Abstract
Previously, we showed that Abl kinases (c-Abl, Arg) are activated downstream of PDGF in a manner dependent on Src kinases and PLC-γ1, and promote PDGF-mediated proliferation and migration of fibroblasts. We additionally demonstrated that Abl kinases bind directly to PDGFR-β via their SH2 domains. In this study, we extend these findings by demonstrating that Abl kinases also are activated downstream of a PDGF autocrine growth loop in glioblastoma cells, indicating that the PDGFR-Abl signaling pathway also is likely to be important in glioblastoma development and/or progression. We recently showed that Abl kinases are highly active in many breast cancer cell lines, and the Her-2 receptor tyrosine kinase contributes to cAbl and Arg kinase activation. In this study, we show that Abl kinase SH2 domains bind directly to Her-2, and like PDGFR-β, Her-2 directly phosphorylates c-Abl. Previously, we demonstrated that PDGFR-β directly phosphorylates Abl kinases in vitro, and Abl kinases reciprocally phosphorylate PDGFR-β. Here, we show that PDGFR-β–phosphorylation of Abl kinases has functional consequences as PDGFR-β phosphorylates Abl kinases on Y245 and Y412, sites known to be required for activation of Abl kinases. Moreover, PDGFR-β phosphorylates Arg on two additional unique sites whose function is unknown. Importantly, we also show that Abl-dependent phosphorylation of PDGFR-β also has functional and biological significances. c-Abl phosphorylates three tyrosine residues on PDGFR-β (Y686, Y934, Y970), while Arg only phosphorylates Y686. Y686 and Y934 reside in PDGFR-β catalytic domains, while Y970 is in the C-terminal tail. Using site-directed mutagenesis, we show that Abl-dependent phosphorylation of PDGFR-β activates PDGFR-β activity, in vitro, but serves to downregulate PDGFR-mediated chemotaxis. These data are exciting as they indicate that Abl kinases not only are activated by PDGFR and promote PDGFR-mediated proliferation and migration, but also act in an intricate negative feedback loop to turn-off PDGFR-mediated chemotaxis.
Keywords: Abl kinases, PDGFR, Her-2, chemotaxis
1. INTRODUCTION
The Abl family of non-receptor tyrosine kinases (Abl kinases) includes two proteins, cAbl and Arg, which are encoded by Abl1 and Abl2 genes, respectively. The two kinases are highly homologous in their N-termini, which contain SH3, SH2 and kinase domains, but are more divergent in their C-termini, as c-Abl contains nuclear localization signals and a DNA binding domain, that are absent in Arg [1]. Both proteins contain myristoylation signals that target the proteins to the plasma membrane. Thus, c-Abl and Arg are both localized to the plasma membrane and cytoplasm, whereas c-Abl also is localized in the nucleus [1]. Subcellular localization of c-Abl is important for its function, as activation of c-Abl in the nucleus induces apoptosis, while activation of the cytoplasmic/membrane pool promotes proliferation and migration [1, 2]. Abl kinases are negatively regulated by intramolecular interactions: the kinase domain binds the myristoyl residue, and the SH3 domain interacts with the interlinker region (between SH2 and kinase domains) [3, 4]. Mutations that disrupt these interactions activate the kinases, producing oncogenic proteins that transform many cell types [4]. c-Abl activity is dramatically increased following purification and high level overexpression, which suggests that a soluble inhibitor keeps c-Abl in an inactive state [4]. In addition, tyrosine phosphorylation of c-Abl in the activation loop of the kinase domain (Y412) and in the interlinker region (Y245) is required for full kinase activity [4,24]. Src family kinases directly phosphorylate these residues and induce activation of Abl kinases [4,24].
The kinase activities of c-Abl and Arg are increased by extracellular stimuli such as cytokines, growth factors and integrins [1]. We showed that activation of PDGFR (platelet-derived-growth factor receptor) and EGFR (epidermal-growth factor receptor) stimulates the kinase activities of the cytoplasmic/membrane pools of c-Abl and Arg in fibroblasts [5]. In addition, we demonstrated that PDGF-mediated activation requires Src kinases, which directly phosphorylate and activate Abl kinases, and PLC-γ1, which may release negative regulation by hydrolyzing a potential Abl inhibitor, PIP2 [5, 6]. Importantly, we showed that activation of Abl kinases downstream of PDGFR has relevant biological consequences, as Abl kinases are required for PDGF-mediated proliferation, membrane ruffling, and migration [5, 6]. Abl kinases promote proliferation by activating Rac/NADPH oxidase (NOX) and SHP-2/ERK-dependent pathways [8, 9], and promote membrane ruffling and PDGF-induced migration in a Rac- or PLC-γ1 dependent manner, respectively [6, 10]. In addition to the requirement of Src kinases and PLC-γ1 in activation of Abl kinases downstream of PDGFR-β, we also showed that PDGFR-β binds directly to Abl kinases and phosphorylates c-Abl and Arg [7]. Interestingly cAbl and Arg also reciprocally phosphorylate PDGFR-β [7]. However, until now, the consequences of bidirectional Abl-PDGFR phosphorylation events have not been elucidated.
Abl kinases are most known for their involvement in human leukemia. Abl1 is translocated next to BCR forming a constitutively active BCR-Abl fusion protein, which drives the development of CML (chronic myelogenous leukemia) [11]. c-Abl and Arg are also translocated next to Tel in other forms of leukemia and myeloproliferative disease, and the cAbl gene is amplified in T-cell acute lymphocytic leukemia (ALL) [11–13]. Recently, we showed that Abl kinases also are activated in solid tumor cell lines, as Abl kinases are highly active in invasive breast cancer cells [14]. Additionally, we demonstrated that the mode of activation of Abl kinases in breast cancer cells is different from their activation in leukemia; c-Abl and Arg are activated downstream of deregulated tyrosine kinases such as EGFR, ErbB2/Her-2, IGF-1R, and Src in breast cancer cells, rather than being activated by chromosomal translocation as they are in leukemia [14]. It is not known whether Abl kinases also are activated downstream of PDGFR in solid tumors. Significantly, we showed that activation of Abl kinases in breast cancer cells promotes proliferation, survival, and invasion [14, 15]. Recently, Arlinghaus and colleagues demonstrated that Abl kinases also are activated in another type of solid tumor, non-small cell lung cancer, via another mechanism; loss of expression of an Abl kinase negative regulator, Fus1 [16].
In this report, we extend our previous results by demonstrating a number of novel findings. First, we show that in addition to binding and being phosphorylated by PDGFR-β in fibroblasts, Abl kinases also are phosphorylated by Her-2 derived from breast cancer cells, and are activated downstream of deregulated PDGFR in glioblastoma cells. Moreover, we demonstrate that the bidirectional phosphorylation of PDGFR-β and Abl kinases in fibroblasts has functional consequences, as PDGFR-β phosphorylates Abl kinases on two sites that regulate c-Abl and Arg activity, and Abl kinases phosphorylate PDGFR-β on several residues. Mutation of Abl-dependent phosphorylation sites results in PDGF receptors that have decreased in vitro catalytic activity. Significantly, Abl-dependent phosphorylation of PDGFR-β has biological consequences, as mutation of the phosphorylation sites results in receptors that have an increased ability to induce chemotaxis, which indicates that Abl-dependent phosphorylation of PDGFR-β acts in a negative feedback loop to downregulate PDGFR-β–mediated chemotaxis.
2. MATERIALS AND METHODS
2.1. Antibodies, Reagents, Plasmids, and Cell Lines
Ph cells, which lack endogenous PDGFR-α, 293T cells, and BT-474 cells have been described [14, 17]. SKBR3 cells were obtained from the University of North Carolina, Tissue Culture Facility (Chapel Hill, NC). SF9 cells were maintained in Grace’s insect medium containing 10% heat-inactivated fetal bovine serum (Invitrogen, Carlesbad, CA). U87 glioblastoma cells transfected with vector or PDGF dominant-negatives were previously described [27]. Antibodies directed against c-Abl (K12-immunoprecipitations, Santa Cruz Biotechnologies, Santa Cruz, CA; 8E9-western blotting, BD Biosciences, Chicago, IL), GST, and Her-2 (Santa Cruz Biotechnologies, Santa Cruz, CA) were purchased commercially. Antibodies directed against the extracellular domain of PDGFR-α (80.8), or C-terminal domain of Arg were described previously [7, 17]. PDGF-AA was obtained from Upstate Biotechnologies (Lake Placid, NY), and EGF was purchased from Roche Corp. (Indianapolis, IN). c-Abl, PDGFR-β (pMT3-PDGFR-β, pMT3-PDGFR-β-K634R), pLXSN2-PDGFR-α/β, and Arg constructs were described previously [6, 7, 17, 19, 20]. GST-Crk and GST fusion proteins containing fragments of c-Abl and Arg were previously described [7, 19, 21].
2.2. Transfection, Retroviral Infection, SF9 Infection
293T cells were transfected for 5–8 hours using calcium phosphate (15 μg DNA, 62 μl calcium chloride, 500 μl Hepes Buffered Saline, and 438 μl water per 60 mm dish) in medium containing 0.1 mM chloroquine. For retroviral infection, 293T cells were transfected with 10μg expression and 5μg pSVψ2 plasmids as above, two days after transfection, the media was filtered through a 0.45μM filter, and placed on fibroblasts for 4 hours in the presence of 4μg/ml polybrene (3ml/100mm dish). SF9 cells were infected as we previously described [19]. Baculovirus for GST-Abl-SH2-SH1-KR and the procedure for infecting SF9 cells was described previously [7, 19, 22].
2.3. Immunoprecipitations, GST-pulldowns, kinase assays, immunoblotting, and far western analyses
Procedures were previously described [5–7, 14]. For immunoprecipitations, kinase assays, GST-pulldowns, and far western blots, cells were lysed in kinase lysis buffer (50mM HEPES pH 7, 150 mM NaCl, 10% glycerol, 1% Triton-X-100, 1.5 mM MgCl2, 1mM EGTA, and inhibitors (1mM PMSF, 1mM sodium orthovanadate, 25mM sodium fluoride, 10 μg/ml leupeptin, aprotinin, pepstatin)), and RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton-X 100, 0.1% SDS, 1% sodium deoxycholate, inhibitors) was used for to prepare lysates for phospho-blots. For kinase assays, c-Abl or Arg immunoprecipitates were washed twice in RIPA buffer, twice in NaCl buffer (10 mM Tris, pH 7.4, 5 mM EDTA, 1% Triton-X-100, 100 mM NaCl, inhibitors), twice in the previous buffer lacking NaCl, and twice in kinase buffer (20 mM Tris pH7.4, 10 mM MgCl2, 1 mM DTT), and incubated for 40′ at room temperature in kinase buffer containing 1 μM cold ATP, 5 μCi 32P-γ-ATP and 1 μg substrate. For GST-pulldowns, 1 μg of GST-tagged fusion proteins was incubated with cellular lysate and glutathione-sepharose, and precipitates were probed with anti-Her-2 antibody. For far westerns, blots containing Her-2 immunoprecipitates, were blocked in 5% BSA in 50mM Tris-HCl, pH 7.4, 150mM NaCl, incubated with GST-fusion proteins (2μg/ml) in 20 mM Hepes, pH 7.2, 150 mM NaCl, 0.1% Triton-X 100, 10% glycerol, 2% BSA, and GST binding was assessed by western blot with anti-GST antibody. Kinase assays were quantitated on a Storm phosphoimager (Molecular Dynamics, GE Healthcare; Piscataway, NJ).
2.4. Mass Spectrometry
For in vitro studies, a PDGFR-β expression construct (pMT3-PDGFR-β)[20] was transfected into 293T cells, PDGFR-β was immunoprecipitated, and incubated with soluble GST-fusion proteins in the presence of 1 mM ATP, at 37°C for 30 minutes [19]. For identifying PDGFR-β phosphorylation sites on Abl kinases in vivo (in cells), SF9 cells were infected with PDGFR-β and GST-Abl-SH2-SH1-K290R (kinase-inactive) baculovirus, and GST-Abl SH2-SH1-K290R was precipitated with glutathione sepharose. To identify phosphorylation sites on PDGFR-β induced by Abl kinases, a kinase-inactive PDGFR-β (K634R) was coexpressed with c-Abl or Arg in 293T cells, and PDGFR-β-K634R was immunoprecipitated from the lysates. Kinase reactions, glutathione sepharose precipitates, and immunoprecipitates were run on SDS-PAGE gels, the gels were Coomassie-stained, bands were removed from the gels, and subjected to mass spectrometry analysis. Mass spectrometry was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ DECA XP quadrupole ion trap mass spectrometer.
2.5. Site-Directed Mutagenesis
pLXSN2-PDGFR-α/β was mutated using Quikchange (Stratagene, La Jolla, CA) using the following mutagenesis primers:
Y686F-forward: GAGTACTGCCGCTTCGGAGACCTGGTG
Y686F-reverse: CACCAGGTCTCCGAAGCGGCAGTACTC
Y934F-forward: CCTCCGACGAGATCTTTGAGATCATGCAGAAG
Y934F-reverse: CTTCTGCATGATCTCAAAGATCTCGTCGGAGG
Y970F-forward: GGTTACAAAAAGAAGTTCCAGCAGGTGGATGAGG
Y970F-reverse: CCTCATCCACCTGCTGGAACTTCTTTTTGTAACC
All constructs were completely sequenced following mutagenesis to confirm that no additional mutations were introduced by the PCR reaction.
2.6. Chemotaxis Assays
Ph cells were infected with undiluted retroviruses, and serum-starved in DMEM (no serum), two days after infection for 24 hours. Both sides of the migration membranes (0.8μm; Thermo Fisher; Waltham, MA) were coated with collagen (100ng/ml) for 2 hours at 37°C. Chambers were washed 2X with DMEM, and cells (5×105 cells/ml; 200μl) in DMEM/1% bovine serum albumin (BSA) were loaded in the top chamber above a bottom chamber containing vehicle or PDGF-AA (100ng/ml) in DMEM/1% BSA. Cells were allowed to migrate for 4–6 hours at 37°C, cells on the upper surface of the membrane were removed, and cells on the undersurface were fixed, stained (Diff-Quik kit; Fisher; Hampton, NH), and mounted on microscope slides. Fifteen to twenty random 40X fields were counted for each membrane.
3. RESULTS
3.1. Abl kinase SH2 domains interact with Her-2/ErbB2, and Her-2 directly phosphorylates c-Abl
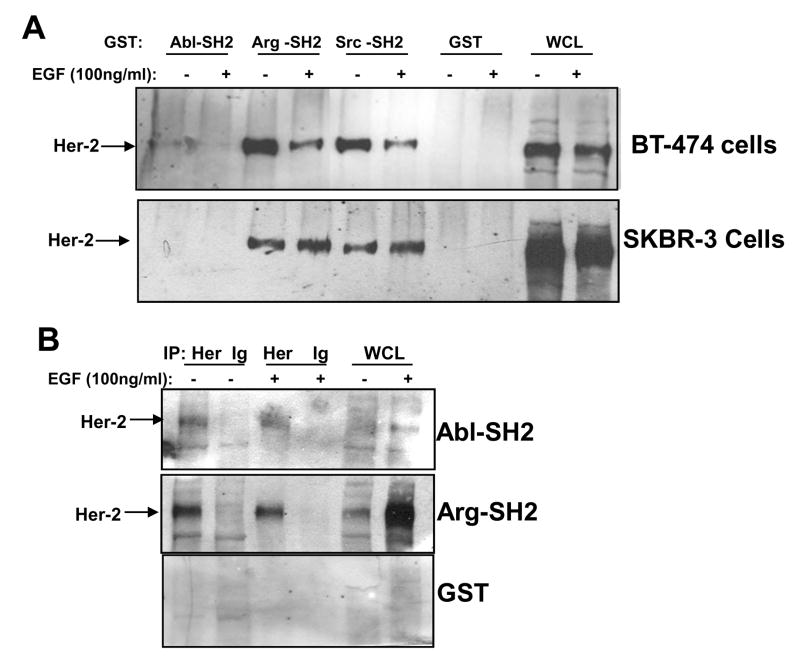
Previously, we showed that Abl kinases form a complex with PDGFR-β in fibroblasts and EGFR in breast cancer cells [7, 14]. Abl kinase SH2 domains bind the phosphorylated intracellular domain of both receptors, and the binding is direct and does not involve intermediate proteins [7, 14]. Her-2/ErbB2 is a receptor tyrosine kinase in the EGFR family that is involved in breast cancer disease progression, and previously, we showed that Abl kinases are activated downstream of Her-2 in breast cancer cells [14]. Thus, we evaluated whether Abl kinase SH2 domains bind Her-2 derived from breast cancer cells. GST-fusion proteins containing fragments of c-Abl or Arg were incubated with lysates obtained from BT-474 (express Her-2) and SKBR3 (express Her-2 and overexpress EGFR) breast cancer cells that naturally express Her-2 [14]. The Arg SH2 domain bound strongly to Her-2 derived from lysates from both cell lines (Fig. 1A). In contrast, the Abl-SH2 domain did not bind Her-2 from SKBR3 lysate, and bound Her-2 derived from BT-474 cells very weakly. Interestingly, binding of the Arg-SH2 domain to Her-2 derived from EGF-stimulated BT-474 cells was decreased as compared to the binding of the SH2 domains to Her-2 isolated from unstimulated cells (Fig. 1A). In contrast, the Arg-SH2 domain interacted better with Her-2 isolated from EGF-stimulated SKBR3 cells as compared to Her-2 from unstimulated SKBR3 lysates (Fig. 1A). Her-2 phosphorylation was decreased in EGF-stimulated BT-474 cell lysates (data not shown), which explains why there may be decreased binding of the Arg-SH2 domain. However, it is unclear why Her-2 phosphorylation is decreased in BT-474 cells following EGF stimulation. To determine whether c-Abl and Arg SH2 domains directly bind Her-2, Far Western analyses were performed. Consistent with the pulldown studies, the Arg SH2 domain bound strongly to immobilized, immunoprecipitated Her-2, whereas binding of the c-Abl SH2 domain to Her-2 was fairly weak (Fig. 1B). Taken together, these data indicate that the Abl kinase SH2 domains directly bind a variety of receptor tyrosine kinases including Her-2.
Figure 1. Abelson kinase SH2 domains interact with ErbB2/Her-2.
(A) BT-474 and SKBR3 cells were serum-starved, stimulated with EGF (100ng/ml) for the indicated times, lysed, lysates were incubated with c-Abl and Arg GST-fusion proteins and glutathione sepharose, and precipitates were probed with a Her-2 antibody (Santa Cruz; sc-294). (B) Her-2 was immunoprecipitated from BT-474 lysates, run on SDS-PAGE gels, and blots were incubated with GST-fusion proteins followed by GST primary antibody, and HRP-conjugated secondary antibody. The experiments shown are representative of three independent experiments.
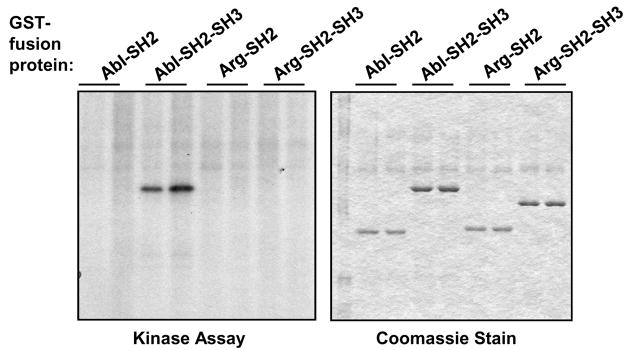
We previously showed that PDGFR-β phosphorylates c-Abl and Arg in vitro [7]. In addition, we showed that Abl kinases are activated downstream of EGFR, Her-2, and IGF-1R in some breast cancer cells [14]. Moreover, Her-2 directly binds Abl kinase SH2 domains (Fig. 1). Hence, we tested whether Her-2 is capable of directly phosphorylating c-Abl and Arg. Her-2 was immunoprecipitated from BT-474 cells, immunoprecipitates were washed with a stringent series of buffers, and incubated with GST-fusion proteins containing fragments of c-Abl or Arg in an in vitro kinase assay. Interestingly, although the Arg SH2 domain bound strongly to Her-2 (Fig. 1A), it was not phosphorylated by Her-2 and neither was an Arg-SH2-SH3 fusion protein (Fig. 2, left). Conversely, a fragment containing the c-Abl-SH2-SH3 domains was efficiently phosphorylated by Her-2, while a fragment containing only the c-Abl-SH2 domain was not phosphorylated (Fig. 2, left). These data indicate that the c-Abl-SH2-SH3 domain is phosphorylated by Her-2, and Arg is either not phosphorylated by Her-2 or is phosphorylated in another region not present in the GST-fusion fragments. Therefore, although the c-Abl SH2 domain did not bind strongly to Her-2 in our in vitro studies, Her-2 efficiently phosphorylated a c-Abl fragment containing the SH2 domain (SH2-SH3), in vitro. In contrast, the Arg SH2 domain bound tightly to Her-2, but fragments containing the SH2 domain were not phosphorylated. Kinase/substrate interactions are short-lived because the substrate is released following phosphorylation, which may explain why the Arg-SH2 domain bound better than the c-Abl-SH2 domain to Her-2; the Arg-SH2 domain binds Her-2 but is not phosphorylated by Her-2 and thus is not released from the receptor. The data obtained with Her-2 are interesting because they differ from the data obtained with PDGFR-β. PDGFR-β phosphorylated GST-fusion proteins containing c-Abl-SH2, c-Abl-SH2-SH1-KR (a fragment containing the SH2 domain and a kinase-inactive kinase domain), c-Abl-SH2-SH3, and Arg-SH2-SH3 but not Arg-SH2 [7]. Taken together, these data indicate that Her-2-dependent phosphorylation of c-Abl differs from PDGFR-β-dependent phosphorylation, as PDGFR-β likely phosphorylates a residue in the Abl-SH2 fragment that is not phosphorylated by Her-2. In addition, Her-2 either does not phosphorylate Arg, or phosphorylates Arg in an area outside the SH2-SH3 domains, whereas PDGFR-β phosphorylates a residue(s) within those domains [7].
Figure 2. Her-2 phosphorylates c-Abl in vitro.
Her-2 was immunoprecipitated from BT-474 lysates (Santa Cruz; sc-294), in duplicate, and incubated in an in vitro kinase assay withvarious GST-fusion proteins, and 32P-γ-ATP. Reaction mixtures were run on SDS-PAGE gel, dried, stained with Coomassie blue (right) and exposed to film (left). The experiment shown is representative of three independent experiments.
3.2. Abl kinases are activated by autocrine PDGF signaling in a glioblastoma cell line
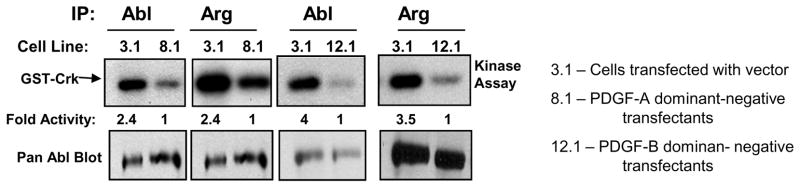
Previously, we showed that Abl kinases are highly activated downstream of ErbB, IGF-1R, and Src kinases in invasive breast cancer cells [14]. In addition, we showed that Her-2 and PDGFR bind and phosphorylate Abl kinases. Thus, to determine whether Abl kinases may also be activated in a different solid tumor, downstream of PDGFR, we assessed whether Abl kinases are activated in a glioblastoma cell line that contains a PDGF autocrine growth loop (U87)[25]. Many brain cancers contain PDGF autocrine growth loops, as they secrete PDGF and simultaneously express PDGF receptors [26]. To determine whether Abl kinases are activated in glioblastoma cells containing deregulated PDGFR, we utilized U87 glioblastoma cells stably transfected with vector (3.1) or dominant-negative forms of PDGF-A (8.1) or PDGF-B (12.1) [27]. Expression of either PDGF dominant-negative ligands blocks PDGF autocrine signaling [27]. Significantly, we found that the activities of both c-Abl and Arg were dramatically reduced in cells expressing PDGF dominant-negatives as compared to vector control cells (Fig. 3). These data indicate that Abl kinases are activated in glioblastoma cells via a similar mechanism to what we observed in breast cancer cells; downstream of a deregulated receptor tyrosine kinase. Taken together, these data indicate that activation of Abl kinases by deregulated receptor tyrosine kinases is a general feature of several types of solid tumors, and is different from the mechanism by which Abl kinases are activated in leukemia (ie. translocation).
Figure 3. Inhibition of constitutive PDGF signaling decreases c-Abl and Arg kinase activity in U87 glioblastoma cells.
U87 cells expressing vector (3.1), a PDGF-A dominant-negative (R159E/K160E/K161E)(8.1), or a PDGF-B dominant-negative (R160E/K161E/K162E) (12.1) were grown for 3 days, lysed, and the activity of c-Abl and Arg were assessed by in vitro kinase assay using GST-Crk substrate (top). Lysates were blotted with a pan-Abl antibody (8E9; bottom). The experiment shown is representative of three independent experiments.
3.3. PDGFR-β phosphorylates Abl kinases on residues necessary for full activation
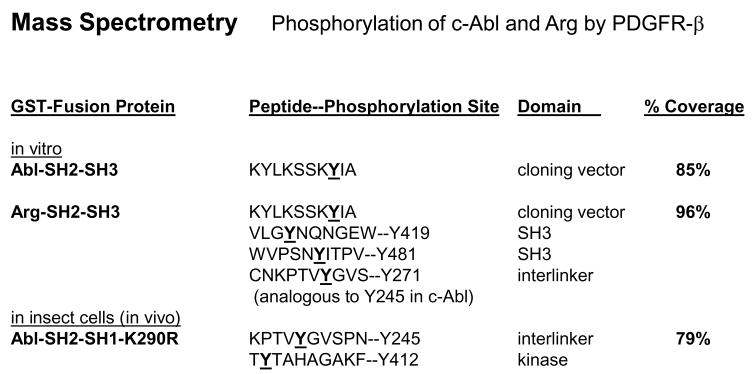
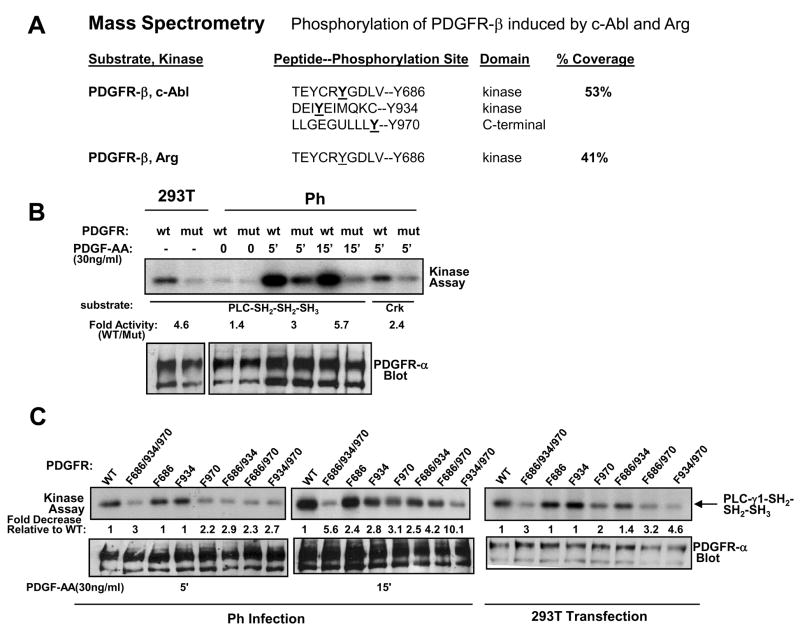
To identify the tyrosine residues on c-Abl and Arg phosphorylated by PDGFR-β, we first mapped the sites on c-Abl- SH2-SH3 and Arg-SH2-SH3 domains, phosphorylated by PDGFR, in vitro. PDGFR-β was highly expressed in 293T cells, immunoprecipitated, and incubated with GST-fusion fragments of c-Abl and Arg in an in vitro kinase assay. Extremely high expression of receptor tyrosine kinases promotes receptor dimerization and activation in the absence of ligand [23]. Phosphorylated GST-fusion proteins were removed from the Coomassie-stained gel and subjected to mass spectrometry. We were unable to detect PDGFR-dependent phosphorylation of the c-Abl-SH2-SH3 fragment by mass spectrometry, except for a tyrosine residue that originated from the pGEX cloning vector (Fig. 4; Supplemental Fig. 1). However, PDGFR-β phosphorylated the Arg-SH2-SH3 protein on Y271, a site analogous to c-Abl Y245, as well as on two unique sites within the Arg-SH3 domain (Fig. 4; Supplemental Fig. 2). To identify the phosphorylation sites on c-Abl and Arg induced by PDGFR-β in cells, we coexpressed PDGFR-β and kinase-inactive forms of c-Abl or Arg (c-Abl-SH2-SH1-K290R, full-length c-Abl-KR or Arg-KR) in SF9 cells. Although full length c-Abl-KR and Arg-KR were phosphorylated in the presence of PDGFR-β [7], we were unable to identify tyrosine phosphorylation sites in the proteins by mass spectrometry (data not shown). However, we were able to detect PDGFR-β-dependent phosphorylation sites on the smaller c-Abl-SH2-SH1-KR fragment, and identified the sites as Y245 and Y412 (Fig. 4; Supplemental Fig. 3). Both Y245 and Y412 are important tyrosine residues in the Abl proteins, as phosphorylation of Y412 followed by phosphorylation on Y245 is required for c-Abl activation [24]. These data are extremely significant, as they indicate that PDGFR-β phosphorylates Abl kinases on tyrosine residues known to activate Abelson activity, which was demonstrated using both in vitro and in vivo (in cells) methods: c-Abl (Y245, Y412; SH2-SH-1-KR; in cells) and Arg (Y271; SH2-SH3; in vitro). In addition, PDGFR-β also phosphorylates Arg on two additional SH3 domain tyrosine residues (Y419, Y481), whose function is unknown.
Figure 4. PDGFR-β phosphorylates Abl kinases on Y245 and Y412.
PDGFR-β was immunoprecipitated, incubated with soluble GST-c-Abl-SH2-SH3 and GST-Arg-SH2-SH3 in an in vitro kinase assay, and phosphorylated GST-fusion proteins were analyzed by mass spectrometry. For the in vivo (in cells) study, SF9 cells were infected with PDGFR-β and GST-c-Abl-SH2-SH1-K290R baculovirus, GST-c-Abl-SH2-SH1-K290R was precipitated with glutathione sepharose, and analyzed by mass spectrometry. Percent coverage refers to the percentage of the protein recovered as peptides.
3.4. Abl kinases induce tyrosine phosphorylation of PDGFR-β
Previously, we showed that Abl kinases directly bind PDGFR-β, and promote proliferation and migration downstream of PDGFR in fibroblasts [5, 28]. We also showed that, in addition to PDGFR-β phosphorylation of Abl kinases, Abl kinases reciprocally induce phosphorylation of PDGFR-β [7]. To determine the sites on PDGFR-β phosphorylated in an Abl kinase-dependent manner, c-Abl and Arg were coexpressed with a kinase-inactive form of PDGFR-β (K634R) in 293T cells. High-level overexpression of c-Abl and Arg in 293T cells activates the kinases [7, 29]. PDGFR-β-K634R was immunoprecipitated, removed from the Coomassie stained gel, and tyrosine phosphorylation sites were mapped by mass spectrometry. We found that c-Abl induced phosphorylation of PDGFR-β on three sites (Y686, Y934, Y970), while Arg induced phosphorylation of PDGFR-β on one site (Y686)(Fig. 5A; Supplementary Figs. 4,5). Two of the phosphorylation sites (Y686, Y934) are located in the catalytic domains of the receptor, while the third phosphorylation site (Y970) resides in the C-terminal autophosphorylation domain. Peptides containing Y934 and Y970 were obtained for PDGFR-β phosphorylated by Arg; however those peptides were not phosphorylated (Supplementary Figs. 4,5), which indicates that Arg may specifically phosphorylate only Y686. However, we cannot rule out the possibility that Y934 and Y970 phosphorylation merely was not detected due to lower overall phosphorylation of the protein (Fig. 5A), and/or due to the decreased number of those peptides isolated.
Figure 5. Abl kinases phosphorylate PDGFR-β and increase its kinase activity.
(A) PDGFR-β-K634R was coexpressed with c-Abl or Arg in 293T cells, PDGFR-β was immunoprecipitated, and subjected to mass spectrometry analysis. Percent coverage refers to the percentage of the protein recovered as peptides. (B, C) Abl-dependent phosphorylation sites in a chimeric PDGFR-α/β (Y686, Y934, Y970) receptor were mutated to phenylalanine by site-directed mutagenesis. Wild-type (wt) and triple mutant (mut) receptors were transfected into 293T cells, and diluted retrovirus (1:3) from the transfected 293T cells was used to infect Ph cells, which lack endogenous PDGFR-α. Infected Ph cells were starved and stimulated with PDGF-AA (30ng/ml) for the indicated times. Chimeric receptors were immunoprecipitated from the lysates using antibody raised against the extracellular domain of PDGFR-α (80.8), and the immunoprecipitates were incubated an in vitro a kinase assay with GST-PLC-SH2-SH2-SH3 or GST-Crk substrates. Lysates were blotted with PDGFR-α (80.8) antibody. Experiments shown are representative of three independent experiments.
3.5. Abl kinase-dependent phosphorylation of PDGFR-β increases its kinase activity
To determine the functional significance of Abl-dependent phosphorylation of PDGFR, we mutated the tyrosine phosphorylation sites in a chimeric PDGFR to phenylalanine by in vitro mutagenesis (triple mutant; Y686F/Y934F/Y970F). Chimeric receptors contain the extracellular domain of PDGFR-α fused to the intracellular domain of PDGFR-β. These receptors are activated by PDGF-AA, due to the presence of the extracellular domain of PDGFR-α, but induce PDGFR-β signaling events because the intracellular domain is derived from PDGFR-β [17]. First, wild-type and triple mutant chimeric receptors were overexpressed in 293T cells, and the activity of the receptors was compared by in vitro kinase assay using a physiological substrate of PDGFR, PLC-γ1. High-level overexpression of the wild-type chimeric receptor in 293T cells induced activation and autophosphorylation of the receptor in the absence of PDGF stimulation (Fig. 5B, left). Mutation of all three Abelson phosphorylation sites (mut) resulted in receptors with 5-fold lower activity as compared to wild-type, when overexpressed in 293T cells (Fig. 5B, left). Next, the chimeric receptors were expressed in fibroblasts lacking PDGFR-α (Ph cells) by retroviral infection using diluted virus (1:3). PDGF-AA stimulation of Ph cells-expressing chimeric receptors activates PDGFR-β signaling via the chimeric receptors without activating endogenous PDGFR-β receptors, since endogenous PDGFR-β cannot be activated by PDGF-AA and requires PDGF-BB for activation. Low-level expression of the wild-type chimeric receptor in Ph fibroblasts did not result in ligand-independent activation; however, PDGF-AA activated the chimeric receptors (Fig. 5B, right). Interestingly, the triple mutant receptor had a decreased capacity to phosphorylate PLC-γ1 following activation by PDGF-AA (3–6 fold less activity), and also had a decreased ability to phosphorylate a second substrate, GST-Crk (Fig. 5B, right). Taken together these data indicate that the triple mutant PDGFR has decreased in vitro activity, which implies that Abl kinase-dependent phosphorylation of PDGFR increases the activity of PDGFR-β.
To determine the effect of each Abl-dependent phosphorylation site on chimeric PDGFR activity, we created single and double mutants in the background of the chimeric PDGFR, and assessed activity of the chimeric receptors as in Fig. 5B. In 293T cells, in unstimulated conditions, and in Ph cells stimulated with PDGF-AA for a short time (5′), only the Y970F single mutant had decreased activity compared to wild-type, while Y686F and Y934F activities were similar to wild-type (Fig. 5C). However, following 15 minutes PDGF-AA stimulation, all of the single-mutants had decreased activity compared to wild-type (Fig. 5C). These data indicate that all three phosphotyrosines are important for sustained PDGF-stimulated activity of PDGFR-β. In addition, mutation of any combination of two residues significantly reduced PDGFR activity when the mutant receptors were expressed in unstimulated 293T cells as well as when they were expressed in Ph cells stimulated with PDGF-AA for 5 or 15 minutes (Fig. 5C). Interestingly, the Y934F/Y970F mutant had the least activity of the double mutants, and mutation of all three tyrosines resulted in a receptor with the least in vitro activity (Fig. 5C). Taken together, these data indicate that all three phosphorylation sites contribute to PDGFR-β activity, in vitro.
3.6. Abl-dependent phosphorylation of PDGFR-β downregulates PDGF-induced chemotaxis
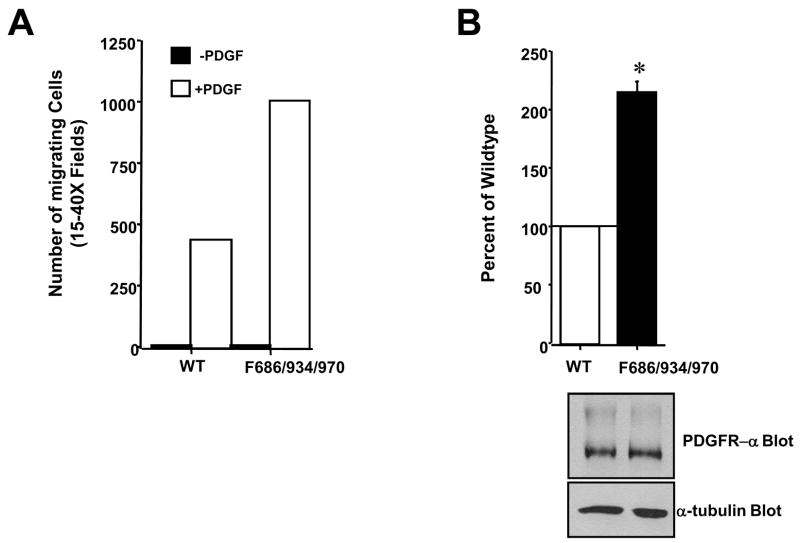
To determine the biological consequence of Abl-dependent phosphorylation of PDGFR-β, we assessed whether mutation of Abl-dependent phosphorylation sites affected PDGF-induced chemotaxis. Unexpectedly, we found that expression of a mutant chimeric receptor lacking the three Abl phosphorylation sites (triple mutant; Y686F/Y934F/Y970F) resulted in cells that had a significantly increased ability to migrate towards PDGF-AA (Fig. 6). These data indicate that although mutation of Abl-dependent phosphorylation sites results in lower in vitro activity, the mutant receptors have an increased ability to induce chemotaxis. In addition, these data suggest that phosphorylation of PDGFR-β by Abl kinases serves as part of a negative feedback loop, downregulating PDGFR-β-mediated chemotaxis.
Figure 6. Abl-dependent phosphorylation of PDGFR inhibits PDGFR-induced chemotaxis.
Ph cells were infected with undiluted retrovirus encoding wild-type or triple mutant (F686/934/970) chimeric receptors, placed in the top well of migration chambers with PDGF-AA (100ng/ml) in the bottom chamber. Cells were allowed to migrate 4–6 hours, and cells on the undersurface of the membrane were fixed, stained, and 15–20 40X fields were counted. (A) One representative experiment is shown. (B) Mean ±s.e.m of three independent experiments. The number of migrated cells from wells lacking PDGF were subtracted from the number of migrated cells from wells containing PDGF, and expressed as a percentage of the number of migrated wild-type receptor-expressing cells. *p=0.002 using a t-test. Cells remaining from set-up of the migration assay were lysed and blotted with antibodies to α-tubulin and PDGFR-α. A representative experiment is shown.
To determine whether particular signaling pathways are differentially activated by the mutant chimeric receptors, we infected Ph cells with diluted virus encoding wild-type and triple mutant receptors, starved and stimulated the cells with PDGF-AA, and blotted with phospho-specific antibodies to ERK1/2, AKT, SHP-2 (Y580), and PLC-γ1 (Y783). We were unable to detect any significant, reproducible differences in phosphorylation of any of these proteins (data not shown). These data indicate that Abl-dependent phosphorylation of PDGFR is likely to affect other, as yet unknown signaling pathways to mediate chemotaxis. To determine whether Abl dependent phosphorylation of PDGFR affects mitogenesis, we tested the ability of wild-type and triple mutant receptors to induce mitogenesis. However, PDGF-AA stimulation of Ph cells expressing wild-type or mutant chimeric receptors was not sufficient to induce mitogenesis, even with high concentrations of PDGF-AA (50ng/ml; data not shown), and thus this biological outcome could not be assessed.
4. DISCUSSION
In this report, we demonstrate that Abl kinases are phosphorylated by PDGFR-β on sites that are required for Abl kinase activation (Y412, Y245). In addition, we show that Abl kinases phosphorylate PDGFR-β on several sites: Y686 is in the proximal catalytic domain, Y934 is in the distal catalytic domain, and Y970 resides in the C-terminus. Other non-receptor tyrosine kinases also have been shown to phosphorylate receptor tyrosine kinases. Src phosphorylates EGFR in C-terminal (Y1101) and catalytic domains (Y845), and phosphorylation of Y845 by Src promotes EGFR-mediated DNA synthesis [30]. Although mutation of Y845 to phenylalanine inhibits DNA synthesis, phosphorylation of downstream proteins SHC and ERK1/2 are unaffected by loss of this phosphorylation site [32]. Additionally, Src kinases phosphorylate PDGFR-β on Y934, which creates a binding site for phosphoinositide 3-kinase (PI-3-kinase), and the Y934F mutant receptor has an increased ability to activate PLC-γ1 in porcine aortic endothelial cells (PAE) [32]. The Y934F mutant receptor has a decreased ability to induce mitogenesis and an increased ability to induce chemotaxis in PAE cells [32]. Interestingly, we found that Abl kinases also phosphorylate Y934 on PDGFR-β, in addition to Y686 and Y970. Although, we were unable to detect a change in AKT or PLC-γ1 activation in Ph cells expressing the mutant chimeric PDGFR receptor that cannot be phosphorylated by the Abl kinases (triple mutant; Y686F/Y934F/Y970F), the mutant receptor induced increased chemotaxis, similar to PDGFR-β-Y934F [32]. It is not clear why the triple mutant receptor, which contains Y934F does not have an altered ability to activate PI-3-kinase or PLC-γ1 signaling pathways in vivo, as was demonstrated for PDGFR-β-Y934F. This may be due to the fact that we utilized fibroblasts (Ph cells) and a chimeric PDGF receptor in our study, whereas Hansen et al. used PAE cells and PDGFR-β [32]. Alternatively, mutation of the three tyrosine residues may affect the binding of PLC-γ1 or PI3-kinase to PDGFR in a manner distinct from mutation of a single residue (Y934F).
Abl and Src family non-receptor kinases phosphorylate some of the same substrates (e.g. cortactin), and therefore, it is not surprising that both kinases phosphorylate Y934 on PDGFR-β. However, unlike Src kinases, Abl kinases also phosphorylate two unique residues on PDGFR-β (Y686 and Y970). Interestingly, mutation of the three Abl-dependent phosphorylation sites affects the basal and PDGF-stimulated in vitro activity of PDGFR-β; however we were unable to detect specific signaling pathways that are altered, in vivo. The reason for this discrepancy is not clear. It is possible that there are other signaling pathways, not yet tested, which are activated in a differential manner by the mutant receptors. In addition, it is not clear why the mutant receptors, which contain significantly lower in vitro kinase activity, have an increased ability to stimulate chemotaxis. It is possible that high activity might favor mitogenesis rather than migration. Alternatively, the mutant receptors might have an increased ability to phosphorylate some substrates (ie. those involved in migration), and a decreased ability to phosphorylate other substrates (such as those involved in proliferation).
Phosphorylation of Y686 and Y970 could affect internalization of PDGFR-β. Following activation, receptor tyrosine kinases, such as EGFR and PDGFR, are internalized in endosomes, ubiquitinated and degraded by the lysosome, or are recycled back to the plasma membrane, and the receptor tyrosine kinases continue to signal in endosomes, activating signaling pathways such as ERK and PI3-kinase/AKT, and promoting biological outcomes such as proliferation [33, 34]. Active Abl kinases have been shown to phosphorylate EGFR on a C-terminal residue, Y1173, which inhibits receptor endocytosis [31]. Future experiments will be required to determine whether phosphorylation of Y686 and Y970 affects PDGFR-β internalization or recycling to the plasma membrane.
5. CONCLUSIONS
In this report, we present a number of novel findings. First, we show that Abl kinases bind receptor tyrosine kinases via a common mechanism; direct binding of the Abl kinase SH2 domains to receptor intracellular domains. Second, we show that Abl kinases are directly phosphorylated by several receptor tyrosine kinases including Her-2 and PDGFR-β. Significantly, we demonstrate that phosphorylation of Abl kinases by PDGFR-β has important functional consequences, as the receptor phosphorylates residues known to be required for full c-Abl and Arg activity (Y412, Y245). Third, we show that in addition to being activated by PDGF stimulation of fibroblasts, Abl kinases also are activated downstream of a PDGF autocrine growth loop in glioblastoma cells. Fourth, we demonstrate that Abl kinases, in turn, phosphorylate PDGFR-β, increasing its activity in vitro, and decreasing its ability to induce chemotaxis in vivo. These data indicate that in addition to acting downstream of receptor tyrosine kinases to promote proliferation and migration, Abl kinases also downregulate PDGF-induced chemotaxis by phosphorylating PDGFR-β.
Supplementary Material
Acknowledgments
We thank Dr. Ann Marie Pendergast (Duke University, Durham, NC) for allowing us to initiate this project in her laboratory; preparation of mass spectrometry samples was performed in her laboratory. We thank Dr. Andrius Kazlauskas (Harvard University, Cambridge, MA) for providing us with the chimeric PDGFR expression system, PDGFR constructs, and PDGFR-α antibody; Dr. Anthony Koleske (Yale, University, New Haven, CT) for Arg expression constructs and GST-Arg fusion proteins; and Dr. Bruce Mayer (University of Connecticut Health Center, School of Medicine, Farmington, CT) for the GST-cAbl-KR baculovirus. We thank Dr. William Lane and the Harvard Mass Spectrometry Facility for performing the mass spectrometry analysis, and Jonathan Sims for reading the manuscript. This work was supported a Concern Foundation Young Investigator Award, American Cancer Society Pilot Grant #85-001-16 IRG, and NIH Grant P20 RR20171 from the National Center for Research Resources to R.P. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- PDGF
platelet-derived-growth factor
- PDGFR
platelet-derived-growth factor receptor
- EGF
epidermal-growth factor
- EGFR
epidermal-growth factor receptor
- SH
Src-homology
- CML
chronic myelogenous leukemia
- IGF-1R
insulin-like growth factor receptor
- ALL
acute lymphocytic leukemia
- PAE
porcine aortic endothelial cells
- PI3K
PI-3-kinase, phosphoinositide 3-kinase
- NOX
NADPH oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sirvent A, Benistant C, Roche S. Biol Cell. 2008:617–631. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- 2.Wang JYJ. Curr Opin Cell Biol. 1998:240–247. doi: 10.1016/s0955-0674(98)80146-4. [DOI] [PubMed] [Google Scholar]
- 3.Nagar B, Hantschel O, Young M, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 4.Pendergast AM. Adv Cancer Res. 2002:51–100. doi: 10.1016/s0065-230x(02)85003-5. [DOI] [PubMed] [Google Scholar]
- 5.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. Genes Dev. 1999:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plattner R, Irvin BJ, Guo S, Blackburn K, Kazlauskas A, Abraham RT, York JD, Pendergast AM. Nat Cell Biol. 2003:309–319. doi: 10.1038/ncb949. [DOI] [PubMed] [Google Scholar]
- 7.Plattner R, Koleske AJ, Kazlauskas A, Pendergast AM. Mol Cell Biol. 2004:2573–2583. doi: 10.1128/MCB.24.6.2573-2583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boureux A, Furstoss O, Simon V, Roche S. J Cell Sci. 2005:3717–3726. doi: 10.1242/jcs.02491. [DOI] [PubMed] [Google Scholar]
- 9.Mitra S, Beach C, Feng GS, Plattner R. J Cell Sci. 2008:3335–3346. doi: 10.1242/jcs.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sini P, Cannas A, Koleske AJ, Di Fiore PP, Scita G. Nat Cell Biol. 2004:268–274. doi: 10.1038/ncb1096. [DOI] [PubMed] [Google Scholar]
- 11.Pendergast AM. In: Chronic Myeloid Leukaemia: Biology and Treatment. Carella AM, Daley GQ, Eaves CJ, Goldman JM, Helmann R, editors. Martin Dunitz, Lt; London: 2001. pp. 19–39. [Google Scholar]
- 12.Scheijen B, Griffin JD. Oncogene. 2002:3314–3333. doi: 10.1038/sj.onc.1205317. [DOI] [PubMed] [Google Scholar]
- 13.Kim HJ, Woo HY, Koo HH, Tak EY, Kim SH. Am J Hematol. 2004:360–363. doi: 10.1002/ajh.20117. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan D, Plattner R. Cancer Res. 2006:5648–5655. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan D, Sims JT, Plattner R. Oncogene. 2008:1095–1105. doi: 10.1038/sj.onc.1210714. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Sun T, Ji L, Deng W, Roth J, Minna J, Arlinghaus R. Oncogene. 2007:6989–6996. doi: 10.1038/sj.onc.1210500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeMali KA, Kazlauskas A. Mol Cell Biol. 1998:2014–2022. doi: 10.1128/mcb.18.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenkranz S, DeMali KA, Gelderloos JA, Bazenet C, Kazlauskas A. J Biol Chem. 1999:28335–28343. doi: 10.1074/jbc.274.40.28335. [DOI] [PubMed] [Google Scholar]
- 19.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. Genes and Development. 1999:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valius M, Bazenet C, Kazlauskas A. Mol Cell Biol. 1993:133–143. doi: 10.1128/mcb.13.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Miller AL, Mooseker MS, Koleske AJ. Proc Natl Acad Sci USA. 2001:14865–14870. doi: 10.1073/pnas.251249298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer B, Baltimore D. Mol Cell Biol. 1994:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleishman SJ, Schlessinger J. N Ben-Tal Proc Natl Acad Sci U S A. 2002:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brasher BB, Van Etten RA. J Biol Chem. 2000:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi H, Kanzawa T, Kondo Y, Kondo S. Br J Cancer. 2004:1069–1075. doi: 10.1038/sj.bjc.6601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Ustach C, Kim HR. J Biochem Mol Biol. 2003:49–59. doi: 10.5483/bmbrep.2003.36.1.049. [DOI] [PubMed] [Google Scholar]
- 27.Schilling D, Reid IJ, Hujer A, Morgan D, Demoll E, Bummer P, Fenstermaker RA, Kaetzel DM. Biochem J. 1998:637–644. doi: 10.1042/bj3330637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plattner R, Pendergast AM. Cell Cycle. 2003:273–274. [PubMed] [Google Scholar]
- 29.Pendergast AM, Muller AJ, Havlik MH, Clark R, McCormick F, Witte ON. Proc Natl Acad Sci USA. 1991:5927–5931. doi: 10.1073/pnas.88.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. J Biol Chem. 1999:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 31.Tanos B, Pendergast AM. J Biol Chem. 2006:32714–32723. doi: 10.1074/jbc.M603126200. [DOI] [PubMed] [Google Scholar]
- 32.Hansen K, Johnell M, Siegbahn A, Rorsman C, Engstrom U, Wernstedt C, Heldin CH, Ronnstrand L. EMBO J. 1996:5299–5313. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Pennock SD, Chen X, Kazlauskas A, Wang Z. J Biol Chem. 2004:8038–8046. doi: 10.1074/jbc.M311494200. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Pennock S, Chen X, Wang Z. Mol Cell Biol. 2002:7279–7290. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.