Abstract
The accumulation of a given element is a complex process controlled by a network of gene products critical for uptake, binding, transportation, and sequestration. Many of these genes and physiological processes affect more than one element. Therefore, to understand how elements are regulated, it is necessary to measure as many of the elements contained in a cell, tissue or organism (the ionome) as possible. The elements which share components of their network vary depending on the species and genotype of the plants that are studied and environment they are grown in. Several recent papers describe high-throughput elemental profiling studies of how the ionome responds to the environment or explore the genetics that control the ionome. When combined with new genotyping technologies, ionomics provides a rapid way to identify genes that control elemental accumulation in plants.
Elements are an essential component of every living cell. As detailed in other parts of this issue, elements are used to regulate the electrochemical balance of cellular compartments, as cofactors in biochemical reactions, and as structural components in biological molecules and complexes. Other elements can have deleterious effects on the cell by disrupting these and other processes. Therefore, the physiologic and genetic networks controlling an elements uptake, transport and accumulation are of great interest to researchers, and have been the subject of intense study.
Alterations in the levels of any given element in a plant can be affected by a wide variety of factors:
Changes in the soil chemical environment, either caused by the plant (acidification or release of organic compounds), or by changes in the environment (rain, drought, animal waste or decay of organic matter).
Changes in the morphology of the plant, including root structure and whether the plant is in the vegetative or reproductive stage of its life cycle.
Changes in the plants uptake capacity brought on by the presence, absence or regulation of channels or transporters, or, for ions that diffuse into the plant, changes in the biochemical structure of the cell walls. Elements have to cross multiple membranes to travel from the soil to the cells in leaves, and each membrane can have a different composition of transporters.
Changes in the accumulation of chelators such as organic acids, peptides or proteins.
Changes in the amount of an element that is sequestered in intracellular compartments such as the ER, mitochondria or vacuole.
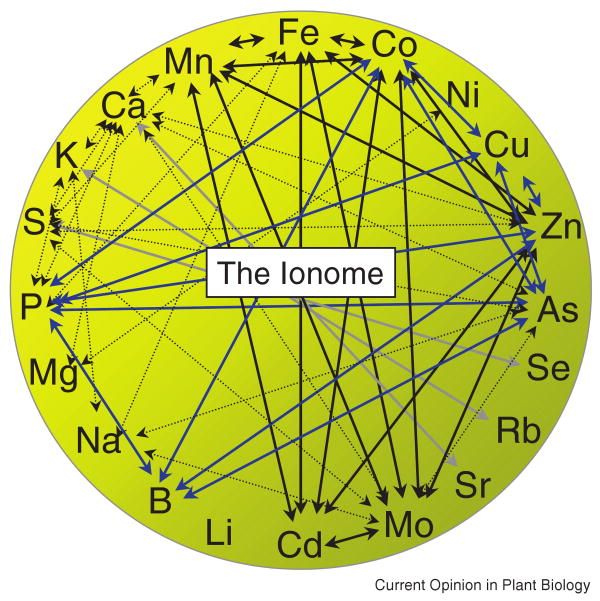
A few of these changes may exclusively affect a single element, for example, changes in a transporter or chelator that has high specificity for that element. But most of the above changes will affect more than one element. Therefore, experiments focused on single elements which do not address total mineral nutrient and trace element content of the plant will not reveal the regulatory networks involved in the homoeostasis of the ionome. These interactions among elements (Figure 1) necessitate an understanding of how the entire elemental composition of a cell, tissue, or organism (the ionome) responds to genetic and environmental perturbations. Fortunately, advances in mass spectroscopy technology enable researchers to measure the levels of dozens of elements in hundreds of samples in a single analysis. This allows us to ask questions about the ionome as a whole, a process termed ionomics [1]. In this review, I will discuss advantages of this approach, what has been learned about relationships between different elements, and how this technique can be combined with conventional and emerging genetics techniques to identify genes which control the ionome in organisms.
Figure 1.
Genetic, physiological and chemical interactions between elements. Arrows denote interactions discussed in the text. It should be noted, however, that this is only a subset of known elemental interactions. Solid Black: Elements in the Fe model from Baxter et al. [3]. Solid Blue: Elements in the P model from Baxter et al. [3]. Solid Grey: Chemical Analogs.
Ionomic Technologies
Inductively coupled plasma spectroscopy, either mass spectroscopy (ICP-MS) or optical emission spectroscopy (ICP-OES) allows for the simultaneous measurement of dozens of elements. Improvements in these techniques over the last few decades have overcome obstacles that prevented previous efforts from detecting all but the most severe ionomic differences [2]. With the appropriate equipment, sample preparation for these techniques is not significantly more onerous than for techniques that measure single elements. Most ionomic studies to date have measured 12-20 elements [3-8] (studies mentioned in this review which measure fewer elements have used other analysis methods). While these do not comprise the complete list of elements that are of interest in plants, they usually comprise the major macronutrients, and many of the micronutrients. As most alterations to the environment, physiology or genetics of a plant will affect multiple elements, one of the elements in these subsets will likely be affected by these alterations. This makes ionomics an excellent tool for detecting alterations in a plant's physiology or its environment.
Even when a researcher is only interested in single elements, ionomics approaches can improve the quality of data by measuring elemental analogs. Chemical analogs of elements of interest like Rb for K, Sr for Ca and Se for S provide independent measurements that are highly correlated with the element of interest. This data can be used to independently confirm that the concentration the elements of interest is elevated or reduced in the sample and that these changes are not due to analytical noise, or in cases where the element of interest is difficult to measure (for example Rb and Se are easier to measure by ICP-MS than K and S) the analog can be used as a proxy. It should be noted however that while these analogs are highly correlated with the elements of interest, some plants may be able to discriminate between them. For example some species of Astragalus can discriminate between S and Se [9].
Elemental to Element relationships
Critical to the understanding of how the ionome is regulated is an understanding of the overlap between networks. While some relationships between elements may exist regardless of the tissue, species or environment under study, many associations will vary with a combination of these factors. For example, an investigation of arsenite uptake in rice discovered that Si transporters are one method of arsenite uptake into plants [10]. This is in contrast to arsenate, which can enter plants via phosphate transporters [11] (for more detail see Verbruggen et al., this volume). The element that As is sharing a transporter with is therefore dependent on the speciation of As, which is a function of the soil environment, mainly pH and oxidative capacity, where the plant is growing.
While some shared elemental network components can be predicted by analysis of properties like the binding and transport properties of a transporter, in practice, living systems are so complex that most relationships cannot be predicted easily from chemical or biological principles (the exception being chemical analogs, described above) and must therefore be determined experimentally. While the interaction of genotype and environment is clearly a critical driver of alterations in the ionome, most studies to date looking at relationships between elements have focused on identifying either genetic or physiological correlations in isolation.
Genetic Interactions
Lahner et al [7] and Chen et al [5] performed large (>2000 plants) ionomic screens of mutagenized populations of Arabidopsis thaliana grown on soil and Lotus japonica grown in liquid culture, respectively. In both studies, the majority of mutants identified differed significantly in more than one element. Chen et al. [4] also performed a screen of 45 natural Lotus japonica accessions. Principle component revealed several groups of positively correlated elements: [As, Fe, Na, Zn and Mo], [Mg and Ni], and [Mn, Ca, and Sr]. Baxter et al. [12] recently cloned one of the mutants identified by Lahner et al. study. Although the molecular function of the gene is yet to be characterized the phenotype demonstrates the complex physiological interactions that affect the ionome and the potential for novel biological discovery via high-throughput elemental screening. Loss of function of the gene causes an increase in the amount of suberin in the roots, likely in the casparian strip. This increase in root suberin causes a decrease in transpiration. The change in permeability and transpiration is accompanied by decreases in leaf Ca, Mn and Zn and increases in S, K, As and Mo. The direction of the change in these elements is interpreted as evidence for which primary path they take to the xylem: elements which enter through the apoplast are less able to pass through the casparian strip while elements which enter through the cell to cell symplastic pathway become more concentrated in the slower moving xylem sap. This demonstrates how a single physiological change in the plant can impact the concentrations of multiple elements in the plant, even if they don't directly share transporters or chelators.
The tissue specificity of ion balance was also recently highlighted in a study of the seed ionome. Vreugdenhil et al. [13] measured Na, Mg, P, K, Ca, Mn, Fe and Zn in seeds of an A. thaliana Cvi × Ler Recombinant Inbred Line (RIL) population. In this experiment the levels of K, Ca, Mg, Mn and Zn were all significantly correlated with P. The strong correlation is expected because the main form of these cations in seeds is as a complex with the P containing compound phytate. In Arabidopsis, phytate is only found in the seeds, suggesting that this particular set of correlated elements is unlikely to be found in other tissues.
Environment Interactions
Baxter et al [3] measured the shoot ionome of Col-0 (the sequenced accession of A. thaliana) under conditions that induced known Fe deficient or P deficient responses. Fe deficient conditions produced significantly elevated concentrations of Mn, Co, Zn and Cd and decreased concentrations of Mo (Figure 1, Black arrows). P deficient conditions produced significantly increased concentrations of B, Zn and As and decreased concentrations of P, Cu and Co (Figure 1 blue arrows). Several known physiological responses to low Fe [14] are likely responsible for the ionomic signature: 1) The rhizospehere is acidified, which makes Mn, Fe, Co, Zn, and Cd more soluble and Mo less soluble, 2) Ferro-chelate reductase activity is upregulated as well as 3) The IRT1 transporter which has a lower specificity for Fe than other transporters and takes up Mn, Co, Zn and Cd. The molecular determinants of the alterations in the ionome caused by fluctuations in P are largely unknown. An increased uptake of As can occur through P transporters which are upregulated by P starvation. Nevertheless, even in the absence of a physiologically specified model, the ionomic signatures were robust enough that logistical regression models using ion levels as parameters could predict the Fe or P nutritional status of the plants. Changing the concentration of other components of the nutrient solution did not cause the models to incorrectly predict the Fe or P status of the growth conditions, suggesting that the models describe signatures are specific to Fe and P deficiency and robust to changes in the environment. This is a breakthrough because it demonstrates the nutritional status of a plant can be measured without measuring the element itself. This approach opens up the possibility of high-throughput profiling of the ionome to determine nutritional status for difficult to measure elements, and other physiological states. The two signatures do share two elements, and due to the formation of FePO4 precipitates, Fe and P homeostasis are intertwined. In fact, one response of A. thaliana to P deficiency (short roots) has recently been shown to actually be the result of Fe toxicity [15].
While these results show great promise for understanding how the ionome is regulated, clearly more work is needed. Changes in the mineral nutritional environment of plants growing in the field are not likely to be confined to a single component. The Fe and P models of Baxter et al., [3] would need to be recalibrated, perhaps adding or dropping elements, if the growth media were changed more significantly (eg. using a completely different type of soil or switching to liquid culture). As we move forward, more complex experiments that alter multiple components of the environment and integrate multiple genotypes will be required to elucidate the molecular underpinnings of these models.
Gene × Environment Interactions
In a recent study of ionomic variation, Neutron activation was used to measure the levels of up to 42 elements in 670 plant species collected from 29 sites [8]. For the macronutrients K, Mg and Ca, genetic variation at the family level had the largest contribution to the variance, while local effects, site to site variation, dominated for most of the other elements. In this study, significant correlations were observed between Ca and Mg as well as the Ca analogs Sr and Ba, and between K and its analogs Rb and Cs. Since these plants were sampled from their native habitat, the environment at each site contributed to the selection of which species were growing there, confounding the contributions of site and phylogeny in the observed variation.
Two recent studies have analyzed the combined effects of genotype and environment on the ionome by profiling structured genetic populations under different environments. Ghandilyian et al [16] measured Mg, P, K, Ca, Mn, Fe and Zn in the seeds of three different Arabidopsis RIL populations. For two of these populations, they also analyzed multiple tissues from plants grown in two different environments: soil and hydroponics. Most of the significant element to element correlations within a RIL population were only found in one environment. They also found very few strong correlations between the same element in different tissues, suggesting that there are tissue-specific factors controlling elements. In the seeds they identified several quantitative trait loci (QTL) hotspots, with QTLs for multiple elements (including one that colocalized with a phytate QTL), however most of the QTL locations in the leaves were for single elements. They also found, consistent with the poor reproducibility of element to element correlations, that there was very little correspondence between the QTLs identified in different environments. Broadley et al [17] attempted to identify gene by environmental interactions by measuring Ca and Mg in 355 cultivars of Brassica oleraeca, 71 F1 hybrid lines and a double haploid mapping population in the field and greenhouse under two different external phosphate conditions. Ca and Mg were highly correlated with each other, and had several colocalized QTL, but were found to not significantly vary between P conditions.
Ca and Mg
Comparison of the above studies points out an interesting contradiction in the inferred relationship between Ca and Mg. As noted above, in survey data Watanabe et al [8] and Broadley et al. observed significant correlations between Ca and Mg. Analysis of ionomic data for A. thaliana accessions and RIL populations in the PiiMS database (www.ionomicshub.org) [18] also shows strong correlations between Ca and Mg. For plants grown in soil therefore, Ca and Mg seem to be highly correlated in the leaves despite the fact that many known Ca transporters do not transport Mg. Contrasting with this result, the mutant screends of Lahner et al., [7] and Chen et al., [5] isolated many mutants where either Ca or Mg, but not both, was significantly altered (for example esb1). One explanation for this contradiction is that while mutant lines with altered Ca and Mg ratios are viable, they may have reduced fitness and fail to survive multiple generations with strongly affected Ca:Mg ratios. The extremely low Ca:Mg ratios of serpentine soils adaptation to these soils have been posited as a driver of speciation [19] suggesting that the Ca:Mg ratio is extremely important for plant viability. Interestingly, in a screen for Arabidopsis mutants that can grow on a serpentine-like Ca:Mg ratio, Bradshaw et al. [20] isolated a line with a T-DNA insertion in a Ca/H+ antiporter, Cax1. The cax1-1 line does not have altered Ca concentrations in leaves when grown on soil [21], but the cellular Ca distribution may be altered.
Identifying Genes Using Ionomics
As shown above, elements are rarely correlated with the same sets of other elements across tissue, environment or genotype. It is therefore unlikely that we will find sets of elements which are controlled by all of the same genes. To fully understand the complex regulation of the ionome, we will need to find the genes that control the accumulation and distribution of each element.
Ionomics should make the process of finding genes responsible for ionomic variation more efficient, as it allows for simultaneous mapping experiments to be conducted on the same segregating populations. In induced mutant populations, where a single mutant locus is mapped in each line, a multi-element phenotype is easier to map because the multiple correlated measurements will increase the confidence in classifying individuals as mutant or wildtype.
Natural variation in elemental levels has also led to gene identification [22-24]. In populations developed to identify natural alleles such as RILs, introduced above, using multiple elements to find loci has not been used. As the parents of these populations are chosen for their diversity, there are likely to be multiple loci controlling each element. As such, each element has been treated as an individual trait and scans for colocalized QTL have been used to determine which elements a given locus controls. This variation, however, creates some difficulty in mapping mutants. Typically mutants are mapped by crossing the mutagenized line to an accession with a large number of genetic differences from the mutant parent. Alleles from the outcross parent often affect one or more of the elements being mapped and make mapping more difficult by adding segregating variance and contributing to poor penetrance of the original mutant ionomic phenotype. For this reason the mapping partner needs to be considered carefully. For example, if an A. thaliana mutant in the Col-0 background were crossed to Ler, Mo could not be used in mutant identification due to the confounding affects of segregation at the Ler MOT1 locus [22]. Fortunately, technological improvements permit simultaneous measurements of thousands of markers, allowing any accession to be used as the outcross partner to rapidly map the causal locus.
Panels of diverse germplasm have been assembled in many plant species. The development of high throughput genotyping platforms and association mapping methodologies [25] has the potential to accelerate the process of identifying new loci which control the ionome. Utilizing panels of natural accessions that have been genotyped to extremely high resolution, association mapping has the potential to identify all of the common alleles which affect a trait, and narrow the mapping interval significantly when compared to QTL mapping. For example, in A. thaliana, due to the decay of linkage disequilibrium, association mapping should narrow the mapping window to ∼20kb [26]. With the narrower intervals, traits with associations that are colocalized are more likely to indicate a single locus controlling both traits than a colocalized QTL. As ionomics moves forward, the ionomic profiling of these diversity panels should begin to identify and describe the molecular basis of natural variation in elemental uptake and homeostasis.
Future Directions
With the exception of the broad survey of Watanabe et al., [8] most of the ionomics studies to date have been limited to a small number of species. Like most areas of study, for a full and useful understanding of the ionome we will need to expand these efforts to more species, as different species and families likely have different elemental uptake mechanisms. Guelke et al [27] showed that the isotope fractionation of Fe changes from stems to leaves to seeds in Fe uptake strategy I plants, but remains stable in strategy II plants. This indicates that there are different numbers of oxidation and reduction cycles in these plants, suggesting that in addition to the different uptake mechanisms, these species have fundamentally different Fe translocation and homeostasis mechanisms. Given how many elements are interrelated with Fe in the strategy II plant A. thaliana, these differences are likely to have implications for the entire ionome. Extending these analysis methodologies to crop plants as in Broadley et al. could allow the direct identification of nutritionally improved lines. From a basic research perspective, the rich dataset linking variety performance with field location from the agronomic discipline can be used to understand gene by environment interactions.
Conclusions
Elemental accumulation is a complex process that impacts almost every aspect of plant growth, development and survival. Mass spectroscopy technology has progressed to the point that multiple elements can easily be measured in a high throughput manner. Measuring multiple elements allows researchers to explore the dynamics of the ionome as a whole, not just individual elements in isolation. It also makes gene identification experiments more efficient, which should allow new genetic mapping techniques to identify hundreds of new loci that control this complex system.
Acknowledgments
The author was supported by the National Institutes of Health, National Institute of General Medicine (R01 GM078536-01A1), the National Science Foundation Arabidopsis 2010 Program (IOB 0419695) and Plant Genome Research Program (DBI 0701119), and the Indiana 21st Century Research and Technology Fund (912010479.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salt DE, Baxter I, Lahner B. Ionomics and the study of the plant ionome. Annu Rev Plant Biol. 2008;59:709–733. doi: 10.1146/annurev.arplant.59.032607.092942. [DOI] [PubMed] [Google Scholar]
- 2.Delhaize E, Randall P, Wallace P, Pinkerton A. Screening Arabidopsis for mutants in mineral nutrition. Plant and Soil. 1993;155/156:131–134. [Google Scholar]
- **3.Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE. The leaf ionome as a multivariable system to detect a plant's physiological status. Proc Natl Acad Sci U S A. 2008;105:12081–12086. doi: 10.1073/pnas.0804175105. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors use 5 and 6 element signatures to detect when the plants are experiencing low Fe or P conditions. The signatures are robust enough to be used to build models that predict the physiological state of the plant.
- 4.Chen Z, Shinano T, Ezawa T, Wasaki J, Kimura K, Osaki M, Zhu Y. Elemental interconnections in Lotus japonicus: a systematic study of the affects of elements additions on different natural variants. Soil Science & Plant Nutrition. 2009;55(1):91–101. [Google Scholar]
- *5.Chen Z, Watanabe T, Shinano T, Okazaki K, Osaki M. Ionomic Study of Lotus japonica. New Phytol. 2008;181(4):795–801. doi: 10.1111/j.1469-8137.2008.02730.x. [DOI] [PubMed] [Google Scholar]; The authors perfromed a large mutant screen in the legume Lotus Japonica and identified many interesting lines. This study demonstrates that a the ionomics approach can be used to identify interesting lines in species other than Arabidopsis.
- 6.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- *8.Watanabe T, Broadley MR, Jansen S, White PJ, Takada J, Satake K, Takamatsu T, Tuah SJ, Osaki M. Evolutionary control of leaf element composition in plants. New Phytol. 2007;174:516–523. doi: 10.1111/j.1469-8137.2007.02078.x. [DOI] [PubMed] [Google Scholar]; The authors conducted a broad survey of species from a variety of locations and measuring 42 elements.
- 9.Sors TG, Ellis DR, Na GN, Lahner B, Lee S, Leustek T, Pickering IJ, Salt DE. Analysis of sulfur and selenium assimilation in Astragalus plants with varying capacities to accumulate selenium. Plant J. 2005;42:785–797. doi: 10.1111/j.1365-313X.2005.02413.x. [DOI] [PubMed] [Google Scholar]
- 10.Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci U S A. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abedin MJ, Feldmann J, Meharg AA. Uptake kinetics of arsenic species in rice plants. Plant Physiol. 2002;128:1120–1128. doi: 10.1104/pp.010733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12.Baxter I, Hosmani P, Rus A, Lahner B, Borevitz J, Muthukumar B, Mickelbart M, Schreiber L, Franke R, Salt DE. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]; By using ionomics to identify and characterize a mutant with increased suberin in the root, the authors demonstrate how a physiological change can have a dramatic effect on the elemental accumulation of a plant.
- 13.Vreugdenhil D, Aarts MGM, Koornneef M, Nelissen H, Ernst WHO. Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ. 2004;27:828–839. [Google Scholar]
- 14.Kim SA, Guerinot ML. Mining iron: iron uptake and transport in plants. FEBS Lett. 2007;581:2273–2280. doi: 10.1016/j.febslet.2007.04.043. [DOI] [PubMed] [Google Scholar]
- **15.Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that a commonly used measure of the response to phosphate deficiency, a short root, is due to Fe toxicity. This highlights the importance of understanding the dynamics of the whole ionome, not just individual elements.
- **16.Ghandilyan A, Ilk N, Hanhart C, Mbengue M, Barboza L, Schat H, Koornneef M, El-Lithy M, Vreugdenhil D, Reymond M, et al. A strong effect of growth medium and organ type on the identification of QTLs for phytate and mineral concentrations in three Arabidopsis thaliana RIL populations. J Exp Bot. 2009;60:1409–1425. doi: 10.1093/jxb/erp084. [DOI] [PubMed] [Google Scholar]; The authors measure multiple elements in multiple tissues of RIL populations grown in different environments and find radically different sets of QTLs. This study demonstrates the complexity and variability of the networks of elemental relationships.
- *17.Broadley MR, Hammond JP, King GJ, Astley D, Bowen HC, Meacham MC, Mead A, Pink DA, Teakle GR, Hayden RM, et al. Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea. Plant Physiol. 2008;146:1707–1720. doi: 10.1104/pp.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using field and greenhouse grown plants, the authors demonstrate that Ca and Mg vary across the subtaxa of Brassica olerace. In a recombinant inbread population, they identify several QTL that control both elements. This is a nice example of applying elemental profiling to field grown plants.
- *18.Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. Purdue Ionomics Information Management System. An integrated functional genomics platform. Plant Physiol. 2007;143:600–611. doi: 10.1104/pp.106.092528. [DOI] [PMC free article] [PubMed] [Google Scholar]; A data management system for collecting ionomics data and the associated metadata. This type of system is essential for ionomics studies due to the large volume of data that they generate.
- 19.Kruckeberg A. California Serpentines: Flora, Vegitation, Geology, Soils and Management Problems. Berkeley, Ca, USA: University of California Press; 1982. [Google Scholar]
- 20.Bradshaw HD., Jr Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol. 2005;167:81–88. doi: 10.1111/j.1469-8137.2005.01408.x. [DOI] [PubMed] [Google Scholar]
- 21.Cheng NH, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, Lahner B, Salt DE, Hirschi KD. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 2005;138:2048–2060. doi: 10.1104/pp.105.061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, et al. Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1) PLoS Genet. 2008;4:e1000004. doi: 10.1371/journal.pgen.1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors use ionomics to identify and characterize a mitochondrial molybdenum transporter in Arabidopsis. A good example of the integration of ionomics with novel genetics and genomics techniques to identify genes.
- 23.Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet. 2007;39:896–900. doi: 10.1038/ng2050. [DOI] [PubMed] [Google Scholar]
- 24.Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006;2:e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao KY, Aranzana MJ, Kim S, Lister C, Shindo C, Tang CL, Toomajian C, Zheng HG, Dean C, Marjoram P, et al. An Arabidopsis example of association mapping in structured samples. Plos Genetics. 2007;3 doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 27.Guelke M, Von Blanckenburg F. Fractionation of stable iron isotopes in higher plants. Environ Sci Technol. 2007;41:1896–1901. doi: 10.1021/es062288j. [DOI] [PubMed] [Google Scholar]