosphorylation sites on a single protein are often differentially regulated with individual functions, quantification of each phosphorylation site in the global signaling network is critically important.
To address the need for site-specific quantification, Zhang et al. [26] performed time-resolved temporal analysis of EGFR signaling network by quantitative MS using iTRAQ. In this study, proteins from whole cell lysate were proteolyzed to peptides and labeled with iTRAQ prior to mixing. Tyrosine phosphorylated tryptic peptides were then enriched, first by IP with an antiphosphotyrosine antibody, and then by IMAC to eliminate nonspecifically retained nonphosphorylated peptides. As a result, 104 tyrosine phosphorylation sites from 76 proteins were identified with temporal phosphorylation profiles at four time points of EGF stimulation. Site specific monitoring of protein phosphorylation in this study provided explicit detail regarding the regulation of proteins within the signaling network, including differential regulation of multiple sites on given proteins, and identification of phosphorylation “modules”, clusters of sites with selfsimilar temporal profiles.
Peptide IP has now been successfully implemented in a variety of phosphoproteomic studies, including a recent large scale analysis to identify phosphotyrosine signaling networks in lung cancer cell lines and tumors [48]. In this study, oncogenic tyrosine kinase signaling was characterized by analysis of tyrosine phosphorylation in 41 nonsmall cell lung cancer (NSCLC) cell lines and over 150 NSCLC tumors, resulting in the identification of a total of 4551 sites of tyrosine phosphorylation on greater than 2700 different proteins. Bioinformatic analysis of the dataset identified a subset of NSCLC tumors and cell lines exhibiting high tyrosine phosphorylation, possibly due to the presence of abnormally activated or overexpressed tyrosine kinases. Potential “driver” tyrosine kinases were identified by a ranking process to identify unusually high levels of tyrosine kinase activity in a subgroup of patients. Among the 18 tumors with highest EGFR rank, nine tumors were confirmed to have an activating mutation in the kinase domain. Based on this success, a similar approach was used to identify other candidate driver tyrosine kinases in the remaining tumors, including the fusion tyrosine kinase EML4-ALK, as also recently reported by Soda et al. [49]. This example demonstrates that MS-based phosphoproteomic discovery capabilities are highly complementary to the genomic cDNA screening technology that was used to originally identify this transforming fusion tyrosine kinase.
7 Global phosphoproteomics
With advances in MS and phosphopeptide separation methodologies, the scale of global phosphoproteomic studies has increased significantly since the first large-scale global analysis of the yeast phosphoproteome [14] only 5 years ago. For instance, Olsen et al. [50] recently quantified global phosphorylation changes in EGF stimulated HeLa cells by combining SILAC for relative quantitation with SCX and TiO2 chromatography for phosphopeptide enrichment. Enriched phosphopeptides were analyzed by more than 100 LC-MS/MS runs to identify over 6600 phosphorylation sites on 2240 proteins with their annotated subcellular (nuclear vs. cytosolic) localization. This information represents the largest global phosphorylation dataset available to date for the EGFR signaling network, and covers a broad spectrum of phosphorylation events, from EGFR autophosphorylation to phosphorylation of terminal effector molecules such as transcription factors. However, even this dataset is still far from comprehensive, as many well characterized phosphorylation sites were not identified in this analysis. For instance, the low number of pTyr sites (103) in the dataset is likely due to the large dynamic range associated with simultaneous analysis of serine, threonine, and tyrosine phosphorylation. Given the large number of previously uncharacterized phosphorylation sites and the massive size of this dataset, extraction of biological hypotheses is not trivial. However, it is likely that improved functional characterization of the EGFR signaling network may arise from linking this dataset to other complementary datasets [51] (e.g., Friedman and Perrimon’s work RNAi-based screening to identify components in the RTK–Erk network [52]).
8 Reproducibility of phosphoproteomics
One of the principal limitations with each of the above studies has been the irreproducibility of MS-based data, such that replicate analyses of the same sample (or analysis of biological replicates) will typically identify only 60–70% of the same phosphorylation sites. Much of this irreproducibility stems from operating the mass spectrometer in a nonbiased “discovery” mode, in which the instrument continuously repeats a cycle consisting of a full-scan mass spectrum, followed by fragmentation of a certain number of the most abundant peaks for peptide and phosphorylation site identification. This mode enables identification of novel phosphorylation sites, but the semiautomated peak selection process is inherently irreproducible (see, for example, a study of peptide/protein identification reproducibility by MS [53]), making it difficult to directly compare multiple datasets. Recently we have developed an approach combining “discovery” mode analysis of selected biological samples with high reproducibility multiple reaction monitoring (MRM)-based “monitoring” mode for quantification of hundreds of selected phosphorylated peptides [45]. This method was applied to investigate the temporal dynamics of 226 phosphorylation sites at seven time points of EGF stimulation of HMECs. Because preselected phosphopeptides are specifically monitored in MRM mode, the number of peptides reproducibly identified from four replicates increased from 34% in discovery mode to 88% in monitoring mode. This combined method should be applicable to a variety of biological systems, and will enable reproducible network-wide quantification of cell perturbation effects across a broad variety of stimulation conditions.
9 Phosphoproteomics and systems biology
Computational and systems biology approaches have become increasingly important in the analysis of phosphoproteomic data. To provide higher meaning to the data, quantification of both the phosphorylation network and the corresponding biological response must be collected. Bioinformatics and mathematical modeling can then be applied to build hypotheses connecting phosphorylation information to cellular phenotypes. In the past, identification of key elements in signaling networks has largely been accomplished in a subjective way through the manual comparison of fold –change phosphorylation and cell behavior. Recently, mathematical modeling methods such as partial least squares regressions (PLSR) have been implemented to objectively correlate phosphoproteomic data with cellular response to stimulation. For instance, Wolf-Yadlin [25] et al. and Kumar et al. [54] applied PLSR to the quantitative MS data describing the effects of HER2 overexpression on phosphotyrosine signaling in HMECs stimulated by EGF or heregulin (HRG). Cell migration and proliferation were collected under the same conditions and PLSR was used to integrate the data types. The final model described a set of signaling molecules that are most relevant for the changes in migration induced by HER2 overexpression. This type of modeling can provide insight into the functionality of unknown proteins, which can be further tested by biological experiments.
As described above, the value of phosphoproteomic datasets significantly increases when these data are used to generate hypotheses as to the function of selected phosphorylation sites, and even more when these hypotheses are experimentally validated. For instance, Kratchmarova et al. [55] interrogated tyrosine-phosphorylation mediated signaling networks following EGF or PDGF stimulation of mesenchymal stem cells. Interestingly, although most of the network responded similarly to these two stimulations, activation of the PI3K pathway was exclusive to PDGF stimulation. Since EGF stimulation of these cells drives osteoblast differentiation, the authors hypothesized that PDGFR-associated PI3K activation could be key to controlling biological response to differential growth factor stimulation. Indeed, this hypothesis was validated by small molecule inhibition of PI3K followed by PDGF stimulation to drive ostoblast differentiation.
Another example of biological validation of MS-based phosphoproteomic data was provided recently by Huang et al. [56] in the quantitative phosphoproteomic analysis of the EGFRvIII signaling pathways in U87MG glioblastoma cell lines. Clustering of phosphorylation data identified previously unknown crosstalk between EGFRvIII and c-Met, a receptor tyrosine kinase that is well known to drive malignancy in various cancers. Since EGFRvIII and c-Met may signal cooperatively to drive tumor growth, U87GM cells expressing EGFRvIII were treated with the EGFR kinase inhibitor AG1478 and the c-Met inhibitor PHA665752. Either compound alone had minimal cytotoxic effect, but the combination of the two compounds significantly increased cytotoxicity at lower doses, indicating that EGFRvIII utilizes other receptor tyrosine kinases to potentiate oncogenic signaling. This finding has recently been corroborated through the analysis of glioblastoma cell lines and tumors using antiphosphotyrosine antibody arrays [57].
10 Phosphoproteomics and drug development
As described above, quantitative MS-based phosphoproteomics has been applied to identify oncogenic kinases which may serve as potential drug targets. To validate this hypothesis, cells are often treated with selected kinase inhibitors with the goal of altering cellular phenotype, but it is often difficult to establish whether the effect was due to on-or off-target effects of the compound. In order to determine the mechanism of action, it may be necessary to quantify the specificity of the inhibitor, a nontrivial task. To address this challenge, Bantscheff et al. [58] developed a kinase capturing bead (“Kinobead”) consisting of multiple immobilized broad-selectivity kinase inhibitors. On application to cell lysate, a large number of kinases (and other purine-binding proteins) are retained on the Kinobeads due to the interaction with the kinase inhibitors. To obtain a quantitative target profile of a selected compound, cells or cell lysate are treated with the compound at varying concentrations prior to affinity isolation with the Kinobead. Kinases inhibited by the selected compound exhibit decreased binding to the Kinobead and therefore yield decreased signal by quantitative (iTRAQ-based) MS. Combining this approach with phosphorylation analysis can yield a profile as to the phosphorylation status of the kinases bound to the selected compound, potentially identifying whether the compound binds to the active or inactive isoform of the kinase. After establishing the specificity of the inhibitor, it will then be possible to regather quantitative phosphoproteomic data to determine the effect on the cell signaling network of inhibiting the selected targets of the inhibitor. The workflow for this approach is outlined schematically in Fig. 3. Following through this iterative process, one could begin to build out downstream signaling networks directly or indirectly affected by a selected kinase in the context of various human pathologies.
Figure 3.
Signaling network analysis by mass spectrometry, drug-target discovery, and kinase inhibition. Mass spectrometry-based phosphoproteomics is used to quantify signaling networks in various cell states resulting from multiple perturbations (e.g., cytokine stimulation, RNAi, small molecule inhibitors). Computational modeling is used to link this information to quantitative phenotypic measurements, thereby identifying key nodes in the signaling network. To validate this model, compounds are developed to target these nodes, and the phenotypic effect is monitored. The specificity of the compounds can then be tested by the Kinobead assay to quantify on- and off-target effects. Changes in the signaling network can then be quantified to establish kinase–substrate relationships given the established targets of the compound.
11 Conclusions
What does the future hold for quantitative phosphoproteomics by MS? The field is in a rapid state of flux, including new enrichment strategies, novel quantification reagents, and new instrumentation. With each improvement it becomes possible to identify and quantify increasing numbers of phosphorylation sites, digging deeper and deeper into the elusive comprehensive phosphoproteome. However, as many of the above applications demonstrate, size of the dataset is not always the most important metric. Instead, understanding the biological implications of many of the phosphorylation sites is critical, since the ultimate goal of most of these studies is to increase insight into cellular signaling and biological control. Linking phosphorylation data to other quantitative phenotypic endpoints is a crucial step in this procedure, and one that has been often ignored in the effort to gather larger data sets. Going forward, the combination of MS, phenotypic characterization, mathematical modeling, and selected perturbations should provide rapid advancement in our understanding of the complexities of cellular signaling network, information that will enable the development of better therapeutic agents with fewer off-target effects.
Abbreviations
- EGFR
epidermal growth factor receptor
- HMEC
human mammary epithelial cells
- IMAC
immobilized metal affinity chromatography
- IP
immunoprecipitation
- NSCLC
nonsmall cell lung cancer
- PLSR
partial least squares regressions
- SCX
strong cation exchange
- SILAC
stable-isotope labeling of amino acids in cell culture
Footnotes
The authors have declared no conflict of interest.
References
- 1.Hunter T. Signaling-2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 3.Huang PH, Cavenee WK, Furnari FB, White FM. Uncovering therapeutic targets for glioblastoma: A systems biology approach. Cell Cycle. 2007;6:2750–2754. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Linding R. Network medicine. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, et al. Analysis of receptor signaling pathways by mass spectrometry: Identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. USA. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 7.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rush J, Moritz A, Lee KA, Guo A, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2004 doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 9.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 10.Hinsby AM, Olsen JV, Bennett KL, Mann M. Signaling initiated by overexpression of the fibroblast growth factor receptor-1 investigated by mass spectrometry. Mol. Cell. Proteomics. 2003;2:29–36. doi: 10.1074/mcp.m200075-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Ibarrola N, Molina H, Iwahori A, Pandey A. A novel proteomic approach for specific identification of tyrosine kinase substrates using 13C-labeled tyrosine. J. Biol. Chem. 2004 doi: 10.1074/jbc.M311714200. [DOI] [PubMed] [Google Scholar]
- 12.Hinsby AM, Olsen JV, Mann M. Tyrosine phosphoproteomics of fibroblast growth factor signaling: A role for insulin receptor substrate-4. J. Biol. Chem. 2004;279:46438–46447. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 14.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 15.Ficarro SB, Salomon AR, Brill LM, Mason DE, et al. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 16.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Gruhler A, Olsen JV, Mohammed S, Mortensen P, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Motoyama A, Xu T, Ruse CI, Wohlschlegel JA, Yates JR., III Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides. Anal. Chem. 2007;79:3623–3634. doi: 10.1021/ac062292d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 21.Tao WA, Wollscheid B, O’Brien R, Eng JK, et al. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 22.Bodenmiller B, Mueller LN, Pedrioli PG, Pflieger D, et al. An integrated chemical, mass spectrometric and computational strategy for (quantitative) phosphoproteomics: Application to Drosophila melanogaster Kc167 cells. Mol. Biosyst. 2007;3:275–286. doi: 10.1039/b617545g. [DOI] [PubMed] [Google Scholar]
- 23.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes. 2006;55:2171–2179. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 24.Kim JE, White FM. Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J. Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 25.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Choe L, D’Ascenzo M, Relkin NR, Pappin D, et al. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 29.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, et al. Proteome- wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 2004;52:1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- 32.Gruhler A, Schulze WX, Matthiesen R, Mann M, Jensen ON. Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol. Cell. Proteomics. 2005;4:1697–1709. doi: 10.1074/mcp.M500190-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, et al. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 34.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., III Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 35.Barnidge DR, Hall GD, Stocker JL, Muddiman DC. Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC-MS/MS. J. Proteome Res. 2004;3:658–661. doi: 10.1021/pr034124x. [DOI] [PubMed] [Google Scholar]
- 36.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 37.Pratt JM, Simpson DM, Doherty MK, Rivers J, et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 38.Mirzaei H, McBee J, Watts J, Aebersold R. Comparative evaluation of current peptide production platforms used in absolute quantification in proteomics. Mol. Cell. Proteomics. 2007;7:813–823. doi: 10.1074/mcp.M700495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Ishihama Y, Oda Y, Tabata T, Sato T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Mallick P, Schirle M, Chen SS, Flory MR, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 42.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Quantitative analysis of complex protein mixtures using isotopecoded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 43.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, et al. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 44.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. USA. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen JV, Macek B, Lange O, Makarov A, et al. Higherenergy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 47.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 48.Rikova K, Guo A, Zeng Q, Possemato A, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 49.Soda M, Choi YL, Enomoto M, Takada S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 50.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 51.Yaffe MB, White FM. Signaling netwErks get the global treatment. Genome Biol. 2007;8:202. doi: 10.1186/gb-2007-8-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 53.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in largescale proteomics investigations. Nat. Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 54.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. Modeling HER2 effects on cell behavior from mass spectrometry phosphotyrosine data. PLoS Comput. Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 56.Huang PH, Mukasa A, Bonavia R, Flynn RA, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 58.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 59.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 60.Chi A, Huttenhower C, Geer LY, Coon JJ, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]