Abstract
HLA-B27- and -B57-positive HIV-infected humans have long been associated with control of HIV replication, implying that CD8+ T cell responses contribute to control of viral replication. In a similar fashion, fifty percent of Mamu-B*08-positive Indian rhesus macaques control SIVmac239 replication and become elite controllers with chronic phase viremia below 1,000 vRNA copies/ml. Interestingly, Mamu-B*08-restricted SIV-derived epitopes appeared to match the peptide binding profile for HLA-B*2705 in humans. We, therefore, defined a detailed peptide-binding motif for Mamu-B*08 and investigated binding similarities between the macaque and human MHC class I molecules. Analysis of a panel of almost 900 peptides revealed that despite substantial sequence differences between Mamu-B*08 and HLA-B*2705, the peptide-binding repertoires of these two MHC class I molecules share a remarkable degree of overlap. Detailed knowledge of the Mamu-B*08 peptide-binding motif enabled us to identify six additional novel Mamu-B*08-restricted SIV-specific CD8+ T cell immune responses directed against epitopes in Gag, Vpr, and Env. All 13 Mamu-B*08-restricted epitopes contain an R at the position 2 primary anchor, and 10 also possess either R or K at the N-terminus. Such dibasic peptides are less prone to cellular degradation. This work highlights the relevance of the Mamu-B*08-positive SIV-infected Indian rhesus macaque as a model to examine elite control of immunodeficiency virus replication. The remarkable similarity of the peptide-binding motifs and repertoires for Mamu-B*08 and HLA-B*2705 suggests that the nature of the peptide bound by the MHC class I molecule may play an important role in control of immunodeficiency virus replication.
Keywords: Immunodeficiency Diseases, MHC, Antigens/Peptides/Epitopes, Cytotoxic T Cells, Antigen Presentation/Processing, AIDS
INTRODUCTION
In light of recent phase II and III HIV vaccine trial failures (1–3), it may be useful to place additional emphasis on understanding the immune correlates involved in control of immunodeficiency virus replication. Human elite controllers (ECs) are a rare population of individuals that control HIV replication to extremely low levels without medical intervention (4). Similarly, a small number of macaques control pathogenic simian immunodeficiency virus (SIV) replication and become ECs (5, 6). These unique animals are a valuable resource to complement human studies in understanding immunodeficiency virus pathogenesis and immunity.
Recently, we identified an MHC class I allele in Indian rhesus macaques, Mamu-B*08, that is enriched in EC cohorts and associated with reduced chronic phase plasma virus concentrations of the pathogenic strain SIVmac239 (6). Over 50% of Mamu-B*08-positive, SIV-infected macaques become ECs. We then identified seven CD8+ T cell responses in SIV-infected EC macaques that were restricted by Mamu-B*08 (7). All seven SIV epitopes contained R at position 2 and L at the C-terminus. Interestingly, these epitopes appeared to match the peptide binding profile of HLA-B27 (8–11), an allele associated with slow disease progression in humans (12–17). Furthermore, in both HLA-B27-positive humans and Mamu-B*08-positive macaques, virus-specific CD8+ T cells have been implicated in controlling immunodeficiency virus replication (7, 13, 15, 18–21).
The apparent similarities between Mamu-B*08 and HLA-B27 warranted a more thorough comparison of these two alleles. We also investigated Mamu-B*03 due to its sequence similarity with Mamu-B*08 (22) as well as its association with slow SIV disease progression (23). While Mamu-B*08 and Mamu-B*03 exhibit sequence similarity in their α1 and α2 domains, the sequences of these macaque alleles and HLA-B*2705 are considerably different (Figure 1). Despite this fact, a previous study demonstrated that Mamu-B*03 could bind several endogenous HLA-B27 ligands (24).
Figure 1.
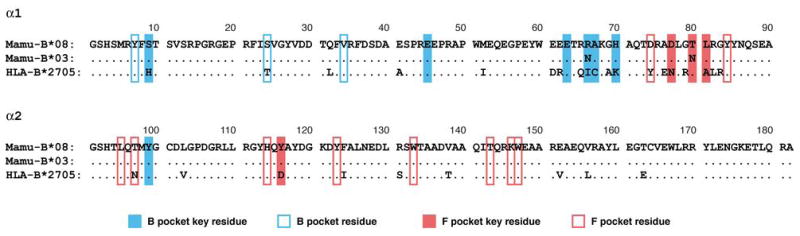
Comparison of the α1 and α2 domains of the MHC class I alleles, Mamu-B*08, Mamu-B*03, and HLA-B*2705. Residues matching the sequence of Mamu-B*08 are indicated by dots (.). Residues predicted to form the B and F binding pockets are indicated based on previous studies (74–76).
In this study, we examined binding similarities between Mamu-B*08, Mamu-B*03, and HLA-B*2705. Using ligand elution and positional scanning combinatorial library (PSCL) approaches, we first defined a detailed peptide-binding motif for Mamu-B*08, along with Mamu-B*03, and compared it to that of HLA-B*2705. We then tested the capacity of a panel of almost 900 peptides to bind these MHC class I molecules. This analysis revealed that despite minimal sequence similarity in the peptide-binding groove between the human molecule, HLA-B*2705, and the Indian rhesus macaque molecules, Mamu-B*08 and Mamu-B*03, these molecules bound the same peptides.
We also examined the chronic phase breadth and magnitude of SIV-specific responses restricted by Mamu-B*08. Using the Mamu-B*08 peptide-binding motif, we found over 200 peptides that bound to Mamu-B*08 with IC50 values ≤500 nM. The peptides were subsequently tested in IFN-γ ELISPOT assays. In addition to the seven previously described Mamu-B*08-restricted epitopes, we identified six novel SIV-specific CD8+ T cell responses restricted by Mamu-B*08. Four epitopes were located in Env, while two were located in Gag and Vpr. Interestingly, the region containing the SIV Gag epitope is conserved between SIVmac239 and HIV-2 and also contains the immundominant HLA-B27-restricted, HIV-1 epitope Gag263–272KK10. However, unlike the situation in HLA-B27-positive HIV-infected humans, Gag263–271YL9 is subdominant and infrequently targeted in Mamu-B*08-positive SIV-infected macaques. In addition, while viral escape is predominantly detected in the HLA-B27-restricted Gag263–272KK10 response in HIV-infected humans (13, 18, 19), we found that viral variation in Mamu-B*08-positive macaques typically occurs in a number of other CD8+ T cell epitopes. Interestingly, most of the SIV variants are located in Vif and Nef. Therefore, while HLA-B*2705 and Mamu-B*08 are similar in function and human and macaques expressing these two MHC class I alleles can control immunodeficiency virus replication, individuals expressing these alleles typically target different areas of the virus. This suggests that control of immunodeficiency virus replication is due to shared characteristics among viral epitopes.
MATERIALS AND METHODS
MHC class I typing of macaques
Indian rhesus macaques (Macaca mulatta) were genotyped for Mamu-B*08 (6) along with the following MHC class I alleles, Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03, -B*04, -B*17, and -B*29 using PCR amplification with sequence-specific primers (PCR-SSP) as previously described (25).
Animals and virus infections
Macaques were infected with the pathogenic molecular clone SIVmac239 (26) (GenBank accession M33262) with the exception of macaque r99006, which was infected with an SIVmac239 recombinant virus bearing escape mutations in three CD8+ T cell epitopes (27). SIV viremia was monitored in the infected macaques by quantitative PCR as previously described (6, 28).
Seven macaques at varying disease courses in the chronic phase of SIV infection (>35 weeks post-SIV infection) were utilized in this study. Four of the macaques were considered ECs with viral loads <1,000 vRNA copies/ml during this study (r00032, r02019, r98016, and r99006). The controller animal, r01027, maintained viral loads <20,000 vRNA copies/ml or a log less than the viral set point of typical progressor macaques (6), while macaque r91003 displayed a typical progressor disease course with viral loads ~0.5 × 106 vRNA copies/ml. Animal r00078 was previously an EC that had steadily increasing viral loads since one year after depletion of its CD8+ cells (28). At the time of the initial ELISPOT assays, the plasma virus concentrations in r00078 was ~30,000 vRNA copies/ml.
All SIV-infected animals were maintained at the National Primate Research Center (University of Wisconsin-Madison, Madison, WI) and cared for according to the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee.
Creation of stable MHC class I transfectants
A soluble Mamu-B*08 transfectant was produced in the MHC class I deficient EBV-transformed B-lymphoblastoid cell line 721.221. Briefly, a pcDNA 3.1− vector (Invitrogen, Carlsbad, CA) encoding soluble Mamu-B*08, was created by removing the cytoplasmic and membrane domains with PCR mutagenesis. This construct was then transfected into the 721.221 cell line (29). A previously created membrane-bound Mamu-B*08 transfectant was also utilized in this study (7).
Mamu-B*08 peptide-binding motif from endogenous ligands
Transfected 721.221 cells producing soluble Mamu-B*08 were cultured in a hollow fiber bioreactor (Toray, Tokyo, Japan) in a CP2500 unit (Biovest International, Minneapolis, MN), and supernatant containing soluble Mamu-B*08 was collected. Approximately 25 mg of soluble Mamu-B*08 was affinity purified from supernatant with W6/32 antibody (30) coupled to Sepharose 4B matrix (GE Healthcare Systems, Piscataway, NJ). Bound peptide was eluted from Mamu-B*08 heavy and light chains with an acid boil and passed through an ultra-filtration stirred cell with a 3 kDa membrane (Millipore, Bedford, Mass). Ten percent of the eluted peptide pool underwent 14 cycles of N-terminal Edman degradation sequencing on an ABI Procise 492 protein sequencer (Applied Biosystems, Foster City, CA). The relative dominance of amino acids at each position in the peptide pool was determined by calculating the fold increase in picomoles of an amino acid over the previous cycle. Amino acids exhibiting a fold increase of 2.0–2.49, 2.5–3.49, and ≥3.5, over the previous round are considered weak, strong, and dominant amino acids, respectively, at a given position in the peptide pool. The remaining peptide pool was fractionated by reversed-phase high-pressure liquid chromatography and sprayed via nanospray on an ESI-quadrupole time-of-flight Q-Star Elite mass spectrometer (Applied Biosystems, Foster City, CA). Approximately 40 peptide-containing fractions were generated. Individual peptide sequences were determined by MS/MS analysis. A peptide motif was compiled using resultant Edman sequencing data and individual peptide sequences (31).
Positional scanning combinatorial library and peptide synthesis
The positional scanning combinatorial library (PSCL) was synthesized as previously described (32). Each pool in the library contains 9-mer peptides with one fixed residue at a single position. With each of the 20 naturally occurring residues represented at each position along the 9-mer backbone, the entire library consisted of 180 peptide mixtures.
Peptides utilized in screening studies were purchased as crude or purified material from Mimotopes (Minneapolis, MN/Clayton, Victoria, Australia), Pepscan Systems B. V. (Lelystad, Netherlands), A and A Labs (San Diego, CA), Genescript Corporation (Piscataway, NJ), or the Biotechnology Center at the University of Wisconsin-Madison. Peptides synthesized for use as radiolabeled ligands were synthesized by A and A Labs and purified to >95% homogeneity by reverse phase HPLC. Purity of these peptides was determined using analytical reverse-phase HPLC and amino acid analysis, sequencing, and/or mass spectrometry. Peptides were radiolabeled with the chloramine T method (33). Lyophilized peptides were re-suspended at 4–20 mg/ml in 100% DMSO, then diluted to required concentrations in PBS +0.05% (v/v) nonidet P40 (Fluka Biochemika, Buchs, Switzerland).
SIV peptide sequences were derived from the SIVmac239 sequence, Genbank accession M33262 (26).
MHC purification and peptide binding assays
MHC class I purification was performed using affinity chromatography as previously described (33, 34). The Mamu-B*08 and Mamu-B*03 molecules were purified from cell lysates of stable membrane-bound MHC class I transfectants using the anti-HLA class I (-A, -B, and -C) antibody W6/32. HLA-B*2705 molecules were purified from the EBV transformed homozygous cell line LG2 using the HLA -B and -C antibody B123.2 (35–37). Protein purity, concentration, and depletion efficiency steps were monitored by SDS-PAGE.
Quantitative assays for peptide-binding to detergent solubilized MHC class I molecules were based on the competitive inhibition of binding of a high affinity radiolabeled standard probe peptide and performed as detailed in prior studies (33, 34, 37, 38). Peptides were tested at six different concentrations covering a 100,000-fold dose range in three or more independent assays. The radiolabeled peptides utilized for the Mamu-B*08, Mamu-B*03, and HLA-B*2705 assays were, respectively, peptide 3130.0006 (sequence RRDYRRGL, an N175 to Y analog of the SIV Vif172–179RL8 epitope), the artificial ligand 3130.0012 (sequence RRAARAEYL), and the human 60s rL28 38–46 peptide (sequence FRYNGLIHR). For each peptide, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was calculated. Under the conditions used, where [radiolabeled probe] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (39).
Control for MHC specificity is implicit by the fact that in each binding assay, only one species of purified MHC class I molecule is utilized. Control wells to measure non-specific (background) binding were also included. Specificity is further implied because not all peptides bound all three molecules, several peptides bound none of the three molecules, and each molecule tested bound a unique set of peptides. In each experiment, a titration of the unlabeled version of the radiolabeled probe was also tested as a positive control for inhibition.
Bioinformatic analysis
Analysis of the PSCL data was performed as described previously (40). Briefly, IC50 nM values for each mixture were standardized as a ratio to the geometric mean IC50 nM value of the entire set of 180 mixtures, and then normalized at each position so that the value associated with optimal binding at each position corresponds to 1. For each position, an average (geometric) relative binding affinity (ARB) was calculated, and then the ratio of the ARB for the entire library to the ARB for each position was derived. We have denominated this ratio, which describes the factor by which the normalized geometric average binding affinity associated with all 20 residues at a specified position differs from that of the average affinity of the entire library, as the specificity factor (SF). As calculated, positions with the highest specificity will have the highest SF value. Primary anchor positions were then defined as those with an SF ≥2.4. This criterion identifies positions where the majority of residues are associated with significant decreases in binding capacity.
Secondary anchor designations were based on the standard deviation (SD) of residue specific values at each position. Dominant secondary anchor positions were defined as those where the SD was >3 and the SF <2.4, as well as positions associated with an SD >2 if the SF is between 1.5 and 2.4. Weak secondary anchors have been defined as positions associated with a SD in the 2.5–3 range with an SF <1.5, or an SF in the 1.5–2.4 range with an SD <2.
To identify predicted binders, all possible 9-mer peptides in SIV sequences were scored using the matrix values derived from the PSCL analysis as described previously (40). The final score for each peptide represents the product of the corresponding matrix values for each peptide residue-position pair. Peptides of 8, 10, or 11 residues in length were also selected using the combinatorial library as described previously (41). Briefly, 8-mers were selected by scoring residues 1–7 with the corresponding residues in the 9-mer matrix, and the C-terminus was scored using the corresponding 9-mer C-terminal value. Similarly, for 10- and 11-mer peptides, residues 1–8 were scored using the corresponding residues of the 9-mer matrix, and the C-terminus with the 9-mer C-terminal residues. Peptides scoring amongst the top 2.5% (n = 82) for each size were selected. An additional set of 50 peptides representative of scores in each 10% range of the lower 97.5% was selected to evaluate the efficacy of the prediction approach.
IFN-γ Enzyme-Linked Immunospot (ELISPOT) assay
We performed ELISPOT assays as previously described (42). Briefly, PBMC were isolated from EDTA-anticoagulated blood using Ficoll-Paque PLUS (GE Healthcare Systems, Piscataway, NJ) and density centrifugation. 1 × 105 PBMC were used per well in precoated ELISpotPLUS kits (MABTECH Inc, Mariemont, OH) according to manufacturer’s instructions for the detection of IFN-γ secreting cells. All tests were performed in duplicate or triplicate using individual peptides at 10 μM. The positive control, Concanavalin A (Sigma, St. Louis, MO), was used at a final concentration of 5 μg/ml. The negative control wells were devoid of any stimulation. The 96-well plates were incubated for 12–18 hours at 37°C in 5% CO2.
Wells were imaged and counted with an AID EliSpot reader version 4.0 (AID, Strassberg, Germany) and analyzed as previously described (38, 42). A response was considered positive if the mean number of spot-forming cells (SFC) from the duplicate (or triplicate) sample wells exceeded background (mean of wells without peptide stimulation) plus two standard deviations (SD). Background levels were subtracted from each well, and assay results are shown as SFC per 1 × 106 PBMC. Responses of <50 SFC per 1 × 106 PBMC were not considered positive.
Two of the 210 peptides binding Mamu-B*08 ≤500 nM were unable to be tested in ELISPOT assays (Supplemental Table I). However, neither the Rev nor Nef peptide would be considered a novel response as both overlapped previously described Mamu-B*08-restricted epitopes by eight of nine amino acids.
Defining CD8+ T cell epitopes using peptide-specific CD8+ T cell lines
Previous methods used to define Mamu-B*08-restricted SIV epitopes were also utilized in this investigation (7). First, we generated peptide-specific CD8+ T cell lines. After several weeks of in vitro culture to increase specificity, we examined these cell lines in IFN-γ/TNF-α intracellular cytokine staining (ICS) assays with Mamu-B*08 transfectants to verify that immunogenic peptides were restricted by Mamu-B*08. These peptide-specific CD8+ T cell lines were used in similar ICS assays to confirm the minimal optimal epitope for each viral region. CD8+ T cell lines were cultured with media containing 100 Units of interleukin-2/ml (NIH AIDS Research and Reference Reagent Program, Germantown, MD).
MHC class I tetramer and surface staining
MHC class I tetramers were constructed for the six novel SIV epitopes with minor modifications as previously described (38, 43). We performed MHC class I tetramer stains on PBMC and CD8+ T cell lines to verify minimal optimal epitopes as previously described (38, 42).
Sequencing of plasma viral RNA (vRNA)
SIVmac239 sequencing data was obtained from other studies involving SIV-infected Mamu-B*08-positive macaques (7, 21). Viral sequencing was performed based on methods previously described (7, 21, 28, 44).
RESULTS
Sequencing of endogenously bound Mamu-B*08 ligands indicates peptide binding similarity to HLA-B27
With the emergence of SIV-infected Mamu-B*08-positive rhesus macaques as a model to study elite control of HIV replication, we decided to characterize a detailed peptide-binding motif of Mamu-B*08. We first defined a preliminary motif for Mamu-B*08 using an endogenous Mamu-B*08 peptide pool purified from Mamu-B*08 transfectants. By subjecting ten percent of the eluted Mamu-B*08 peptide pool to 14 cycles of N-terminal Edman degradation, we determined the dominance of particular amino acids at each position in the peptide pool. The pool sequencing analysis identified strong signals for R in position 2 and L at the C-terminus (data not shown). We then fractioned the remaining peptide pool by reverse-phase HPLC into approximately 40 peptide-containing fractions. From the various fractions spanning the HPLC chromatogram, ten Mamu-B*08 ligands were selected and sequenced by MS/MS fragmentation (Table I). In agreement with the binding motif determined by Edman sequencing, all ten peptides contained an R at position 2, and eight of the ten possessed L at the C-terminus. The majority (8/10) of the peptides sequenced were nonamers, while the remaining two ligands were decamers.
Table I.
Representative Mamu-B*08 endogenous ligand sequences also bind Mamu-B*03 and HLA-B*2705
| Source protein | Length | Sequencea | MHC class I binding affinity (IC50 nM)b | ||
|---|---|---|---|---|---|
| Mamu-B*08 | Mamu-B*03 | HLA B*2705 | |||
| Vacuolar protein pump subunit SFD alpha domain | 9 | RRYNIIPVL | 1.7 | 11 | 7.9 |
| RTP801 | 10 | SRLWPKIQGL | 6.7 | 100 | 17 |
| Fibronectin receptor alpha subunit | 10 | RRPPLLPLLL | 9.7 | 11 | 100 |
| SEC23B protein | 9 | RRSPFLQVF | 11 | 43 | 14 |
| Proteosome Subunit B2 | 9 | SRTPYHVNL | 15 | 22 | 22 |
| Peroxisome biogenesis factor 1 | 9 | SRLEILNVL | 16 | 275 | 80 |
| Ribosomal protein S15a | 9 | SRQFGFIVL | 16 | 22 | 128 |
| Tektin 4 | 9 | TRYPTILQL | 17 | 66 | 97 |
| Ribosomal protein S25 | 9 | ARAALQELL | 106 | 135 | 137 |
| Non-lens beta gamma crystallin-like protein | 9 | SRYDPSISF | 165 | 2438 | 254 |
Dominant residues from pool sequencing are indicated in bold.
IC50 values >500 nM are not considered binders and are indicated in italics.
We also tested the panel of sequenced ligands for their capacity to bind to Mamu-B*08. The ten endogenous peptides bound Mamu-B*08 with IC50 values ≤165 nM, including eight that bound with much higher affinities of 17 nM or better (Table I). All ten of these natural ligands also bound HLA-B*2705 with affinities ≤500 nM, indicating that the binding similarities of Mamu-B*08 and HLA-B*2705 extend beyond epitope sequence similarity.
Determination of Mamu-B*08 peptide-binding motif using a positional scanning combinatorial peptide library
To complement the pool sequencing analysis and derive a more detailed quantitative motif for Mamu-B*08, we next tested the capacity of a positional scanning combinatorial library (PSCL) to bind purified Mamu-B*08 molecules. The Mamu-B*08 binding capacity (IC50 nM) for each mixture was determined, and then normalized, as described in the Materials and Methods, to generate the resulting Mamu-B*08 matrix shown in Table II. Analysis of the matrix using previous established criteria (40) confirmed position 2 and the C-terminus as the main anchor positions for Mamu-B*08 binding. The second position was especially critical for binding with a specificity factor (SF) of 28.4.
Table II.
Positional scanning combinatorial library (PSCL) derived matrix describing 9-mer binding to Mamu-B*08a
| Position |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| A | 0.176 | 0.004 | 0.604 | 0.446 | 0.451 | 0.067 | 0.265 | 0.296 | 0.049 |
| C | 0.258 | 0.003 | 0.454 | 0.376 | 0.426 | 0.176 | 0.188 | 0.284 | 0.018 |
| D | 0.034 | 0.003 | 0.118 | 1.0 | 0.2 | 0.065 | 0.071 | 0.16 | 0.015 |
| E | 0.034 | 0.003 | 0.39 | 0.39 | 0.286 | 0.131 | 0.214 | 0.242 | 0.033 |
| F | 0.197 | 0.003 | 0.861 | 0.58 | 1.0 | 1.0 | 0.245 | 0.378 | 1.0 |
| G | 0.281 | 0.003 | 0.246 | 0.449 | 0.347 | 0.136 | 0.229 | 0.39 | 0.031 |
| H | 0.518 | 0.003 | 0.281 | 0.451 | 0.466 | 0.097 | 0.279 | 1.0 | 0.021 |
| I | 0.182 | 0.003 | 0.686 | 0.451 | 0.41 | 0.174 | 0.362 | 0.379 | 0.375 |
| K | 0.394 | 0.003 | 0.685 | 0.523 | 0.288 | 0.079 | 0.153 | 0.302 | 0.118 |
| L | 0.27 | 0.003 | 0.672 | 0.41 | 0.27 | 0.186 | 0.23 | 0.527 | 0.673 |
| M | 0.375 | 0.010 | 1.0 | 0.431 | 0.089 | 0.188 | 0.144 | 0.378 | 0.107 |
| N | 0.076 | 0.003 | 0.445 | 0.502 | 0.305 | 0.189 | 0.307 | 0.048 | 0.031 |
| P | 0.034 | 0.003 | 0.38 | 0.541 | 0.383 | 0.079 | 1.0 | 0.349 | 0.024 |
| Q | 0.034 | 0.037 | 0.829 | 0.572 | 0.264 | 0.151 | 0.5 | 0.318 | 0.033 |
| R | 1.0 | 1.0 | 0.587 | 0.439 | 0.367 | 0.231 | 0.425 | 0.445 | 0.089 |
| S | 0.312 | 0.008 | 0.469 | 0.434 | 0.311 | 0.161 | 0.212 | 0.387 | 0.023 |
| T | 0.111 | 0.003 | 0.584 | 0.731 | 0.253 | 0.238 | 0.221 | 0.285 | 0.014 |
| V | 0.106 | 0.003 | 0.335 | 0.405 | 0.266 | 0.145 | 0.168 | 0.398 | 0.079 |
| W | 0.067 | 0.003 | 0.749 | 0.562 | 0.173 | 0.235 | 0.13 | 0.274 | 0.010 |
| Y | 0.17 | 0.003 | 0.91 | 0.51 | 0.376 | 0.163 | 0.254 | 0.448 | 0.032 |
|
| |||||||||
| Average | 0.152 | 0.005 | 0.506 | 0.496 | 0.312 | 0.156 | 0.239 | 0.322 | 0.051 |
| SD | 2.7 | 4.0 | 1.68 | 1.26 | 1.60 | 1.82 | 1.73 | 1.77 | 3.62 |
| SF | 0.989 | 28.4 | 0.296 | 0.302 | 0.48 | 0.963 | 0.627 | 0.466 | 2.93 |
The PSCL was tested for binding to Mamu-B*08, the data analyzed, and primary and secondary anchor positions defined, as described in the Materials and Methods. Values shown represent the average relative binding (ARB) of the corresponding library relative to other pools with the same fixed position. SD indicates the standard deviation between the ARB of pools at the same position. SF is the specificity factor, calculated as described in the Materials and Methods, representing the ratio of the average binding of the entire library to the average of pools at the indicated position. At the primary anchor positions (SF ≥2.4), the most preferred residues, associated with an ARB >0.1 are highlighted by bold font. The library average binding for Mamu-B*08 was 0.150.
In agreement with the pool sequencing derived motif, at position 2 only R was preferred (Table II). All other residues were associated with normalized average relative binding (ARB) values <0.05. At the C-terminus, a broader range of residues was permitted. The PSCL analysis identified the aromatic residue F as the most preferred followed by the hydrophobic aliphatic residues L and I with ARB values of 0.673 and 0.375, respectively. The basic residue K and the hydrophobic residue M were tolerated with ARB values in the 0.1–0.12 range. All other residues were associated with affinities more than 10-fold lower than the optimal residue, F.
Influences on binding at other positions tended to be minor, reflected by relatively low SF and standard deviations (SD). In fact, none of the positions met the criterion we have utilized to define dominant secondary anchors (see (40) and the Materials and Methods), and only position 1, associated with a SD of 2.7, appears to be a weak secondary anchor. In position 1, the basic residues R, H, and K, as well as M and S, were the most preferred, while the acidic residues D and E were two of the most deleterious.
In summary, we used the PSCL to define a detailed motif for Mamu-B*08. This motif was characterized by primary anchor specificity for the basic residue R in position 2 and a preference for hydrophobic (F, L, I, and M) and basic (K) residues at the C-terminus (Figure 2).
Figure 2.

Similarity of primary and secondary anchor positions between Mamu-B*08, Mamu-B*03, and HLA-B*2705. Primary and secondary anchor positions were defined using the PSCL matrix data (Table II and Supplemental Table II), as described in the Materials and Methods. Primary anchor positions are identified by bold font and blue shading. Dominant secondary positions are indicated by bold font and green shading, while weak secondary positions are highlighted by orange shading. Also shown are the most preferred residues at the corresponding position. In the case of the primary anchors, residues associated with an ARB >0.1 are shown. At secondary positions, residues with an ARB >0.3 are shown.
Identification of SIV-derived Mamu-B*08 binding peptides
PSCL-based matrices offer an efficient means to identify MHC class I binding peptides and the ability to identify non-motif binders (40, 41, 45–49). To identify SIV-derived Mamu-B*08 binders, we utilized the Mamu-B*08 PSCL matrix to score all 9-mer peptides in the SIVmac239 proteome. After synthesizing the 9-mer peptides scoring in the upper 2.5% range (n=82), we tested them for binding. An additional set of 50 lower scoring peptides, randomly selected to provide five peptides to represent each successive 10% scoring range, were also synthesized and tested. All of the SIV peptides and their accompanying binding data can be found in Supplemental Table I. We discovered that 49 (60%) of the 82 peptides selected bound Mamu-B*08 with an affinity of ≤500 nM, including 16 peptides that bound with affinities of 10 nM or better. By contrast, none of the 50 peptides with scores beyond the upper 2.5% bound Mamu-B*08 with affinities ≤500 nM.
Both the endogenous ligands (Table I) as well as the previously described Mamu-B*08-restricted SIV epitopes (7) demonstrated that Mamu-B*08 is not restricted to binding only nonamers. Thus, we applied the PSCL binding matrix, as described in the Materials and Methods, to predict SIV-derived 8-mer, 10-mer, and 11-mer binders. For each size (8-, 10-, and 11-mer), the upper 2.5% scoring peptides (n=82 for each size), plus an additional set of 50 peptides from poorer scoring ranges, were synthesized and tested for binding. In total 35 (42.7%), 58 (70.7%), and 37 (45.1%) of the 8-, 10-, and 11-mer peptide sets, respectively bound with affinities of ≤500 nM. As with the 9-mer peptide set, none of the peptides from the lower scoring ranges bound with affinities ≤500 nM. Overall, considering all four size sets together, 179 of the 328 (54.6%) peptides scoring in the top 2.5% range were Mamu-B*08 binders. In the course of subsequent analyses probing lower scoring ranges, we found several additional Mamu-B*08 binders. In total, we identified 210 SIV-derived Mamu-B*08 binders (IC50 ≤500 nM) from the 862 SIV-derived peptide candidates (Supplemental Table I).
Remarkable overlap in the peptide binding repertoires of Mamu-B*08 in comparison with HLA-B*2705
Given the similarities between epitopes restricted by Mamu-B*08, Mamu-B*03, and HLA-B*2705, we next sought to compare their respective PSCL-derived motifs, as well as investigate the extent, if any, that their repertoires overlapped. For this comparison, we created a PSCL matrix describing Mamu-B*03 binding (Supplemental Table II-A). We also utilized the previously reported PSCL matrix-based motif for HLA-B*2705 (40)(Supplemental Table II-B).
The PSCL binding matrices indicated that Mamu-B*08, Mamu-B*03, and HLA-B*2705 share very similar primary anchor specificities. Position 2 was identified as the dominant anchor position for all three MHC class I molecules with an overwhelming preference for the positive residue R (Figure 2). Unlike Mamu-B*08, a second main anchor position was not identified for either Mamu-B*03 or HLA-B*2705. Instead, the C-terminus of Mamu-B*03 was defined as a dominant secondary anchor. In this position, the most preferred residues were L, F, and I, representing a specificity pattern very similar to that of Mamu-B*08. Position 1 was also identified as a dominant secondary anchor and comparable to the Mamu-B*08 specificity, a preference for basic residues was noted. For HLA-B*2705, positions 1, 3, and the C-terminus were identified as weak secondary anchor positions. At the C-terminus, the most preferred residues for HLA-B*2705 were F, R, K, L, I, V, and M, representing an overall pattern that resembles Mamu-B*08 (and Mamu-B*03) specificity, as well as previously published motifs for HLA-B*2705 (8–11). Comparison of the specificity at the N-terminus (P1) also revealed similarity to the preferences observed for Mamu-B*08 and -B*03 (Figure 2).
To determine if these similar motifs would translate into overlapping binding repertoires, we assessed the binding patterns using 899 peptides. The 899 peptides tested included the SIV-derived peptides (Supplemental Table I), the Mamu-B*08 endogenous ligands (Table I), and a number of known HLA-B27-restricted T cell epitopes and endogenous ligands described in the literature (15, 50–54) (Table III). We evaluated each peptide for its capacity to bind Mamu-B*08, Mamu-B*03, and HLA-B*2705 (Table IV). Of the 899 peptides, 363 (40.4%) bound at least one of the three MHC class I molecules with an IC50 ≤500 nM. The degree of cross-reactivity between binders (IC50 ≤500 nM) was exceptionally high, as 241 (66.4%) bound more than one of the MHC class I molecules tested, including 167 (46%) that bound all three molecules. We also discovered the cross-reactivity pattern to be fairly uniform across the three MHC class I molecules. Specifically, between 63% and 85% of the repertoire of any allele overlapped with the repertoire of another allele. These levels of overlap are remarkable, and even higher than those typically seen among alleles within the same HLA supertypes (34, 55–58). Correspondingly, monospecificity was low with only 7% of the Mamu-B*08 binders, 14% of the Mamu-B*03 binders, and 25% of the HLA-B*2705 binders failing to bind to another of the three MHC class I molecules.
Table III.
Known HLA-B27 T cell epitopes and endogenous ligands also bind to Mamu-B*08 and Mamu-B*03
| Organism (source protein) | Length | Sequence | MHC class I binding affinity (IC50 nM)a | # of alleles bound | Reference | ||
|---|---|---|---|---|---|---|---|
| HLA-B*2705 | Mamu-B*08 | Mamu-B*03 | |||||
| HIV (Gag p24) | 10 | RRWIQLGLQK | 0.0027 | 14 | 12 | 3 | 15 |
| HIV (Gag p24) | 10 | KRWIILGLNK | 5.4 | 25 | 14 | 3 | 15 |
| endogenous HLA-B*2705 ligand (ATP-dependent RNA helicase) | 9 | RRSKEITVR | 5.5 | 2698 | 2622 | 1 | 50 |
| C. trachomatis (efflux protein) | 9 | YRLLLTRVL | 5.6 | 5.9 | 14 | 3 | 52 |
| Influenza (NP) | 9 | SRYWAIRTR | 6.0 | 82 | 347 | 3 | 54 |
| endogenous HLA-B*2705 ligand (Histon H3.3) | 9 | RRYQKSTEL | 6.2 | 4.1 | 6.1 | 3 | 50 |
| EBV (EBNA 3C) | 9 | RRIYDLIEL | 6.6 | 7.1 | 5.7 | 3 | 54 |
| C. trachomatis (putative outer membrane protein) | 9 | NRFSVAYML | 9.8 | 444 | 2865 | 2 | 52 |
| endogenous HLA-B*2705 ligand (26S proteasome regulatory subunit S2) | 9 | SRFPEALRL | 12 | 8.4 | 65 | 3 | 53 |
| C. trachomatis (hypothetical protein) | 9 | IRMFKILPL | 13 | 7.4 | 12 | 3 | 52 |
| endogenous HLA-B*2705 ligand (60S ribosomal protein L28) | 9 | FRYNGLIHR | 18 | 799 | 3512 | 1 | 50 |
| endogenous HLA-B*2705 ligand (NADH-ubiquinone oxidoreductase) | 9 | RRISGVDRY | 21 | 403 | 148 | 3 | 50 |
| C. trachomatis (invasin repeat-family-phosphatase) | 9 | IRSSVQNKL | 37 | 109 | 462 | 3 | 52 |
| endogenous HLA-B*2705 ligand (Aggrecan) | 9 | SRHHAFCFR | 42 | 379 | 1345 | 2 | 54 |
| HIV (Gag p17) | 9 | IRLRPGGKK | 45 | 2199 | 9358 | 1 | 15 |
| C. trachomatis (NADH-ubiquinone-oxidoreductase-α-chain) | 9 | MRDHTITLL | 52 | 91 | 380 | 3 | 52 |
| C. trachomatis (hypothetical protein) | 9 | ARKLLLDNL | 74 | 316 | 7004 | 2 | 52, 54 |
| HIV (Nef) | 10 | RRQDILDLWI | 99 | 46 | 53 | 3 | 15 |
| HIV (Env gp41) | 10 | GRRGWEALKY | 129 | 1879 | 6503 | 1 | 15 |
| endogenous HLA-B*2705 ligand (60S ribosomal protein L13) | 9 | ARLFGIRAK | 133 | 6.7 | 148 | 3 | 50 |
| C. trachomatis (hypothetical protein) | 9 | KRLAETLAL | 165 | 55 | 29 | 3 | 52 |
| C. trachomatis (exodeoxyribonuclease-V γ-chain) | 9 | DRLALLANL | 181 | 110 | 871 | 2 | 52 |
| C. trachomatis (protease-ATPase) | 9 | NRAKQVIKL | 324 | 282 | 524 | 2 | 52 |
| Y. enterocolitica (Hsp 60-kDa) | 9 | KRVVINKDT | 406 | 12467 | 1055 | 1 | 51 |
| C. trachomatis (hypothetical protein) | 9 | ERFLAQEQL | 728 | 3760 | 5839 | 0 | 52 |
| C. trachomatis (ATP-dependent zinc protein) | 9 | EREQTLNQL | 2694 | 1747 | 10195 | 0 | 52 |
| C. trachomatis (hypothetical protein) | 9 | NRELIQQEL | 5789 | 54 | 738 | 1 | 52 |
IC50 values >500 nM are not considered binders and are indicated in italics.
Table IV.
Mamu-B*08, Mamu-B*03, and HLA-B*2705 exhibit remarkable overlap in their peptide-binding repertoiresa
| MHC class I molecules | Bindersb | % of binders that are cross-reactive with an IC50 ≤500 nM |
||||
|---|---|---|---|---|---|---|
| Mamu-B*08 | Mamu-B*03 | HLA-B*2705 | Both | Neither | ||
| Mamu-B*08 | 240 | - | 78.3 | 84.6 | 69.6 | 6.7 |
| Mamu-B*03 | 239 | 78.7 | - | 77 | 69.9 | 14.2 |
| HLA-B*2705 | 292 | 69.5 | 63 | - | 57.2 | 24.7 |
Binding capacity to Mamu-B*08, Mamu-B*03, and HLA-B*2705 was tested for 899 peptides (10 Mamu-B*08 endogenous ligands from Table I, 27 HLA-B27-restricted T cell epitopes and endogenous ligands from Table III, and 862 SIV-derived peptides from Supplemental Table I).
Binders were defined as peptides with IC50 values <500 nM.
Of the previously reported HLA-B27 epitopes, we found that 19 of the 24 peptides that bound HLA-B*2705 with an affinity ≤500 nM also bound Mamu-B*08 (Table III). Fourteen of these epitopes bound both Mamu-B*03 and -B*08, again demonstrating the similar peptide binding specificity between Mamu-B*08, Mamu-B*03, and HLA-B*2705. Overall, this shared specificity is reflected in an extraordinary degree of repertoire overlap between the three MHC class I molecules.
Defining six novel SIV epitopes restricted by Mamu-B*08
The 500 nM threshold has been used to define candidate epitopes in previous SIV epitope screens (38, 59–62) and has also been shown to be associated with in vivo T cell recognition in human, macaque, and murine systems (39, 63–65). On this basis, we examined whether the SIV-derived Mamu-B*08-binding peptides identified in the binding studies above were recognized in vivo using fresh PBMC from SIV-infected Mamu-B*08-positive macaques. Using IFN-γ ELISPOT assays, we tested seven macaques at varying disease courses in the chronic phase of SIV infection (>35 weeks post-SIV infection). No Mamu-B*03-positive macaques were available for SIV immunogenicity testing due to the low frequency (<1%) of this allele in Indian rhesus macaque colonies (25).
We evaluated the antigenicity of 208 SIV-derived peptides in duplicate wells of two independent experiments. Seventy-five of these peptides (36%) demonstrated functional reactivity in at least one of the seven SIV-infected macaques (Supplemental Table I). We also assessed two uninfected Mamu-B*08-positive macaques in IFN-γ ELISPOT assays. None of the peptides demonstrated reactivity in either of these control animals (data not shown).
We found a number of responses against previously characterized Mamu-B*08 epitopes in several macaques. Responses against Vif123–131RL9, Vif172–179RL8, Rev12–20KL9 and Nef137–146RL10 were the most prevalent and detected in at least five of the seven SIV-infected macaques (Figure 3). The Nef137–146RL10-specific CD8+ T cell response was typically one of the strongest responses and exceeded 800 SFC/106 PBMC in three macaques in agreement with previous data (7). In comparison, Rev44–51RL8, Nef8–16RL9, and Nef246–254RL9 appeared subdominant during the chronic phase of SIV infection. We detected these responses in less than half of the macaques and at much lower levels, typically <200 SFC/106 PBMC.
Figure 3.

Ex vivo whole PBMC IFN-γ ELISPOT results of Mamu-B*08-restricted minimal optimal CD8+ T-cell epitopes. Freshly isolated PBMC from seven Mamu-B*08-positive SIV-infected macaques were tested with Mamu-B*08 binding peptides in IFN-γ ELISPOT assays. Animals were tested during the chronic phase of SIV infection (>35 weeks post-SIV infection). Results shown represent the 13 SIVmac239 epitopes known to be restricted by Mamu-B*08. Mean values and SDs from triplicate wells were calculated for each assay. Background (the mean of wells without peptide) levels were subtracted from each well. Mean responses <50 SFC per 1 × 106 PBMC were not considered positive. Assay results are shown as SFC per 1 × 106 PBMC. Nef RL9* refers to the epitope at positions 8–16 in Nef, while Nef RL9 refers to the Nef epitope at positions 246–254. The SDs of Nef RL10 in r02019 (1,522 SFC/106 PBMC) and Nef RL10 in r99006 (1,753 SFC/106 PBMC) were 197 and 140, respectively.
Several responses were also directed against viral regions that did not contain epitopes known to be restricted by Mamu-B*08, warranting further investigation. After excluding the peptides that had considerable overlap (at least six amino acids) with the seven previously defined Mamu-B*08-restricted epitopes, 14 immunogenic viral regions, located in Gag, Pol, Vpx, Vpr, Env, and Nef, remained. We generated CD8+ T cell lines against 13 of the 14 novel regions. A peptide-specific CD8+ T cell line could not be generated against Nef16–24LR9, likely due to its low frequency in a small number of macaques (data not shown).
Using the peptide-specific CD8+ T cell lines in conjunction with Mamu-B*08 transfectants in IFN-γ/TNF-α ICS assays, we established that six of the fourteen regions with reactivity in ELISPOT assays were CD8+ T cell responses restricted by Mamu-B*08. Given that Indian rhesus macaques express upwards of 12 or more MHC class I alleles (22, 66), it was not surprising that several of the novel responses detected in ELISPOT assays were restricted by other MHC class I molecules. We then used the Mamu-B*08-restricted, peptide-specific CD8+ T cell lines to identify the minimal optimal epitopes for cases where more than one peptide was reactive for that region. Minimal confirmation occurred by testing the functional avidity on peptide-specific CD8+ T cell lines and constructing MHC class I tetramers (data not shown). As with the previous seven epitopes, all six novel responses contained an R at position 2. Of the six novel epitopes, one was located in Gag (Gag263–271YL9), one in Vpr (Vpr62–70IF9), and four in Env (Env524–532KF9, Env573–581KL9, Env717–725LF9, and Env868–876RL9). Like the Nef137–146RL10 epitope, the Env573–581KL9 epitope was previously identified in Mamu-B*03-positive SIV-infected macaques (23, 67).
Similar to the previously identified SIV epitopes, the newly identified epitopes bound with extremely high affinity to Mamu-B*08. Overall, 10 of the 13 Mamu-B*08-restricted epitopes bound Mamu-B*08 with affinities ≤50 nM (Table V). Interestingly, all 13 Mamu B*08 epitopes identified to date were amongst the top 0.58% scoring peptides corresponding to their size in the PSCL matrix based predictions described above. For instance, all ten of the SIV 9-mer peptides restricted by Mamu-B*08 would have been found by generating just 20 of the ~3,330 possible 9-mers. Of the 13 Mamu-B*08-restricted epitopes, nine and ten peptides, respectively, were found to bind HLA-B*2705 and Mamu-B*03 with affinities ≤50 nM. Overall, only one peptide failed to bind all three MHC class I molecules with an IC50 ≤ 500 nM.
Table V.
SIV epitopes restricted by Mamu-B*08 bind both macaque and human MHC class I molecules
| Protein | Amino acid positions | Length | Sequence | Short namea | MHC class I binding affinity (IC50 nM)b | ||
|---|---|---|---|---|---|---|---|
| Mamu-B*08 | Mamu-B*03 | HLA-B*2705 | |||||
| Gag | 263–271 | 9 | YRRWIQLGL | YL9 | 1.5 | 14 | 2.7 |
| Nef | 246–254 | 9 | RRLTARGLL | RL9 | 3.2 | 8.0 | 9.9 |
| Rev | 12–20 | 9 | KRLRLIHLL | KL9 | 3.2 | 12 | 1.1 |
| Env | 868–876 | 9 | RRIRQGLEL | RL9 | 5.5 | 7.9 | 13 |
| Vpr | 62–70 | 9 | IRILQRALF | IF9 | 6.5 | 171 | 14 |
| Env | 524–532 | 9 | KRGVFVLGF | KF9 | 7.2 | 32 | 13 |
| Vif | 123–131 | 9 | RRAIRGEQL | RL9 | 7.5 | 15 | 28 |
| Nef | 137–146 | 10 | RRHRILDIYL | RL10 | 11 | 9.6 | 1.8 |
| Env | 573–581 | 9 | KRQQELLRL | KL9 | 12 | 9.7 | 69 |
| Rev | 44–51 | 8 | RRRWQQLL | RL8 | 20 | 4.6 | 12 |
| Nef | 8–16 | 9 | RRSRPSGDL | RL9 | 105 | 22 | 351 |
| Vif | 172–179 | 8 | RRDNRRGL | RL8 | 125 | 523 | 175 |
| Env | 717–725 | 9 | LRQGYRPVF | LF9 | 217 | 114 | 52 |
Novel epitopes identified in this study are indicated in bold.
IC50 values >500 nM are not considered binders and are indicated in italics.
In comparison to responses directed against Vif123–131RL9, Vif172–179RL8, Rev12–20KL9 and Nef137–146RL10, the six novel responses appeared to be subdominant during the chronic phase of SIV infection (Figure 3). We detected these responses in up to three animals and typically at lower magnitudes. Interestingly, the SIV epitope in Gag p27 (Gag263–271YL9) overlaps with an immunodominant CD8+ T cell epitope found in HLA-B*27-positive humans infected with HIV-1 (Gag263–272KK10) and HIV-2 (Gag263–272RK10) (68–70). However unlike the immunodominance of this Gag epitope observed in HIV-infected individuals (13, 15, 18–20), we found Gag263–271YL9-specific CD8+ T cells in only two of the seven SIV-infected animals (r99006 and r00078) at frequencies of 80 and 305 SFC/106 PBMC, respectively (Figure 3).
Impact of viral variation in Mamu-B*08-restricted CD8+ T cell epitopes on Mamu-B*08 binding capacity
Previously, it has been shown that Mamu-B*08-specific CD8+ T cells selected for viral variation in several SIV epitopes (7, 21). We, therefore, tested the Mamu-B*08 binding capacity of a number of in vivo viral variants to determine if they reduced the MHC class I peptide binding to Mamu-B*08 (Table VI).
Table VI.
Binding capacity of SIV variants within Mamu-B*08-restricted epitopes
| Mamu-B*08-restricted SIV epitopes | Amino acid sequence | Mamu-B*08 binding affinity (IC50 nM)a | Fold reduction relative to wildtype |
|---|---|---|---|
| Vif123–131RL9 | RRAIRGEQL | 7.5 | - |
| ...V.....b | 36 | 4.9 | |
| ..V....R.c | 63 | 8.4 | |
| ..VV.....c | 100 | 13 | |
| ..V....R.c | 125 | 17 | |
| ..V......c, d | 158 | 21 | |
| T........e | 517 | 69 | |
| T.......Qc | 1474 | 197 | |
| ........Qb | 1517 | 203 | |
| .K.......b | 2901 | 387 | |
| .KV......c | 3325 | 444 | |
|
| |||
| Vif172–179RL8 | RRDNRRGL | 125 | - |
| .......Fc | 167 | 1.3 | |
| G.......c, d | 325 | 2.6 | |
| .....G.Fc | 557 | 4.5 | |
| .G......c | 594 | 4.8 | |
| G......Pc | 38985 | 313 | |
| GG......c | >70000 | >560 | |
|
| |||
| Rev12–20KL9 | KRLRLIHLL | 3.2 | - |
| .....L...c | 1.6 | - | |
| .....V...e | 5.1 | 1.6 | |
| ..K......c | 14 | 4.5 | |
|
| |||
| Rev44–51RL8 | RRRWQQLL | 20 | - |
| ......I.c | 59 | 2.9 | |
| ....E...c | 127 | 6.3 | |
|
| |||
| Nef8–16RL9 | RRSRPSGDL | 105 | - |
| ....L....c | 148 | 1.4 | |
| ....Q....c | 428 | 4.1 | |
| K...Q....c | 502 | 4.8 | |
| K........c | 892 | 8.5 | |
|
| |||
| Nef137–146RL10f | RRHRILDIYL | 11 | - |
| .......T..c, d | 11 | - | |
| G.........e | 107 | 9.7 | |
|
| |||
| Nef246–254RL9 | RRLTARGLL | 3.2 | - |
| ...A...I.c | 2.2 | - | |
| K........c | 4.8 | 1.5 | |
| .......I.e | 5.0 | 1.6 | |
| K..V.....c | 6.2 | 2.0 | |
| K..A.....c | 10 | 3.0 | |
| ...A.....b | 11 | 3.4 | |
| ........Ib | 22 | 6.8 | |
| .......IIc | 77 | 24 | |
| E........c | 172 | 54 | |
| E..A.....c | 195 | 62 | |
| ........Pd | 477 | 151 | |
|
| |||
| Env524–532KF9 | KRGVFVLGF | 7.2 | - |
| ........Ye | 321 | 44 | |
|
| |||
| Env573–581KL9 | KRQQELLRL | 12 | - |
| ...H.....g | 71 | 5.9 | |
| ........Mg | 397 | 33 | |
Peptides with IC50 values >500 nM are not considered binders and are indicated in italics.
Variant not found in vivo at time of this submission.
Variant found in vivo during the chronic phase of SIV infection (7).
Variant found in vivo ≤18 weeks post SIV-infection (21).
Previously unpublished viral variant (Loffredo et al.).
Predominant viral variation is one amino acid upstream of this epitope, position 136: A to P (7, 21).
Previously published variant in Mamu-B*03-positive macaques escape (23).
In general, the variant peptides studied were associated with substantial decreases in Mamu-B*08 binding capacity, with only a few having affinities within three-fold of the wild type epitope. Many of these peptides did not exhibit variation at position 2 and the C-terminus, the primary anchor positions for Mamu-B*08 binding, and bound with a biologically relevant affinity of ≤500 nM (Table VI). When variation was identified at the primary anchor positions, these peptides typically bound weakly, if at all, with the majority having affinities >500 nM. We tested a number of peptides with variation in the primary anchor positions in IFN-γ ELISPOT assays. In comparison to the wildtype peptides, we found the variants to be greatly reduced in magnitude, and in some instances, no reactivity was observed (data not shown). However, ELISPOT is limited in its ability to test CD8+ T cell recognition of viral variants (71, 72). In the future, more extensive testing will be necessary to thoroughly examine the impact of these SIV variants on cellular recognition.
While variation was found in the Nef137–146RL10 epitope, these mutations did not greatly affect the binding capacity. In this case, as previously reported (7), an alanine to proline substitution at the residue immediately preceding the N-terminus of the Nef137–146RL10 epitope appears to affect peptide processing. A similar alanine to proline substitution immediately preceding the N-terminal residue of the epitope has also been shown to inhibit processing of an HLA-B57-restricted peptide in Gag of HIV (73).
The majority of the previously defined epitopes exhibited a wide range of viral variation. However, at the time of this study, we detected viral variation in only one of the newly discovered epitopes, Env524–532KF9 (Table VI). In this case, an F to Y substitution at the C-terminus of Env524–532KF9 reduced binding over 40-fold from 7.2 to 321 nM. The lack of viral variation was particularly surprising for the Env573–581KL9 epitope. Viral escape from Env573–581KL9-specific CD8+ T lymphocytes was previously found in Mamu-B*03-positive SIV-infected macaques during the chronic phase of SIV infection (23). However, we observed no such variation in our SIV-infected Mamu-B*08-positive infected macaques, despite detecting Env573–581KL9-specific CD8+ T cells in three of the seven macaques tested during the chronic phase of infection. Two of these responses were >600 SFC/106 PBMC (Figure 3).
DISCUSSION
Characteristics of a successful immune response against HIV remain unknown. Investigating MHC class I alleles that are implicated in slow disease progression may provide some insight. HLA-B27 is one such allele that has been associated with elite control of HIV infection (12–17). Recently, it has been established that Mamu-B*08-positive macaques control SIVmac239 replication in >50% of infected animals (6). Furthermore, Mamu-B*08-restricted SIV epitopes fit the HLA-B27 peptide binding motif (7). We, therefore, sought to investigate the binding similarities between Mamu-B*08 and HLA-B27.
In the present study, we found a remarkably high degree of overlap in the peptide-binding repertoires of Mamu-B*08 and HLA-B*2705, as well as Mamu-B*03. The considerable overlap between Mamu-B*08 and Mamu-B*03 is not surprising given their high level of sequence similarity (22). These two alleles differ by only two residues in the α1 and α2 domains (Figure 1). However, the extent to which the binding specificity of the Mamu- alleles cross-react with HLA-B*2705 is very surprising even in light of previous observations (7, 24). HLA-B*2705 differs from Mamu-B*08 and Mamu-B*03 at four of the seven key residues predicted to form the B pocket, thought to determine the residue specificity at position 2 of peptide ligands (74–76). In addition, differences are apparent in key residues predicted to form the F pocket, believed to dictate the C-terminal peptide specificity. HLA-B*2705 differs from Mamu-B*08 at three of the four key F pocket residues. Overall, HLA-B*2705 differs from Mamu-B*08 and Mamu-B*03 at 28 and 29 residues, respectively, throughout the α1 and α2 domains (Figure 1).
Despite these sequence differences, the overlapping binding motifs and peptide repertoires of these alleles are astounding. The summary motifs exhibit the same narrow specificity at position 2, and partially overlapping, although very similar, preferences at the C-terminus (Figure 2). The fact that the motifs of each molecule are not identical is reflected in the observation that while the overlaps in the peptide-binding repertoires observed are unusually high, they are not absolute. Each molecule is associated with its own unique repertoire and specificity. However, when considered in the context of other HLA- and Mamu- MHC class I alleles whose binding specificities have been characterized, the parallels between the molecules studied herein remain remarkable. Mamu-B*08, Mamu-B*03, and HLA-B*2705 share a preference for hydrophobic residues at the C-terminus. In addition, Mamu-B*08 and HLA-B*2705 share a strong propensity for basic residues, which are also well tolerated by Mamu-B*03. This pattern of specificity is relatively rare amongst HLA class I alleles, being largely limited to those in the HLA-B27-supertype. At position 2, all three MHC class I molecules have an identical preference for R, a specificity that has not been widely observed amongst alleles whose motifs have been described in detail (8, 77). The rates of overlap in the peptide-binding repertoires, ranging between 63% and 85% depending on pairing (Table IV), are higher than those typically observed amongst alleles within an HLA supertype or even amongst very closely related alleles of the same serological family (34, 55–58). In a large fraction of cases, these molecules also bind the same peptides with very similar IC50 values (nM).
Binding cross-reactivity was evident not only in the case of SIV-derived peptides, but also when comparing the binding of known epitopes and ligands previously associated with HLA-B27 (15, 50–54) (Table III). Of the 24 selected sequences that bound HLA-B*2705 with an IC50 ≤500 nM, 19 of them bound Mamu-B*08. In addition, when searching the Immune Epitope Database (IEDB) (78) and SYFPEITHI databases (8), we found that two of the Mamu-B*08 endogenous ligands, RRYNIIPVL and SRTPYHVNL (Table I), were previously identified as HLA-B27 ligands. Such functional overlap between Mamu-B*08 and HLA-B27, in spite of substantial sequence diversity, may be best explained by convergent evolution (79).
Using a systematic epitope screening process similar to that used to investigate SIV epitopes restricted by Mamu-A*01, -A*02, -A*11, -B*01, and -B*17 (36, 38, 59–62), we identified 210 SIV-derived peptides that bound to Mamu-B*08 with an IC50 of ≤500 nM (Supplemental Table I). From these 210 peptides, 75 peptides, spanning 21 viral regions, elicited responses in IFN-γ ELISPOT assays. Seven of the 21 regions contained Mamu-B*08-restricted epitopes previously identified in SIV-infected EC macaques that were depleted of their CD8+ cells (28). An additional six regions contained novel epitopes restricted by Mamu-B*08. Interestingly, a majority of the thirteen Mamu-B*08-restricted epitopes had a number of overlapping peptides that bound Mamu-B*08 and were immunogenic in IFN-γ ELISPOT assays (Supplemental Table I). With the strong preference for R at position 2 and a number of consecutive arginines within these viral regions, it is possible that overlapping minimal optimal epitopes may be present in these areas too. It should also be noted that our approach might fail to detect CD8+ T lymphocyte responses with low binding affinities, as those with an IC50 >500 nM were not tested in IFN-γ ELISPOT assays.
In each of the seven SIV-infected macaques, we identified between four and twelve Mamu-B*08-restricted responses in the chronic phase of infection (Figure 3). CD8+ T cells directed against Vif123–131RL9, Vif172–179RL8, Rev12–20KL9 and Nef137–146RL10 typically represented the strongest and most frequent immune responses. The six novel SIV-specific epitopes, located in Gag, Vpr, and Env, appeared subdominant during the chronic phase of infection (Figure 3).
Gag263–271YL9 was the one of two regions containing an SIV epitope in which an HLA-B27-restricted HIV epitope has also been identified. The previously defined Mamu-B*08-restricted epitope Nef137–146RL10 also appears to share homology with the HIV epitope Nef105–114RI10 that is restricted by HLA-B*2705. Interestingly, the region containing the Gag epitope is completely conserved between SIVmac239 and HIV-2 and contains the immunodominant HLA-B*2705-restricted epitope Gag263–272KK10 in HIV-1 (13, 15, 18–20, 68–70). However, unlike in HIV-infected HLA-B27-positive humans, the seven Mamu-B*08-positive SIV-infected macaques rarely targeted this Gag epitope (Figure 3). Tetramer stains of both acute and chronic phase PBMC samples confirmed this finding (data not shown). Therefore, while Mamu-B*08 and HLA-B*2705 are functionally similar, the two MHC class I molecules largely target different epitopes in SIV and HIV, respectively.
This observation suggests that it may not be the epitopes themselves that contribute to the viral control associated with these alleles, but rather other as yet unidentified characteristics that they share. For instance, Mamu-B*08-restricted SIV epitopes, like HLA-B27-restricted epitopes, commonly display a characteristic dibasic peptide motif at the N-terminus. It has previously been shown that these peptides are relatively resistant to peptidase activity, are more stable in the cytosol, and hence are more efficiently presented by HLA-B27 (80). In the case of the 13 Mamu-B*08-restricted SIV epitopes, ten displayed this dibasic motif with either R or K at the N-terminus of the peptide. This suggests Mamu-B*08, like HLA-B27, may be able to present peptides from antigens when far fewer copies of the epitope are present and therefore may eliminate virally infected cells more efficiently.
We also investigated the impact of viral escape variants on Mamu-B*08 peptide binding. Overall, less than a third of viral variants identified in Mamu-B*08-restricted epitopes changed primary anchor residues, resulting in a drastic reduction in binding (Table VI). Variation at other positions typically did not abrogate binding to Mamu-B*08, suggesting that the observed viral variations more likely alters TCR engagement to disrupt recognition by CD8+ T cells. Interestingly, viral variation was evident at position 1 in five of the Mamu-B*08-restricted epitopes. Position 1 appears to play some role in MHC class I binding (Table II and Figure 2). However, the effect of these mutations may also be in the disruption of the dibasic peptide motifs discussed above, resulting in the alteration of peptide processing and/or stability.
The SIV-infected Indian rhesus macaque is the best animal model available for HIV/AIDS research. Mamu-B*08-positive macaques are especially important due to high frequency of animals that become ECs (>50%) and the striking functional similarity of this MHC class I molecule to the human EC allele, HLA-B27. This study provided a detailed comparison of three alleles associated with slow disease progression. In total, thirteen Mamu-B*08-restricted responses, located in Gag, Vpr, Vif, Rev, Env, and Nef have now been identified, enabling more comprehensive pathogenesis studies. Dissecting the mechanisms of elite control in Mamu-B*08-positive macaques will be vitally important to guide HIV vaccine design in the future.
Supplementary Material
Acknowledgments
We thank the MHC Genotyping Core at the WNPRC (William Rehrauer, Chrystal Glidden, Gretta Borchardt, and Debi Fisk) for genotyping our Indian rhesus macaques. We also appreciate Jason Reed, Emma Gostick, and David A. Price for assistance with the construction of MHC class I tetramers, Jessica Furlott for immunological assay assistance, and Carrie Moore, Sandy Ngo, and Amiyah Steen for performing MHC-peptide binding assays. We are grateful to Clemencia Pinilla for providing us with the combinatorial peptide library. Laura Valentine provided helpful discussions. We also thank the Virology, Genetics, Immunology, and Animal core laboratories as well as Research Support Services at the National Primate Research Center, University of Wisconsin-Madison (WNPRC) for technical assistance.
The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: IL-2, human (item #136) from Hoffman-La Roche, Inc.
This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
This research was supported by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Disease contracts N01-AI-40023, N01-AI-40024, and HHSN266200400006C (to A.S.), and HHSN266200400088C (to D.I.W.) as well as NIH grants R01 AI049120, R01 AI052056, R24 RR015371, and R24 RR016038 to D.I.W. Additionally, this publication was made possible in part by grant number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the NIH, awarded to the WNPRC, University of Wisconsin-Madison. This work was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459 and RR020141 (WNPRC).
References
- 1.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O’Connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loffredo JT, Friedrich TC, Leon EJ, Stephany JJ, Rodrigues DS, Spencer SP, Bean AT, Beal DR, Burwitz BJ, Rudersdorf RA, Wallace LT, Piaskowski SM, May GE, Sidney J, Gostick E, Wilson NA, Price DA, Kallas EG, Piontkivska H, Hughes AL, Sette A, Watkins DI. CD8+ T Cells from SIV Elite Controller Macaques Recognize Mamu-B*08-Bound Epitopes and Select for Widespread Viral Variation. PLoS ONE. 2007;2:e1152. doi: 10.1371/journal.pone.0001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 9.Jardetzky TS, Lane WS, Robinson RA, Madden DR, Wiley DC. Identification of self peptides bound to purified HLA-B27. Nature. 1991;353:326–329. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 10.Rotzschke O, Falk K, Stevanovic S, Gnau V, Jung G, Rammensee HG. Dominant aromatic/aliphatic C-terminal anchor in HLA-B*2702 and B*2705 peptide motifs. Immunogenetics. 1994;39:74–77. doi: 10.1007/BF00171803. [DOI] [PubMed] [Google Scholar]
- 11.Lopez de Castro JA, Alvarez I, Marcilla M, Paradela A, Ramos M, Sesma L, Vazquez M. HLA-B27: a registry of constitutive peptide ligands. Tissue Antigens. 2004;63:424–445. doi: 10.1111/j.0001-2815.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaslow RA, Rivers C, Tang J, Bender TJ, Goepfert PA, El Habib R, Weinhold K, Mulligan MJ. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J Virol. 2001;75:8681–8689. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 14.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O’Brien S, Andrieu JM, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson GW, Zagury JF. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 15.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, Yu XG, Meier A, Rockstroh JK, Allen TM, Jessen H, Rosenberg ES, Carrington M, Walker BD. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Bashirova A, Iversen AK, Phair J, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Altfeld M, O’Brien SJ, Carrington M. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 17.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 18.Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, Walker BD, Goulder PJ. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts MR, Exley B, Price DA, Bansal A, Camacho ZT, Teaberry V, West SM, Ambrozak DR, Tomaras G, Roederer M, Kilby JM, Tartaglia J, Belshe R, Gao F, Douek DC, Weinhold KJ, Koup RA, Goepfert P, Ferrari G. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc Natl Acad Sci U S A. 2005;102:4512–4517. doi: 10.1073/pnas.0408773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Streeck H, Lichterfeld M, Alter G, Meier A, Teigen N, Yassine-Diab B, Sidhu HK, Little S, Kelleher A, Routy JP, Rosenberg ES, Sekaly RP, Walker BD, Altfeld M. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J Virol. 2007;81:7725–7731. doi: 10.1128/JVI.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loffredo JT, Bean AT, Beal DR, Leon EJ, May GE, Piaskowski SM, Furlott JR, Reed J, Musani SK, Rakasz EG, Friedrich TC, Wilson NA, Allison DB, Watkins DI. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82:1723–1738. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, Watkins DI. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- 23.Evans DT, O’Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 24.Dzuris JL, Sidney J, Appella E, Chesnut RW, Watkins DI, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000;164:283–291. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- 25.Kaizu M, Borchardt GJ, Glidden CE, Fisk DL, Loffredo JT, Watkins DI, Rehrauer WM. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8(+) T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- 26.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 27.Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothe BR, Sidney J, Sette A, Kunstman K, Wolinsky S, Piatak M, Lifson J, Hughes AL, Wilson N, O’Connor DH, Watkins DI. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 28.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, Leon EJ, Soma T, Napoe G, Capuano SVr, Wilson NA, Watkins DI. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prilliman K, Lindsey M, Zuo Y, Jackson KW, Zhang Y, Hildebrand W. Large-scale production of class I bound peptides: assigning a signature to HLA-B*1501. Immunogenetics. 1997;45:379–385. doi: 10.1007/s002510050219. [DOI] [PubMed] [Google Scholar]
- 30.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 31.Wahl A, Hildebrand WH. Methods in Molecular Biology. Modulation of Antigen Presentation During Infection. Humana Press; 2009. Changes in MHC Class I Peptide Epitopes Following Viral Infection. In press. [Google Scholar]
- 32.Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- 33.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol Chapter. 2001;18:Unit 18.3. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 34.Sidney J, Southwood S, Sette A. Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires. Immunogenetics. 2005;57:393–408. doi: 10.1007/s00251-005-0004-2. [DOI] [PubMed] [Google Scholar]
- 35.Sidney J, del Guercio MF, Southwood S, Engelhard VH, Appella E, Rammensee HG, Falk K, Rotzschke O, Takiguchi M, Kubo RT, et al. Several HLA alleles share overlapping peptide specificities. J Immunol. 1995;154:247–259. [PubMed] [Google Scholar]
- 36.Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, DeMars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 37.Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothe BR, Kuntzen T, Oniangue-Ndza C, Trocha A, Yu XG, Brander C, Sette A, Walker BD, Allen TM. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loffredo JT, Sidney J, Wojewoda C, Dodds E, Reynolds MR, Napoe G, Mothe BR, O’Connor DH, Wilson NA, Watkins DI, Sette A. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J Immunol. 2004;173:5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- 39.Sette A, Sidney J, del Guercio MF, Southwood S, Ruppert J, Dahlberg C, Grey HM, Kubo RT. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994;31:813–822. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 40.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidney J, Peters B, Moore C, Pencille TJ, Ngo S, Masterman KA, Asabe S, Pinilla C, Chisari FV, Sette A. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A*0301 and A*0401 using a combinatorial peptide library. Immunogenetics. 2007;59:745–751. doi: 10.1007/s00251-007-0243-5. [DOI] [PubMed] [Google Scholar]
- 42.Loffredo JT, Burwitz BJ, Rakasz EG, Spencer SP, Stephany JJ, Vela JP, Martin SR, Reed J, Piaskowski SM, Furlott J, Weisgrau KL, Rodrigues DS, Soma T, Napoe G, Friedrich TC, Wilson NA, Kallas EG, Watkins DI. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J Virol. 2007;81:2624–2634. doi: 10.1128/JVI.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchinson SL, Wooldridge L, Tafuro S, Laugel B, Glick M, Boulter JM, Jakobsen BK, Price DA, Sewell AK. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J Biol Chem. 2003;278:24285–24293. doi: 10.1074/jbc.M300633200. [DOI] [PubMed] [Google Scholar]
- 44.O’Connor DH, McDermott AB, Krebs KC, Dodds EJ, Miller JE, Gonzalez EJ, Jacoby TJ, Yant L, Piontkivska H, Pantophlet R, Burton DR, Rehrauer WM, Wilson N, Hughes AL, Watkins DI. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J Virol. 2004;78:14012–14022. doi: 10.1128/JVI.78.24.14012-14022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. Decrypting the structure of major histocompatibility complex class I-restricted cytotoxic T lymphocyte epitopes with complex peptide libraries. J Exp Med. 1995;181:2097–2108. doi: 10.1084/jem.181.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, Buus S. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur J Immunol. 1996;26:1911–1918. doi: 10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- 47.Udaka K, Wiesmuller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- 48.Lauemoller SL, Holm A, Hilden J, Brunak S, Holst Nissen M, Stryhn A, Ostergaard Pedersen L, Buus S. Quantitative predictions of peptide binding to MHC class I molecules using specificity matrices and anchor-stratified calibrations. Tissue Antigens. 2001;57:405–414. doi: 10.1034/j.1399-0039.2001.057005405.x. [DOI] [PubMed] [Google Scholar]
- 49.Peters B, Bui HH, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, Wilson SS, Sidney J, Lund O, Buus S, Sette A. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ugrinovic S, Mertz A, Wu P, Braun J, Sieper J. A single nonamer from the Yersinia 60-kDa heat shock protein is the target of HLA-B27-restricted CTL response in Yersinia-induced reactive arthritis. J Immunol. 1997;159:5715–5723. [PubMed] [Google Scholar]
- 52.Kuon W, Holzhutter HG, Appel H, Grolms M, Kollnberger S, Traeder A, Henklein P, Weiss E, Thiel A, Lauster R, Bowness P, Radbruch A, Kloetzel PM, Sieper J. Identification of HLA-B27-restricted peptides from the Chlamydia trachomatis proteome with possible relevance to HLA-B27-associated diseases. J Immunol. 2001;167:4738–4746. doi: 10.4049/jimmunol.167.8.4738. [DOI] [PubMed] [Google Scholar]
- 53.Alvarez I, Marti M, Vazquez J, Camafeita E, Ogueta S, Lopez de Castro JA. The Cys-67 residue of HLA-B27 influences cell surface stability, peptide specificity, and T-cell antigen presentation. J Biol Chem. 2001;276:48740–48747. doi: 10.1074/jbc.M108882200. [DOI] [PubMed] [Google Scholar]
- 54.Appel H, Kuon W, Kuhne M, Hulsmeyer M, Kollnberger S, Kuhlmann S, Weiss E, Zeitz M, Wucherpfennig K, Bowness P, Sieper J. The solvent-inaccessible Cys67 residue of HLA-B27 contributes to T cell recognition of HLA-B27/peptide complexes. J Immunol. 2004;173:6564–6573. doi: 10.4049/jimmunol.173.11.6564. [DOI] [PubMed] [Google Scholar]
- 55.Sidney J, Southwood S, Pasquetto V, Sette A. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J Immunol. 2003;171:5964–5974. doi: 10.4049/jimmunol.171.11.5964. [DOI] [PubMed] [Google Scholar]
- 56.Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum Immunol. 2001;62:1200–1216. doi: 10.1016/s0198-8859(01)00319-6. [DOI] [PubMed] [Google Scholar]
- 57.Lamberth K, Roder G, Harndahl M, Nielsen M, Lundegaard C, Schafer-Nielsen C, Lund O, Buus S. The peptide-binding specificity of HLA-A*3001 demonstrates membership of the HLA-A3 supertype. Immunogenetics. 2008;60:633–643. doi: 10.1007/s00251-008-0317-z. [DOI] [PubMed] [Google Scholar]
- 58.Sylvester-Hvid C, Nielsen M, Lamberth K, Roder G, Justesen S, Lundegaard C, Worning P, Thomadsen H, Lund O, Brunak S, Buus S. SARS CTL vaccine candidates; HLA supertype-, genome-wide scanning and biochemical validation. Tissue Antigens. 2004;63:395–400. doi: 10.1111/j.0001-2815.2004.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen TM, Mothe BR, Sidney J, Jing P, Dzuris JL, Liebl ME, Vogel TU, O’Connor DH, Wang X, Wussow MC, Thomson JA, Altman JD, Watkins DI, Sette A. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mothe BR, Sidney J, Dzuris JL, Liebl ME, Fuenger S, Watkins DI, Sette A. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- 61.Sette A, Sidney J, Bui HH, del Guercio MF, Alexander J, Loffredo J, Watkins DI, Mothe BR. Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics. 2005;57:53–68. doi: 10.1007/s00251-004-0749-z. [DOI] [PubMed] [Google Scholar]
- 62.Loffredo JT, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, Reed J, Peters B, Hickman-Miller HD, Bardet W, Rehrauer WM, O’Connor DH, Wilson NA, Hildebrand WH, Sette A, Watkins DI. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–5997. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- 63.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 64.Vitiello A, Yuan L, Chesnut RW, Sidney J, Southwood S, Farness P, Jackson MR, Peterson PA, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 65.van der Most RG, Murali-Krishna K, Whitton JL, Oseroff C, Alexander J, Southwood S, Sidney J, Chesnut RW, Sette A, Ahmed R. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology. 1998;240:158–167. doi: 10.1006/viro.1997.8934. [DOI] [PubMed] [Google Scholar]
- 66.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans DT, Jing P, Allen TM, O’Connor DH, Horton H, Venham JE, Piekarczyk M, Dzuris J, Dykhuzen M, Mitchen J, Rudersdorf RA, Pauza CD, Sette A, Bontrop RE, DeMars R, Watkins DI. Definition of five new simian immunodeficiency virus cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I molecules: evidence for an influence on disease progression. J Virol. 2000;74:7400–7410. doi: 10.1128/jvi.74.16.7400-7410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nixon DF, Townsend AR, Elvin JG, Rizza CR, Gallwey J, McMichael AJ. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 69.Nixon DF, Huet S, Rothbard J, Kieny MP, Delchambre M, Thiriart C, Rizza CR, Gotch FM, McMichael AJ. An HIV-1 and HIV-2 cross-reactive cytotoxic T-cell epitope. AIDS. 1990;4:841–845. doi: 10.1097/00002030-199009000-00002. [DOI] [PubMed] [Google Scholar]
- 70.Rowland-Jones S, Colbert RA, Dong T, McAdam S, Brown M, Ariyoshi K, Sabally S, Whittle H, McMichael A. Distinct recognition of closely-related HIV-1 and HIV-2 cytotoxic T-cell epitopes presented by HLA-B*2703 and B*2705. AIDS. 1998;12(11):1391–1393. doi: 10.1097/00002030-199811000-00023. [DOI] [PubMed] [Google Scholar]
- 71.Valentine LE, Piaskowski SM, Rakasz EG, Henry NL, Wilson NA, Watkins DI. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J Virol. 2008;82:575–581. doi: 10.1128/JVI.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bennett MS, Ng HL, Ali A, Yang OO. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J Infect Dis. 2008;197:390–397. doi: 10.1086/525281. [DOI] [PubMed] [Google Scholar]
- 73.Draenert R, Verrill CL, Tang Y, Allen TM, Wurcel AG, Boczanowski M, Lechner A, Kim AY, Suscovich T, Brown NV, Addo MM, Walker BD. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J Virol. 2004;78:630–641. doi: 10.1128/JVI.78.2.630-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 75.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 76.Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 77.Rapin N, Hoof I, Lund O, Nielsen M. MHC motif viewer. Immunogenetics. 2008;60:759–765. doi: 10.1007/s00251-008-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger S, Stewart S, Surko P, Way S, Wilson S, Sette A. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sette A, Sidney J, Livingston BD, Dzuris JL, Crimi C, Walker CM, Southwood S, Collins EJ, Hughes AL. Class I molecules with similar peptide-binding specificities are the result of both common ancestry and convergent evolution. Immunogenetics. 2003;54:830–841. doi: 10.1007/s00251-002-0530-0. [DOI] [PubMed] [Google Scholar]
- 80.Herberts CA, Neijssen JJ, de Haan J, Janssen L, Drijfhout JW, Reits EA, Neefjes JJ. Cutting edge: HLA-B27 acquires many N-terminal dibasic peptides: coupling cytosolic peptide stability to antigen presentation. J Immunol. 2006;176:2697–2701. doi: 10.4049/jimmunol.176.5.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


