Abstract
The magnitude of bacterial diarrhea in developing countries is largely unknown since affordable detection methods are not available. We have developed a PCR-based assay for use in areas with limited resources to screen for diarrheagenic strains from clinical isolates. To simplify the assay and minimize reagents, our method implemented the use of plasmids rather than bacteria as template controls, and the use of bacterial suspensions or crude DNA preparations rather than purified genomic DNA as template DNA. The assay consisted of 3 PCR reactions using 3 groups of 5–6 primer pairs to identify the 11 most common bacterial diarrheogenic pathogens. The three-reaction multiplex PCR amplifies DNA targets specific for each one of the six E. coli diarrheogenic strains and the five non-E. coli diarrheogenic strains, including Salmonella spp, Shigella spp, Campylobacter spp., Yersinia enterocolitica, and Vibrio cholerae. The assay may provide an important epidemiological tool to investigate the role of diarrheagenic bacterial pathogens in areas of the world with limited resources.
Keywords: Multiplex PCR, E. coli, surveillance, Diarrhea
INTRODUCTION
Infectious diarrheal disease is a leading cause of morbidity and mortality in children in developing countries (Guerrant et al., 2002; Huilan et al., 1991; Kosek et al., 2003). Travelers, including children, from industrialized nations to developing countries, are also affected by traveler’s diarrhea (Adachi et al., 2002). While the etiological agents and their mechanisms of pathogenesis have been elucidated, information on the prevalence of these agents in developing countries is largely unknown. Assays for identification of these pathogens are limited to research laboratories and affordable identification assays are not commercially available. Most epidemiological data on diarrheal pathogens from developing countries is scattered and generally does not include E. coli as a causative agent, since no approved testing system is available for the identification of the six known diarrheogenic E. coli strains (Reither et al., 2007). The only method available in most developing nations for detection of bacterial diarrheal pathogens is conventional bacterial culture from stool. This method requires a minimum of 48 h for identification of E. coli and non-E. coli species. It is unable to discriminate between nonpathogenic and pathogenic E. coli strains. Furthermore, no commercially available methods exist to differentiate among the six different E. coli pathotypes associated with diarrhea, including enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), diffuse adherent E. coli (DAEC), and enteroinvasive E. coli (EIEC) (Levine, 1987).
A sensitive, specific, and affordable test for rapid identification of bacterial diarrhea is necessary to determine the impact of diarrheal bacterial pathogens on childhood morbidity and mortality in developing countries. This method may be instrumental for epidemiological surveillance of diarrheal diseases, evaluation of food and water for human consumption, and possibly for rapid diagnosis of severe diarrhea.
The most common bacterial strains associated with diarrhea include E. coli, Salmonella, Shigella, Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. (Huilan et al., 1991). Among E. coli, six strains are capable of inducing diarrheal disease by well defined virulence genes and different mechanisms of pathogenesis. To our knowledge, no polymerase chain reaction (PCR)-based method has been published on the use of a comprehensive assay for detection of E. coli and non-E. coli enteropathogens from environmental sources, clinically isolated sources, or stool samples. Several multiplex PCR assays for detection of diarrheogenic E. coli have been reported since 1995 (Stacy-Phipps, 1995); all are based on amplification of gene targets specific for each E. coli pathotype, as shown in Table 1. Most of the assays are able to detect E. coli from clinical isolates (Aranda et al, 2007; Brandal et al., 2007; Nguyen et al.,. 2005; Rapelli et al., 2005; Rich et al., 2001; Toma et al., 2003; Vidal et al., 2005), and only one from both clinical isolates and directly from stools (Stacy-Phipps et al., 1995). While most assays detect amplified DNA on agarose gels stained with ethidium bromide or an equivalent intercalating agent, some assays use fluorescent primer probes (Brandal et al., 2007; Watterworth et al., 2005). Independently, multiplex PCR assay have been developed for detection of non-E. coli enteropathogens from stools, food, water, and animals. Detection of Y. enterocolitica was described by the use of the Yersinia heat-stable enterotoxin gene (yst) PCR (Ibrahim et al., 1997). Detection of Salmonella species relies on amplification of the internal transcribed spacer region of the 16S-23S rRNA gene (Chiu et al., 2005; Park et al, 2006). Campylobacter spp. detection is based on the amplification of the hippuricase gene (hipO) (Persson and Olsen, 2005). V. cholerae detection uses the toxin-coregulated pilus (TCP) and the cholera toxin genes (RTX-A and RTX-B) (Gubala, 2006).
Table 1.
Target genes for PCR amplification. Gene size, location, and strain of origin.
| Primer Mix | Gene target | PCR Size | Location | Strain | Primer Sequence | Orientation | Source |
|---|---|---|---|---|---|---|---|
| M1 | VT | 518 | Chromosome | EHEC | 5′-GAGCGAAATAATTTATATGTG-3′
5′-TGATGATGGCAATTCAGTAT-3′ |
Forward
Reverse |
Aranda et al., 2007 |
| eae | 917 | Chromosome | EHEC, EPEC | 5′-CTGAACGGCGATTACGCGAA-3′
5′-CGAGACGATACGATCCAG-3′ |
Forward
Reverse |
Aranda et al., 2007 | |
| bfpA | 326 | Plasmid | EPEC | 5′-AATGGTGCTTGCGCTTGCTGC-3′
5′-GCCGCTTTATCCAACCTGGTA-3′ |
Forward
Reverse |
Aranda et al., 2007 | |
| aggR | 254 | Plasmid | EAEC | 5′-GTATACACAAAAGAAGGAAGC-3′
5′-ACAGAATCGTCAGCATCAGC-3′ |
Forward
Reverse |
Aranda et al., 2007 | |
| M2 | LT | 218 | Plasmid | ETEC | 5′-GCACACGGAGCTCCTCAGTC-3′
5′-TCCTTCATCCTTTCAATGGCTTT-3′ |
Forward
Reverse |
Vidal et al., 2005 |
| ST | 147 | Plasmid | ETEC | 5′-GCTAAACCAGTAGAG(C)TCTTCAAAA-3′
5′-CCCGGTACAG(A)GCAGGATTACAACA-3′ |
Forward
Reverse |
Nguyen et al., 2005 | |
| daaE | 542 | Plasmid | DAEC | 5′-GAACGTTGGTTAATGTGGGGTAA-3′
5′-TATTCACCGGTCGGTTATCAGT-3′ |
Forward
Reverse |
Vidal et al., 2005 | |
| virF | 618 | Chromosome | EIEC | 5′-AGCTCAGGCAATGAAACTTTGAC-3′
5′-TGGGCTTGATATTCCGATAAGTC-3′ |
Forward
Reverse |
Vidal et al., 2005 | |
| ipaH | 933 | Plasmid | EIEC | 5′-CTCGGCACGTTTTAATAGTCTGG-3′
5′-GTGGAGAGCTGAAGTTTCTCTGC-3′ |
Forward
Reverse |
Vidal et al., 2005 | |
| ITS | 312 | Chromosome | Salmonella spp | 5′-TATGCCCCATCGTGTAGTCAGAAC-3′
5′-TGCGGCTGGATCACCTCCTT-3′ |
Forward
Reverse |
Park et al., 2006 | |
| M3 | YST | 145 | Chromosome | Y. enterocolitica | 5′-GTTAATGCTGTCTTCATTTGGAGC-3′
5′-GACATCCCAATCACTACTGACTTC-3′ |
Forward
Reverse |
This study |
| RTX-A | 120 | Chromosome | V cholerae | 5′-AGCAAGAGCATTGTTGTTCCTACC-3′
5′-ACTTCCCTGTACCGCACTTAGAC-3′ |
Forward
Reverse |
Gubala et al., 2006 | |
| hipO | 344 | Chromosome | C. jejuni | 5′-GACTTCGTGCAGATATGGATGCTT-3′
5′-GCTATAACTATCCGAAGAAGCCATCA-3′ |
Forward
Reverse |
Persson & Olsen, 2005 | |
| virF | 618 | Chromosome | Shigella spp | 5′-AGCTCAGGCAATGAAACTTTGAC-3′
5′-TGGGCTTGATATTCCGATAAGTC-3′ |
Forward
Reverse |
Vidal et al., 2005 | |
| ipaH | 933 | Plasmid | Shigella spp | 5′-CTCGGCACGTTTTAATAGTCTGG-3′
5′-GTGGAGAGCTGAAGTTTCTCTGC-3′ |
Forward
Reverse |
Vidal et al., 2005 |
Conventional PCR assays imply the need for expensive equipment (freezers) and supplies (molecular grade solutions, genomic DNA isolation), as well as trained personnel in molecular biology techniques. These methods use prototype bacterial isolates for PCR controls, which assume the need for −70-°C freezers for bacterial strain storage. Furthermore, these techniques rely on genomic DNA as DNA templates for amplification, which requires the use of multiple molecular-grade reagents and technical expertise. All of these elements—equipment, techniques, reagents, and expertise—are simply not available in many geographical settings in the developing world.
In this study we described a comprehensive assay capable of detecting not only E. coli diarrheogenic strains but also Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter jejuni enteropathogens. DNA templates used in this assay include DNA plasmid controls rather than bacterial strain controls, and bacterial suspensions or crude DNA preparations rather than purified genomic DNA. Three sets of 5 to 6 plasmids containing cloned genes unique for each diarrheagenic bacterial pathogen will be used to provide the positive control template for reference PCR amplicons. By using plasmid controls we obviate the need of bacterial strains that require expensive freezing equipment for storage. By using crude DNA extracts as template DNA we avoid the need for equipment and reagents essential to process bacteria for isolation of highly purified genomic DNA. The emphasis of this assay is simplification, speed of detection, sensitivity, specificity, and capability of implementation in limited-resource areas of the world where this clinical epidemiologic technology is needed the most.
MATERIALS AND METHODS
Strains and plasmids
Control bacterial strains employed in this study are listed in Table 2. E. coli K-12 strain DH5α was used for cloning experiments. Plasmids used in this study are listed in Table 3. E. coli, Shigella, Salmonella, and Y. enterocolitica strains were grown on McConkey agar or Eosin Methylene blue agar, Luria agar or Luria broth at 37 °C overnight, unless otherwise specified. Campylobacter spp. were grown on blood agar and microaerophilic environment using Pack-MicroAero (Mitsubishi Gas Chemical America, New York, NY). All strains were grown at 37 °C overnight from a frozen stock aliquot.
Table 2.
E. coli and non-E. coli reference strains used in this study
| Name | Species | Strain/Biotype | Serotype | Gene Targetsa | Source |
|---|---|---|---|---|---|
| DH5alpha | E. coli | K12 | None | Laboratory collection | |
| 2060–004 | E. coli | EHEC | O157:H7 | VT, eaeA | UIHC b |
| E2348/69 | E. coli | EPEC | O127:H6 | eae, bfpA | (Gomez-Duarte & Kaper, 1995) |
| JM221 | E. coli | EAEC | O78:H33 | aggR | (Nataro et al., 1995) |
| E9034A | E. coli | ETEC | O8:H9 | LT, ST | (Levine et al., 1984) |
| C1845 | E. coli | DAEC | O75:NM | daaE | (Bilge et al., 1989) |
| EC-12 | E. coli | EIEC | - | ipaH, virF | U. of Washingtonc |
| SH24 | S. flexneri | - | - | ipaH, virF | UIHC |
| 1999–002 | S. enterica | - | Type B | ITS | UIHC |
| 1992–045 | Y. enterocolitica | - | - | YST | UIHC |
| 33291 | C. jejuni | - | - | hipO | UIHC |
| N16961 | V. cholerae | El Tor, Inaba | O1 | RTX | (Levine et al., 1988) |
Gene targets: VT, EHEC verotoxin genes 1 or 2. eae, EPEC-EHEC E. coli effacement-attaching gene. bfpA, EPEC bundle-forming pilus structural subunit gene. aggR, EAEC aggregative adherence fimbriae regulator. LT, ETEC heat-labile toxin gene. ST, ETEC heat-stable toxin gene. daaE, DAEC diffuse adherence pilus structural subunit gene. virF, Shigella virulence regulatory gene. ipaH, Shigella invasion plasmid antigen gene. ITS, Salmonella internal transcribed spacer region of 16S–23S rRNA. YST, Yersinia heat-stable enterotoxin gene. RTX-A, V. cholerae cholera toxin subunit A gene. hipO, C. jejuni hippuricase O gene.
Microbiology Laboratory, University of Iowa Hospital and Clinics, Iowa City, IA, kindly provided by Dr. S. Richter.
Department of Microbiology, University of Washington. Kindly provided by Dr. S. Moseley.
Table 3.
Plasmids used or generated in this study.
| Control Group | Plasmid | Description | Reference |
|---|---|---|---|
| P1 | pCR2.1 | Cloning vector (Apr, Kmr) | Invitrogen, Carlsbad, CA |
| pSC-A | Cloning vector (Apr, Tetr) | Stratagene, La Jolla, CA | |
| pOG401 | pSC-A with a 518 bp VT PCR insert from the 2060-004EHEC strain | This study | |
| pOG390 | pSC-A with 482 bp eae gene fragment from the E2348/69 EPEC strain | This study | |
| pOG394 | pSC-A with a 300 bp bfpA gene fragment from the E2348/69 EPEC strain | This study | |
| pOG395 | pSC-A with a 630 bp aggR gene fragment from the EAEC JM221 virulence plasmid | This study | |
| pWD299 | pBR313 with 1.8 kb insert containing LT genes A and B | (Dallas et al., 1979) | |
| P2 | pSLM004 | pBR322 with 0.8 kb TaqI insert containing the ST gene | (Moseley et al., 1983) |
| pOG391 | pSC-A with a 542 bp daaE gene fragment from the C1845 DAEC strain | This study | |
| pOG392 | pSC-A with a 933 bp ipaH gene fragment from the EC-12 EIEC strain | This study | |
| pOG393 | pSC-A with a 618 bp virF gene fragment from the EC-12 EIEC strain | This study | |
| P3 | pOG396 | pCR2.1 with a 120 bp RTX-A gene fragment from the N16961 V. cholerae strain | This study |
| pOG397 | pCR2.1 with a 312 bp ITS gene fragment from the 1999-02 S. enterica strain | This study | |
| pOG398 | pSC-A with a 145 bp YST gene fragment from the 1992-045 Y. enterocolitica strain | This study | |
| pOG400 | pSC-A with a 344 bp hipO gene fragment from the 33291 C. jejuni strain | This study |
DNA templates processing for PCR assays
Bacterial suspensions and crude DNA preparations were used as DNA templates for PCR reactions. Bacterial suspensions were made from each strain by harvesting bacteria from agar plates into a 1.5-ml Eppendorf tubes containing TE buffer (10 mM Tris-HCL pH 8.0, 5 mM EDTA). Bacterial suspensions were diluted to an optical density of 0.1 at 600nm (OD600) and used immediately for PCR. Preparation of crude DNA preparations required 1 ml of bacterial suspension in TE buffer adjusted to approximately 2.0, OD600. The bacterial suspension was then vortexed for 1 min at maximum speed, and centrifuged at maximum speed in an Eppendorf Microfuge for 2 min. The supernatant containing crude genomic DNA was transferred to a new Eppendorf tube for use as DNA template. The crude DNA preparations were stored at 4° C until used.
DNA amplification of target genes
Thirteen gene targets from E. coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter jejuni diarrheogenic strains were amplified using oligonucleotide primer pairs listed in Table 1. One microliter of crude DNA preparation was mixed with 24 μl of a pre-made mix containing primers at a 0.2-μM final concentration and Platinum Blue PCR SuperMix polymerase (Invitrogen, Carlsbad, CA). Combination of 5 to 6 pairs of primers per primer group, and designated M1, M2, and M3, are described in Table 1. Primer mixes were evaluated at different concentrations and the 0.2 μM final concentration for individual primers was chosen for all reactions. The PCR program used for amplification consisted of 2 min at 94 °C of denaturin g temperature, followed by 40 cycles of 30 s at 92 °C of denaturing temperature, 30 s at 59 °C of annealing temperature, and 30 s at 72 °C of extension temperatu re. At the end of the 40 cycles, a 5-min extension at 72 °C was used before samples were r eady for analysis or PCR DNA fragment purification.
Cloning of target genes into plasmids
DNA fragments amplified by PCR from prototype strains were column-purified using Invitrogen PureLink PCR purification kits and following supplier recommendations. Gene target-purified DNA fragments were ligated into plasmid vectors pSC-A (Stratagene) or pCR2.1 (Invitrogen) following conventional molecular biology techniques (Sambrook et al., 2001). Ligation mixes were introduced into E. coli competent cells by transformation as recommended by suppliers. Bacterial clones carrying each one of the cloned gene targets (see Table 3) were stored in a −80-°C freezer for further use. DNA plasmids cont aining each one of the 12 gene targets were isolated from E. coli K12 strains using a HiPure midiprep kit (Invitrogen) as recommended by suppliers (see Table 3). Cloned DNA targets from all plasmids were sent for DNA sequencing to confirm that the cloned DNA corresponded to the gene of interest. DNA sequences obtained from the DNA facility (University of Iowa) were analyzed by the BLAST soft ware from the National Center for Biotechnology Information website. All DNA sequences analyzed corresponded to the gene targets of interest (data not shown).
DNA gel electrophoresis and image recording
DNA amplified by PCR was separated onto a 2.0% agarose gel electrophoresis containing ethidium bromide and using Tris-acetate-EDTA (TAE) as running buffer (Sambrook et al., 2001). Images of DNA separation from gel exposed to UV light were captured with a digital camera. Images were transferred to a PowerPoint software program for analysis, storage, and printing. Material contaminated with ethidium bromide was disposed according to local guidelines. An alternative method for staining of DNA gels includes SYBR green stain (Invitrogen), a reagent not classified as a toxic agent and disposable with conventional waste.
RESULTS
Three-sample Multiplex PCR assay
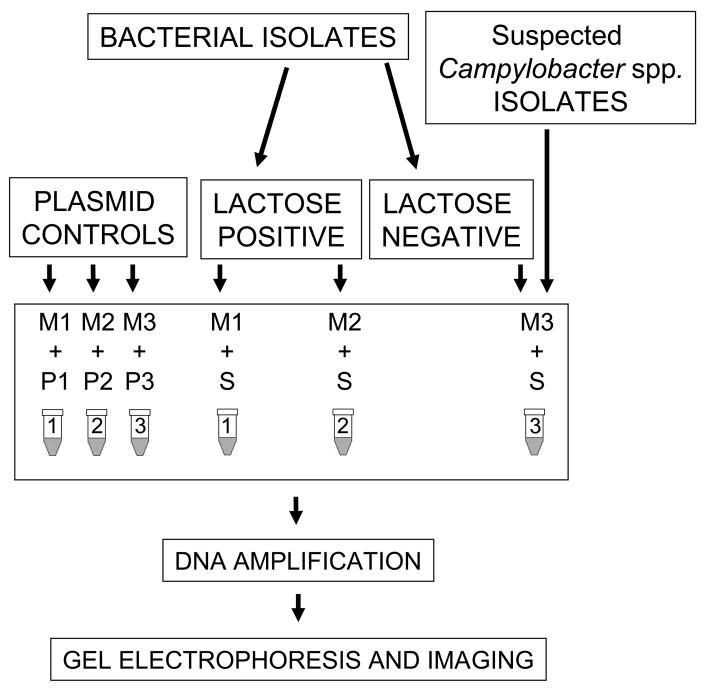
A three-sample, multiplex PCR was designed for identification of the 11 most common bacterial strains associated with diarrhea in a single PCR program run. The six E.coli strains included EHEC, EPEC, EAEC, ETEC, DAEC, and EIEC, and the five non-E. coli strains included Shigella spp, Salmonella, Y. enterocolitica, Campylobacter spp., and V. cholerae. The multiplex PCR consisted of three individual PCR reactions, containing a premade primer mix (M) of Taq polymerase enzyme, enzyme buffer, nucleotides, and a set number of specific primer pairs and the DNA template control (P) or DNA template sample (S) (Figure 1). All clinical isolates were screened for lactose fermentation before being tested with the three-sample multiplex PCR. The assay required the addition of 1 μl of DNA template to the 24-μl premade solution mix. PCR samples 1 to 3 (M1, M2, and M3) had all reagents except DNA templates. M1 mix contained primers for amplification of five gene targets including: verotoxins (VTs) 1 and 2 from EHEC, intimin (eae) from EHEC and EPEC, bundle-forming pilus structural subunit (bfpA) from EPEC, and regulatory gene aggR from EAEC. M2 mix contained primers for amplification of the heat-labile toxin (LT) and the heat-stable toxin (ST) genes from ETEC, for the diffuse adherence structural subunit gene (daaE) from DAEC, for the invasion plasmid antigen H (ipaH), and for the virulence invasion factor (virF) genes from EIEC (or Shigella spp.). M3 contained primers for amplification of non-E. coli genes including cholera toxin (RTX1) from V. cholerae, YST from Y. enterocolitica, hippurinase gene (hipO) from Campylobacter spp., 16S ribosomal RNA region specific for Salmonella spp (ITS), invasion plasmid antigen H (ipaH), and virulence invasion factor (virF) from Shigella spp. (or EIEC). Primer pair mixes are described and shown in Table 1. Primers for YST of Yersinia were modified from previously described primers (Ibrahim et al., 1997). Three nucleotides were added at the 5-prime end of each forward and reverse primer to make the melting time temperature similar to the remaining primers, as indicated in Table 1. Each reaction had a total volume of 25 μl. PCR reaction mixes were amplified in a thermocycler using a single program and processed as described in materials and methods.
Figure 1. Three-sample multiplex PCR diagram.
Diagram shows the sequential steps on the PCR assay beginning with the lactose fermentation screening of bacterial isolates, followed by the addition of the DNA templates (S or P) to the PCR reaction mixes (M), PCR amplification, and the DNA fragment separation onto a 2% agarose gel electrophoresis, and digital imaging capture for further analysis. Mixes M1, M2, and M3 contain all reagents except DNA template. P1, P2, and P3 correspond to plasmid controls templates. S corresponds to sample DNA templates, which may be crude DNA preparations or diluted bacterial suspensions. Tube 1 containing M1 and P1 reagents will amplify gene targets specific for EHEC, EPEC and EAEC. Tube 2 will identify specific targets for ETEC, DAEC, and EIEC. Tube 3 will amplify targets specific for Campylobacter spp., Salmonella spp., Y. enterocolitica, V. cholerae, and Shigella spp.
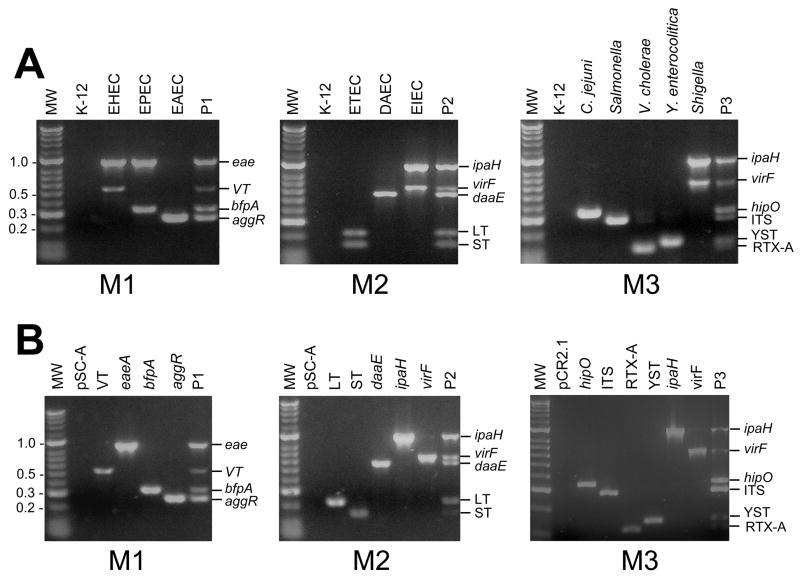
Gene target controls for the three-sample multiplex PCR
Because long term storage of bacterial control strains are not possible in many geographic regions, we have implemented the use of plasmid DNA as gene target controls. Gene targets amplified from control strains were cloned in conventional plasmid vectors, and purified plasmids carrying individual gene targets were used as gene target controls. Gene targets from control strains and from control plasmids were amplified by using the three-sample multiplex PCR. As shown in Figure 2 identical DNA bands were amplified from both, plasmid controls and bacterial control strains. The molecular masses for each gene target was as expected (see Table 3). Plasmid controls were stored dried or in solution at 4 °C. They did not require freezing, and were ready to use anytime. The control reaction P1 contained plasmids pOG390 (eae), pOG394(bfpA), pOG395 (aggR), and pOG401 (VT); the control template P2 contained plasmids pSLM004 (ST), pWD299 (LT), pOG391 (daaE), pOG392 (ipaH) and pOG393 (virF); and the control template P3 contained plasmids pOG392 (ipaH) and pOG393 (virF), pOG396 (RTX-A), pOG397 (ITS), pOG398 (YST), and pOG400 (hipO). Plasmid vectors pSC-A and pCR2.1 used for cloning of gene targets did not amplify DNA bands when used as DNA template on the multiplex PCR assay (Figure 2).
Figure 2. Three-sample multiplex PCR assay using plasmid DNA carrying gene target controls.
Ethidium bromide stained 2% agarose gel electrophoresis showing amplified products from crude DNA preparations from control strains and plasmid controls. M1, M2, and M3 correspond to primer mixes. M1 contains primers specific for EPEC, EHEC, and EAEC gene targets. M2 contains primers for ETEC, DAEC, and EIEC gene targets. M3 contains primers for Campylobacter spp., Salmonella spp., V. cholerae, Y. enterocolitica, and Shigella spp. gene targets (Table 1). Panel A. Three-reaction Multiplex PCR of control strains using crude DNA preparations. Strains used are described in table 2. P1, P2 and P3 correspond to plasmid control mixes. The K12 HB101 E. coli strain is included as a negative control. Panel B. Three-reaction Multiplex PCR of individual plasmid DNA templates described in Table 2 representing individual gene targets. pSC-A plasmid vector; VT: pOG401; eae: pOG390; bfpA: pOG394; aggR: pOG395; P1: pOG401, pOG390, pOG394, and pOG395 plasmids; LT: pWD299 ; ST: pSL0004; daaE: pOG391; ipaH: pOG392; virF: pOG393; P2: pWD299, pSL0004, pOG391, pOG392, and pOG393 plasmids; pCR2.1: plasmid vector; hipO: pOG400; ITS: pOG397; RTX-1: pOG396; YST: pOG398; and P3: pOG400, pOG392, pOG393, pOG397, pOG396, and pOG398 plasmids. MW represents: molecular weight marker (Hyperladder II, BIOLINE).
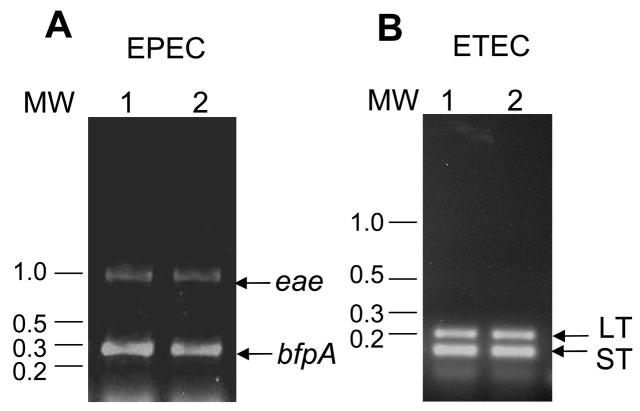
Bacterial suspensions versus crude DNA preparations as DNA templates
To simplify the multiplex PCR and facilitate its use in clinical laboratories with limited resources, we evaluated the efficiency of the multiplex PCR assay by comparing two different DNA templates including bacterial suspensions and crude DNA preparations isolated by the vortex method. Both DNA templates were able to amplify the expected DNA fragments. As shown in Figure 3, both EPEC DNA templates amplified the eae and bfpA gene targets, and the ETEC DNA templates amplified the LT and ST gene targets. The DNA template did not amplify unspecific bands. All strains were tested at decreasing DNA template concentrations, and they were still able to amplify gene targets when the DNA was diluted to 1:1000 or more from the original concentration (data not shown).
Figure 3. Crude DNA preparations versus bacterial suspension templates for use with the PCR assay.
Crude DNA preparations or bacterial suspensions from E2348/69 EPEC and E9034A ETEC bacterial strains were tested on the multiplex PCR assay. Panel A shows a PCR reaction using M1 PCR reaction mix and EPEC DNA template. Lane 1, crude DNA preparation; lane 2, bacterial suspension. Panel B shows multiplex PCR assay using M2 PCR reaction mix and ETEC DNA template. Lane 1, crude DNA preparation; Lane 2, diluted bacterial suspension. Arrows in panel A indicate eae and bfpA amplified DNA fragments. Arrows in panel B indicate LT and ST. MW represents molecular weight markers.
The three-sample multiplex PCR is sensitive
Three-sample multiplex PCR was performed using control template plasmid. All three samples had the Blue mix taq polymerase, primer mix (either M1, M2, or M3) and plasmid mix (either P1, P2, or P3). Plasmid mixes P1, P2, and P3 were sequentially diluted from a concentration starting with 10 mg/ml to as low as 0.0001 ng/ml. The multiplex PCR was able to detect specific target DNA at concentration as low as 0.001 ng/ml, which corresponds to a detection level of 1 to 10 molecules of target DNA per reaction sample (data not shown). While PCR assays reported in the literature use a single sample for detection of all five of six E. coli strains (Nguyen et al., 2005; Vidal et al., 2005), we observed decreased sensitivity and specificity when more than 6 pairs of primer pairs were present in a single PCR reaction. This phenomenon has been previously described (Watterworth et al., 2005). To minimize the presence of non-specific amplified DNA, we used a three-sample PCR for detection of all six E. coli pathotypes and all five non-E. coli enteropathogens.
Validation of the Multiplex PCR method
A collection of previously characterized bacterial strains from different countries including the US was tested with the three-sample PCR assay to validate the method. E. coli, Shigella spp., Salmonella spp., Y. enterocolitica, Campylobacter spp., and V. cholerae strains were initially classified into lactose fermenters and non-lactose fermenters. All strains were tested with the three-sample multiplex PCR and each strain was recognized based on the DNA banding pattern on the agarose gel as E. coli enteropathogens, Shigella spp, Salmonella spp, Y. enterocolitica, Campylobacter spp., or V. cholerae. EIEC and Shigella spp. strains in this study were differentiated by the lactose fermenting phenotype present in all E. coli and absent in all Shigella spp. strains. Negative control strains, including normal flora E. coli strains, Klebsiella spp, and Proteus spp., were also tested with the three-sample multiplex PCR. No amplified DNA bands where observed from these negative controls (See Table 4). Furthermore, there was no cross-reactivity observed and among the E. coli or non-E. coli templates indicating that the assay was specific. While primers tested in this assay have been validated independently for E. coli pathotypes, Shigella spp, Salmonella, Y. enterocolitica, Campylobacter spp., and V. cholerae, we were able to validate the assay as a three-sample multiplex PCR assay.
Table 4.
Bacterial strains tested with the three-sample multiplex PCR
| Number of strains positive by PCR/Number of strains tested | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | VT | eae | bfpA | aggR | LT | ST | daaE | virF | ipaH | hipO | ITS | RTX | YST | Lactose fermenter |
| EHEC | 5/5 | 2/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| EPEC | 0/5 | 5/5 | 5/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| EAEC | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 |
| ETEC | 0/21 | 0/21 | 0/21 | 0/21 | 16/20 | 17/20 | 0/21 | 0/21 | 0/21 | 0/21 | 0/21 | 0/21 | 0/21 | 21/21 |
| DAEC | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| EIEC | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 | 5/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| Negative E. coli | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 10/10 |
| C. jejuni | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | ND |
| Salmonella spp. | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 7/7 | 0/7 | 0/7 | 0/7 |
| Shigella spp. | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 7/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/5 |
| V. cholerae | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 11/11 | 0/11 | 0/11 |
| Y. enterocolitica | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 2/2 | 0/2 |
| Klebsiella spp. | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| Proteus spp. | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
ND: not determined.
DISCUSSION
Diarrhea is a major cause of morbidity and mortality in children in the developing world and an important cause of morbidity to travelers from industrialized nations to developing countries. While rapid, specific, sensitive, affordable, and commercially available methods exist for the identification of enteroviral agents such as Rotavirus, no equivalent method exists for identification of bacterial diarrheal pathogens. Current methods of identification are slow, expensive, time consuming, and require a high level of expertise. Furthermore, identification of any of the six E. coli pathotypes associated with diarrhea can not be done by conventional clinical microbiology. These pathogens are identified in research laboratories, as no approved method of identification is commercially available.
Evaluation of the epidemiology of infectious diarrhea in developing countries requires affordable methods for identification of bacterial agents. Information on incidence, endemicity, and epidemics of diarrhea in developing countries is at present vastly unknown. Rapid identification methods may provide important information regarding the prevalence of bacterial diarrheal pathogens associated to diarrhea, and the type of bacterial contaminants of drinkable water and food products. This information may facilitate epidemiological surveillance and public health measures leading to improved prevention measures, and ultimately decrease infant mortality due to infectious diarrhea. The methods may potentially be used for diagnosis and better medical management of children with severe infectious diarrhea. Technology transfer will be necessary to provide formal training on the use, processing, and interpretation of the rapid PCR test to local health authorities in developing countries.
In the present manuscript we show that a rapid and affordable PCR-based method may be use for the identification of the most common bacteria associated with diarrhea in developing countries, including the six pathotypes of E. coli and the five most common non-E. coli enteropathogens including Shigella spp, Salmonella spp, Y. enterocolitica, Campylobacter spp., and V. cholerae. The PCR assay allowed the specific detection of each bacterial strain, except Shigella spp. and EIEC which were differentiated based on the lactose fermentation phenotype which was only expressed in the E.coli strains tested. The lactose fermentation pre-screening of bacterial samples may decrease the number of assays required per strain as lactose fermenter strains may only be tested with reaction 1 and 2 for detection of E. coli pathotypes while lactose non-fermenter isolates may be tested only with PCR reaction 3 for detection of Shigella spp, Salmonella spp, Y. enterocolitica, Campylobacter spp., and V. cholerae (Figure 1). All E. coli strains tested in our assay were lactose fermenters and all non-E. coli isolates, except Klebsiella spp and Campylobacter spp., were lactose non-fermenters. While lactose non-fermenting E. coli strains have been reported in some geographical areas (Colonna et al., 1992) a decision for testing lactose non-fermenter strains with all three-sample PCR assay should be made if local epidemiological information suggest the presence of these uncommon E. coli strains.
The assay is also capable to detect atypical enteropathogenic E. coli strains, which carry the locus for E. coli effacement (LEE) and lack BFP. These strains may be recognized by the presence of the eae gene and the absence of the bfpA gene. This pattern of detection is important as atypical EPEC strains are considered emerging pathogens in several developing countries as well as Europe (Alikhani et al., 2006; Jenkins et al., 2006; Moreno et al., 2008; Wani et al., 2006)
To our knowledge this is the first description of a PCR-based method used for detection of eleven E. coli and non-E. coli enteropathogens in a single assay. Prior reports on multiplex PCR assays have concentrated on either identification of E. coli pathotypes (Brandal et al., 2007; Nguyen et al., 2005; Matar et al., 2002) or non-E. coli strains (Ibrahim et al., 1997; Park et al., 2006; Gubala, 2006). Our assays have modified the PCR methodology from a single PCR reaction to a three-sample reaction. This method was employed to increase the specificity of the reaction, as this may decrease when more that five pairs of primers are used in a single reaction (Watterworth et al., 2005). The assay is sensitive, as demonstrated by the level of detection to almost single molecule. It is specific, as multiple PCR primers were specific for each target, and no cross-reactivity was detected with heterologous DNA. Specificity was also conserved within a significant template concentration range. Validation of the PCR method confirmed that approximately 100 strains previously characterized were correctly identified by the three-sample PCR method. The assay is rapid, as in 4 hours it is possible to isolate crude DNA preparations from a bacterial suspension, amplify the DNA, separate the DNA fragments in an agarose gel, and have a visual report of the banding pattern.
The PCR assay has been simplified to facilitate its use in limited settings. When testing pure bacterial isolates, the assay can use bacterial suspensions or crude DNA preparations rather than purified genomic DNA. It includes pre-made reactions already containing all reagents necessary for the PCR except the template. It also includes plasmid controls to obviate the need to have −70-°C freezing conditions for bacterial storage, not feasible in many areas with limited resources. After amplification, samples are separated in agarose gel electrophoresis, DNA bands visualized under UV light, and images recorded by a conventional digital camera. While ethidium bromide is a relatively inexpensive reagent for DNA gel staining, it has human and environmental safety concerns. Ethidium bromide is a toxic and carcinogenic compound that requires specific disposal condition and decontamination protocols. To avoid contamination in the work place or the environment, strict safety regulations must be followed or alternative DNA gel staining reagents are advised.
The minimal requirement for the assay’s implementation include access to electricity, water, and refrigeration (−10 to 4 °C). These conditi ons are met in most hospitals and reference laboratories in developing nations. The assay is also flexible with respect to the number of primer pairs and type of pathogens to identify. Detection of V. cholerae, for instance, may be more relevant for Africa and Asia than for Latin America. Furthermore, the assay may potentially identify enteropathogens from environmental sources, clinical isolates, or stool samples. In collaboration with Latin-American research institutions we have been successful in implementing the system in Cartagena and Bogota, Colombia and have found that ETEC is the most prevalent pathogen associated with diarrhea in children less than five years of age. ETEC was also identified as the most common contaminant of food products obtained from public markets in several Colombian cities (our unpublished results).
We believe that the multiplex PCR assay may provide valuable information on the prevalence of bacterial pathogens associated with diarrhea in developing nations. Similarly, it may facilitate epidemiological surveillance of water and edible products for human consumption to determine the risk of acute diarrheal disease at any single time in any densely populated geographic region. Implementation of this assay may not only improve epidemiological information on the dimension of childhood diarrhea in the developing world, but more importantly it may facilitate the implementation of preventive measures to decrease morbidity and mortality due to bacterial diarrhea in children living in the developing world.
Acknowledgments
This work was supported in part by NIH K12 mentored research award to O.G.G.-D. and by the Children’s Miracle Network—University of Iowa Children’s Hospital grant 1892–2007 to O.G.G.-D.
We thank Dr. Sandra Richter, Microbiology Laboratory, University of Iowa Hospital and Clinics, and Dr. James B. Kaper, Department of Microbiology, University of Maryland Baltimore, for kindly providing bacterial strains used in this study. We are grateful with Paul Cassella and Bharani Pandrangi for reading of the manuscript and providing helpful advice. We are thankful to Raquel E. Gomez for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi JA, Ericsson CD, Jiang ZD, DuPont MW, Pallegar SR, DuPont HL. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J Infect Dis. 2002;185:1681–1683. doi: 10.1086/340419. [DOI] [PubMed] [Google Scholar]
- Alikhani MY, Mirsalehian A, Aslani MM. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol. 2006;55:1159–1163. doi: 10.1099/jmm.0.46539-0. [DOI] [PubMed] [Google Scholar]
- Aranda KR, Fabbricotti SH, Fagundes-Neto U, Scaletsky IC. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol Lett. 2007;267:145–150. doi: 10.1111/j.1574-6968.2006.00580.x. [DOI] [PubMed] [Google Scholar]
- Bilge SS, Clausen CR, Lau W, Moseley SL. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal LT, Lindstedt BA, Aas L, Stavnes TL, Lassen J, Kapperud G. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J Microbiol Methods. 2007;68:331–341. doi: 10.1016/j.mimet.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Chiu TH, Chen TR, Hwang WZ, Tsen HY. Sequencing of an internal transcribed spacer region of 16S-23S rRNA gene and designing of PCR primers for the detection of Salmonella spp. in food. Int J Food Microbiol. 2005;97:259–265. doi: 10.1016/j.ijfoodmicro.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Colonna B, Ranucci L, Fradiani PA, Casalino M, Calconi A, Nicoletti M. Organization of aerobactin, hemolysin, and antibacterial resistance genes in lactose-negative Escherichia coli strains of serotype O4 isolated from children with diarrhea. Infect Immun. 1992;60:5224–5231. doi: 10.1128/iai.60.12.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas WS, Gill DM, Falkow S. Cistrons encoding Escherichia coli heat-labile toxin. J Bacteriol. 1979;139:850–858. doi: 10.1128/jb.139.3.850-858.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duarte OG, Kaper JB. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubala AJ. Multiplex real-time PCR detection of Vibrio cholerae. J Microbiol Methods. 2006;65:278–293. doi: 10.1016/j.mimet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Guerrant RL, Kosek M, Moore S, Lorntz B, Brantley R, Lima AA. Magnitude and impact of diarrheal diseases. Arch Med Res. 2002;33:351–355. doi: 10.1016/s0188-4409(02)00379-x. [DOI] [PubMed] [Google Scholar]
- Huilan S, Zhen LG, Mathan MM, Mathew MM, Olarte J, Espejo R, Khin Maung U, Ghafoor MA, Khan MA, Sami Z. Etiology of acute diarrhoea among children in developing countries: a multicentre study in five countries. Bull World Health Organ. 1991;69:549–555. [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A, Liesack W, Griffiths MW, Robins-Browne RM. Development of a highly specific assay for rapid identification of pathogenic strains of Yersinia enterocolitica based on PCR amplification of the Yersinia heat-stable enterotoxin gene (yst) J Clin Microbiol. 1997;35:1636–1638. doi: 10.1128/jcm.35.6.1636-1638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C, Smith HR, Lawson AJ, Willshaw GA, Cheasty T, Wheeler JG, Tompkins DS. Serotypes, intimin subtypes, and antimicrobial resistance patterns of atypical enteropathogenic Escherichia coli isolated in England from 1993 to 1996. Eur J Clin Microbiol Infect Dis. 2006;25:19–24. doi: 10.1007/s10096-005-0075-x. [DOI] [PubMed] [Google Scholar]
- Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- Levine MM, Kaper JB, Herrington D, Losonsky G, Morris JG, Clements ML, Black RE, Tall B, Hall R. Volunteer studies of deletion mutants of Vibrio cholerae O1 prepared by recombinant techniques. Infect Immun. 1988;56:161–167. doi: 10.1128/iai.56.1.161-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements ML, Cheney C. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect Immun. 1984;44:409–420. doi: 10.1128/iai.44.2.409-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matar GM, Abdo D, Khneisser I, Youssef M, Zouheiry H, Abdelnour G, Harakeh HS. The multiplex-PCR-based detection and genotyping of diarrhoeagenic Escherichia coli in diarrhoeal stools. Ann Trop Med Parasitol. 2002;96:317–324. doi: 10.1179/000349802125001032. [DOI] [PubMed] [Google Scholar]
- Moreno AC, Filho AF, Gomes TD, Ramos ST, Montemor LP, Tavares VC, Filho LD, Irino K, Martinez MB. Etiology of childhood diarrhea in the northeast of Brazil: significant emergent diarrheal pathogens. Diagn Microbiol Infect Dis. 2008 doi: 10.1016/j.diagmicrobio.2008.03.017. In press. [DOI] [PubMed] [Google Scholar]
- Moseley SL, Echeverria P, Seriwatana J, Tirapat C, Chaicumpa W, Sakuldaipeara T, Falkow S. Identification of enterotoxigenic Escherichia coli by colony hybridization using three enterotoxin gene probes. J Infect Dis. 1982;145:863–869. doi: 10.1093/infdis/145.6.863. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Deng Y, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis. 1995;171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–760. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YS, Lee SR, Kim YG. Detection of Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus and Listeria monocytogenes in kimchi by multiplex polymerase chain reaction (mPCR) J Microbiol. 2006;44:92–97. [PubMed] [Google Scholar]
- Persson S, Olsen KE. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol. 2005;54:1043–1047. doi: 10.1099/jmm.0.46203-0. [DOI] [PubMed] [Google Scholar]
- Rappelli P, Folgosa E, Solinas ML, Dacosta JL, Pisanu C, Sidat M, Melo J, Cappuccinelli P, Colombo MM. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo, Mozambique. FEMS Immunol Med Microbiol. 2005;43:67–72. doi: 10.1016/j.femsim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Reither K, Ignatius R, Weitzel T, Seidu-Korkor A, Anyidoho L, Saad E, Djie-Maletz A, Ziniel P, Amoo-Sakyi F, Danikuu F, Danour S, Otchwemah RN, Schreier E, Bienzle U, Stark K, Mockenhaupt FP. Acute childhood diarrhoea in northern Ghana: epidemiological, clinical and microbiological characteristics. BMC Infect Dis. 2007;7:104. doi: 10.1186/1471-2334-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich C, Alfidja A, Sirot J, Joly B, Forestier C. Identification of human enterovirulent Escherichia coli strains by multiplex PCR. J Clin Lab Anal. 2001;15:100–103. doi: 10.1002/jcla.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning a Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Stacy-Phipps S, Mecca JJ, Weiss JB. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054–1059. doi: 10.1128/jcm.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, Rivas M, Iwanaga M. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol. 2003;41:2669–2671. doi: 10.1128/JCM.41.6.2669-2671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Kruger E, Duran C, Lagos R, Levine M, Prado V, Toro C, Vidal R. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol. 2005;43:5362–5365. doi: 10.1128/JCM.43.10.5362-5365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani SA, Nabi A, Fayaz I, Ahmad I, Nishikawa Y, Qureshi K, Khan MA, Chowdhary J. Investigation of diarrhoeic faecal samples for enterotoxigenic, Shiga toxin-producing and typical or atypical enteropathogenic Escherichia coli in Kashmir, India. FEMS Microbiol Lett. 2006;261:238–44. doi: 10.1111/j.1574-6968.2006.00354.x. [DOI] [PubMed] [Google Scholar]
- Watterworth L, Topp E, Schraft H, Leung KT. Multiplex PCR-DNA probe assay for the detection of pathogenic Escherichia coli. J Microbiol Methods. 2005;60:93–105. doi: 10.1016/j.mimet.2004.08.016. [DOI] [PubMed] [Google Scholar]